20:
212:
191:
may separate two loci, resulting in the loss of synteny between them. Conversely, translocations can also join two previously separate pieces of chromosomes together, resulting in a gain of synteny between loci. Stronger-than-expected shared synteny can reflect selection for functional relationships
23:
Synteny (in the modern sense) between human and mouse chromosomes. Colors in the human chromosomes indicate regions homologous with parts of the mouse chromosome of the same color. For instance, sequences homologous to mouse chromosome 1 are primarily on human chromosomes 1 and 2, but also 6, 8, and
304:
Shared synteny between different species can be inferred from their genomic sequences. This is typically done using a version of the MCScan algorithm, which finds syntenic blocks between species by comparing their homologous genes and looking for common patterns of collinearity on a chromosomal or
265:
could be used as a model to find genes or genetic markers of interest which could be used in wheat breeding and research. In this context, synteny was also essential in identifying a highly important region in wheat, the Ph1 locus involved in genome stability and fertility, which was located using
83:
Genomic sequencing and mapping have enabled comparison of the general structures of genomes of many different species. The general finding is that organisms of relatively recent divergence show similar blocks of genes in the same relative positions in the genome. This situation is called synteny,
149:
frequencies between them. In contrast, any loci on the same chromosome are by definition syntenic, even if their recombination frequency cannot be distinguished from unlinked loci by practical experiments. Thus, in theory, all linked loci are syntenic, but not all syntenic loci are necessarily
84:
translated roughly as possessing common chromosome sequences. For example, many of the genes of humans are syntenic with those of other mammals—not only apes but also cows, mice, and so on. Study of synteny can show how the genome is cut and pasted in the course of evolution.
219:
The term is currently (since ~2000) more commonly used to describe preservation of the precise order of genes on a chromosome passed down from a common ancestor, despite more "old school" geneticists rejecting what they perceive as a misappopriation of the term, preferring
169:
Students of (classical) genetics employ the term synteny to describe the situation in which two genetic loci have been assigned to the same chromosome but still may be separated by a large enough distance in map units that genetic linkage has not been demonstrated.
254:
in
England and the National Institute of Agrobiological Research in Japan demonstrated that the much smaller rice genome had a similar structure and gene order to that of wheat. Further study found that many cereals are syntenic and thus plants such as
249:
allows the presence and possibly function of genes in a simpler, model organism to infer those in a more complex one. For example, wheat has a very large, complex genome which is difficult to study. In 1994 research from the
783:
Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH (November 1998). "Zebrafish hox clusters and vertebrate genome evolution".
317:
is used to select the best scoring path of shared homologous genes between species, taking into account potential gene loss and gain which may have occurred in the species' evolutionary histories.
231:
of genomic regions in different species. Additionally, exceptional conservation of synteny can reflect important functional relationships between genes. For example, the order of genes in the "
919:
Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G (February 2006). "Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat".
128:"band". This can be interpreted classically as "on the same chromosome", or in the modern sense of having the same order of genes on two (homologous) strings of DNA (or chromosomes).
1112:
A web based visualisation tool allowing to navigate on genomes and visualizing the
Synteny conservation among several datasets (cereals, dicotyledons, animals, a Wheat-based one...)
765:
426:
Moreno-Hagelsieb G, Treviño V, Pérez-Rueda E, Smith TF, Collado-Vides J (2001). "Transcription unit conservation in the three domains of life: a perspective from
288:
relationships among several species, and even to infer the genome organization of extinct ancestral species. A qualitative distinction is sometimes drawn between
227:
The analysis of synteny in the gene order sense has several applications in genomics. Shared synteny is one of the most reliable criteria for establishing the
71:, i.e. conservation of blocks of order within two sets of chromosomes that are being compared with each other. These blocks are referred to as
154:, the genetic loci on a chromosome are syntenic regardless of whether this relationship can be established by experimental methods such as
183:
Shared synteny (also known as conserved synteny) describes preserved co-localization of genes on chromosomes of different species. During
1106:
Server for
Synteny Identification and Analysis of Genome Rearrangement—the Identification of synteny and calculating reversal distances.
192:
between syntenic genes, such as combinations of alleles that are advantageous when inherited together, or shared regulatory mechanisms.
1082:
NIH's
National Library of Medicine NCBI link to Gene Homology resources, and Comparative Chromosome Maps of the Human, Mouse, and Rat.
1021:
Wang, Y; Tang, H; Debarry, JD; Tan, X; Li, J; Wang, X; Lee, TH; Jin, H; Marler, B; Guo, H; Kissinger, JC; Paterson, AH (April 2012).
835:
Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M (1994). "Conservation of genome structure between rice and wheat".
1094:
NIH's
National Library of Medicine NCBI (National Center for Biotechnology Information) link to a tremendous number of resources.
414:
277:, syntenic genes encode a large number of essential cell functions and represent a high level of functional relationships.
972:"Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales"
119:
242:
and which interact with each other in critical ways, is essentially preserved throughout the animal kingdom.
1100:
A free browser-based tool to compare and visualize microsynteny across multiple genomes for a set of genes.
970:
Guerrero, G; Peralta, H; Aguilar, A; Díaz, R; Villalobos, MA; Medrano-Soto, A; Mora, J (17 October 2005).
1130:
1085:
358:"Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms"
667:"Gene order and recombination rate in homologous chromosome regions of the chicken and a passerine bird"
79:
The
Encyclopædia Britannica gives the following description of synteny, using the modern definition:
683:
199:, this conservation of gene content and linkage without preservation of order has also been termed
188:
1125:
331:
228:
678:
107:
1023:"MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity"
310:
146:
1109:
928:
793:
336:
246:
8:
314:
932:
797:
1079:
1047:
1022:
998:
971:
952:
901:
852:
817:
747:
642:
615:
591:
566:
542:
517:
493:
466:
392:
357:
41:
888:
871:
443:
1076:— Probably the most used synteny software program used in comparative genomics.
1052:
1003:
944:
893:
809:
739:
698:
647:
596:
547:
498:
447:
397:
379:
274:
251:
905:
856:
821:
751:
145:: Linkage between two loci is established by the observation of lower-than-expected
1042:
1034:
993:
983:
956:
936:
883:
844:
801:
729:
688:
637:
627:
586:
578:
537:
529:
518:"Genomic regulatory blocks underlie extensive microsynteny conservation in insects"
488:
478:
439:
387:
369:
270:
49:
805:
16:
Co-localization of genetic loci on a chromosome, or the conservation of gene order
567:"Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes"
326:
281:
142:
467:"A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi"
159:
155:
632:
1119:
483:
383:
163:
101:
693:
666:
465:
Hane, JK; Rouxel, T; Howlett, BJ; Kema, GH; Goodwin, SB; Oliver, RP (2011).
1056:
1007:
988:
948:
743:
702:
651:
600:
551:
502:
451:
401:
374:
285:
261:
1038:
897:
813:
415:
heredity (genetics) : Microevolution - Britannica Online
Encyclopedia
1073:
940:
848:
665:
Dawson DA, Akesson M, Burke T, Pemberton JM, Slate J, Hansson B (2007).
309:
scale. Homologies are usually determined on the basis of high bit score
1088:
More information on synteny and its use in comparative cereal genomics.
582:
533:
425:
53:
19:
239:
184:
131:
97:
24:
18. The X chromosome is almost completely syntenic in both species.
232:
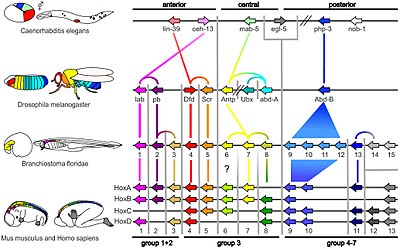
151:
29:
734:
717:
516:
Engström PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B (2007).
292:, preservation of synteny in large portions of a chromosome, and
280:
Patterns of shared synteny or synteny breaks can also be used as
57:
1097:
1091:
306:
236:
872:"Cereal genome evolution. Grasses, line up and form a circle"
211:
1103:
515:
266:
664:
256:
206:
969:
296:, preservation of synteny for only a few genes at a time.
918:
834:
715:
616:"The determinants of gene order conservation in yeasts"
245:
Synteny is widely used in studying complex genomes, as
269:
Synteny is also widely used in microbial genomics. In
716:
Passarge E, Horsthemke B, Farber RA (December 1999).
464:
313:
hits that occur between multiple genomes. From here,
215:
Synteny of Hox gene clusters; lines indicate homology
195:
In light of the more recent shift in the meaning of
1086:Graham Moore group research page - cereal genomics
173:
1020:
869:
782:
1117:
870:Moore G, Devos KM, Wang Z, Gale MD (July 1995).
356:Sinha, Amit U.; Meller, Jaroslaw (2007-03-08).
912:
613:
564:
863:
776:
709:
658:
607:
558:
458:
355:
828:
299:
48:describes the physical co-localization of
1046:
997:
987:
887:
733:
692:
682:
641:
631:
590:
541:
492:
482:
391:
373:
210:
207:Current concept: gene-order preservation
18:
187:, rearrangements to the genome such as
1118:
235:", which are key determinants of the
141:The classical concept is related to
766:"Genomics and Comparative Genomics"
718:"Incorrect use of the term synteny"
13:
14:
1142:
1067:
137:: co-localization on a chromosome
36:refers to two related concepts:
1014:
963:
758:
509:
419:
408:
349:
100:meaning "on the same ribbon";
1:
1074:ACT (Artemis Comparison Tool)
889:10.1016/S0960-9822(95)00148-5
806:10.1126/science.282.5394.1711
614:Poyatos JF, Hurst LD (2007).
444:10.1016/S0168-9525(01)02241-7
342:
565:Heger A, Ponting CP (2007).
88:
7:
320:
162:, physical localization or
10:
1147:
770:www.integratedbreeding.net
118:
106:
633:10.1186/gb-2007-8-11-r233
189:chromosome translocations
976:BMC Evolutionary Biology
484:10.1186/gb-2011-12-5-r45
67:more commonly refers to
56:within an individual or
332:Ultra-conserved element
300:Computational detection
1027:Nucleic Acids Research
989:10.1186/1471-2148-5-55
375:10.1186/1471-2105-8-82
216:
150:linked. Similarly, in
86:
25:
694:10.1093/molbev/msm071
214:
81:
22:
837:Nature Biotechnology
337:Comparative genomics
247:comparative genomics
63:In current biology,
1039:10.1093/nar/gkr1293
941:10.1038/nature04434
933:2006Natur.439..749G
849:10.1038/nbt0394-276
798:1998Sci...282.1711A
315:dynamic programming
1131:Classical genetics
1110:PlantSyntenyViewer
583:10.1101/gr.6249707
534:10.1101/gr.6669607
432:Trends in Genetics
362:BMC Bioinformatics
217:
174:Across evolution:
42:classical genetics
26:
275:Enterobacteriales
252:John Innes Centre
134:Classical concept
1138:
1080:Comparative Maps
1061:
1060:
1050:
1018:
1012:
1011:
1001:
991:
967:
961:
960:
927:(7077): 749–52.
916:
910:
909:
891:
867:
861:
860:
832:
826:
825:
792:(5394): 1711–4.
780:
774:
773:
762:
756:
755:
737:
713:
707:
706:
696:
686:
662:
656:
655:
645:
635:
611:
605:
604:
594:
562:
556:
555:
545:
528:(12): 1898–908.
513:
507:
506:
496:
486:
462:
456:
455:
428:Escherichia coli
423:
417:
412:
406:
405:
395:
377:
353:
271:Hyphomicrobiales
179:
178:
136:
135:
122:
110:
1146:
1145:
1141:
1140:
1139:
1137:
1136:
1135:
1116:
1115:
1070:
1065:
1064:
1019:
1015:
968:
964:
917:
913:
876:Current Biology
868:
864:
833:
829:
781:
777:
764:
763:
759:
722:Nature Genetics
714:
710:
684:10.1.1.330.3005
671:Mol. Biol. Evol
663:
659:
612:
608:
577:(12): 1837–49.
563:
559:
514:
510:
463:
459:
424:
420:
413:
409:
354:
350:
345:
327:Ridge (biology)
323:
302:
209:
181:
176:
175:
143:genetic linkage
139:
133:
132:
116:"along with" +
91:
73:syntenic blocks
17:
12:
11:
5:
1144:
1134:
1133:
1128:
1126:Bioinformatics
1114:
1113:
1107:
1104:Synteny server
1101:
1095:
1092:NCBI Home Page
1089:
1083:
1077:
1069:
1068:External links
1066:
1063:
1062:
1013:
962:
911:
862:
843:(3): 276–278.
827:
775:
757:
708:
677:(7): 1537–52.
657:
606:
557:
508:
471:Genome Biology
457:
438:(4): 175–177.
418:
407:
347:
346:
344:
341:
340:
339:
334:
329:
322:
319:
301:
298:
208:
205:
180:
177:shared synteny
172:
160:genome walking
156:DNA sequencing
138:
130:
90:
87:
77:
76:
61:
15:
9:
6:
4:
3:
2:
1143:
1132:
1129:
1127:
1124:
1123:
1121:
1111:
1108:
1105:
1102:
1099:
1098:SimpleSynteny
1096:
1093:
1090:
1087:
1084:
1081:
1078:
1075:
1072:
1071:
1058:
1054:
1049:
1044:
1040:
1036:
1032:
1028:
1024:
1017:
1009:
1005:
1000:
995:
990:
985:
981:
977:
973:
966:
958:
954:
950:
946:
942:
938:
934:
930:
926:
922:
915:
907:
903:
899:
895:
890:
885:
881:
877:
873:
866:
858:
854:
850:
846:
842:
838:
831:
823:
819:
815:
811:
807:
803:
799:
795:
791:
787:
779:
771:
767:
761:
753:
749:
745:
741:
736:
735:10.1038/70486
731:
727:
723:
719:
712:
704:
700:
695:
690:
685:
680:
676:
672:
668:
661:
653:
649:
644:
639:
634:
629:
625:
621:
617:
610:
602:
598:
593:
588:
584:
580:
576:
572:
568:
561:
553:
549:
544:
539:
535:
531:
527:
523:
519:
512:
504:
500:
495:
490:
485:
480:
476:
472:
468:
461:
453:
449:
445:
441:
437:
433:
429:
422:
416:
411:
403:
399:
394:
389:
385:
381:
376:
371:
367:
363:
359:
352:
348:
338:
335:
333:
330:
328:
325:
324:
318:
316:
312:
308:
297:
295:
291:
287:
284:to infer the
283:
278:
276:
272:
267:
264:
263:
259:or the grass
258:
253:
248:
243:
241:
238:
234:
230:
225:
223:
213:
204:
202:
198:
193:
190:
186:
171:
167:
165:
161:
157:
153:
148:
147:recombination
144:
129:
127:
123:
121:
115:
111:
109:
103:
99:
95:
85:
80:
74:
70:
66:
62:
59:
55:
51:
47:
43:
39:
38:
37:
35:
31:
21:
1030:
1026:
1016:
979:
975:
965:
924:
920:
914:
882:(7): 737–9.
879:
875:
865:
840:
836:
830:
789:
785:
778:
769:
760:
725:
721:
711:
674:
670:
660:
626:(11): R233.
623:
619:
609:
574:
570:
560:
525:
521:
511:
474:
470:
460:
435:
431:
427:
421:
410:
365:
361:
351:
303:
294:microsynteny
293:
290:macrosynteny
289:
286:phylogenetic
279:
268:
262:Brachypodium
260:
244:
226:
222:collinearity
221:
218:
200:
196:
194:
182:
168:
140:
125:
117:
113:
105:
93:
92:
82:
78:
72:
68:
64:
52:on the same
50:genetic loci
45:
33:
27:
620:Genome Biol
233:Hox cluster
201:mesosynteny
164:hap-mapping
158:/assembly,
69:colinearity
32:, the term
1120:Categories
1033:(7): e49.
728:(4): 387.
571:Genome Res
522:Genome Res
477:(5): R45.
343:References
282:characters
54:chromosome
679:CiteSeerX
384:1471-2105
240:body plan
229:orthology
224:instead.
185:evolution
98:neologism
89:Etymology
1057:22217600
1008:16229745
949:16467840
906:11732225
857:41626506
822:46098685
752:31645103
744:10581019
703:17434902
652:17983469
601:17989258
552:17989259
503:21605470
452:11275307
402:17343765
321:See also
152:genomics
30:genetics
1048:3326336
999:1276791
957:4407272
929:Bibcode
898:7583118
814:9831563
794:Bibcode
786:Science
643:2258174
592:2099592
543:2099597
494:3219968
393:1821339
197:synteny
94:Synteny
65:synteny
58:species
46:synteny
34:synteny
1055:
1045:
1006:
996:
982:: 55.
955:
947:
921:Nature
904:
896:
855:
820:
812:
750:
742:
701:
681:
650:
640:
599:
589:
550:
540:
501:
491:
450:
400:
390:
382:
368:: 82.
307:contig
237:animal
126:tainiā
120:ταινία
953:S2CID
902:S2CID
853:S2CID
818:S2CID
748:S2CID
311:BLAST
102:Greek
96:is a
1053:PMID
1004:PMID
945:PMID
894:PMID
810:PMID
740:PMID
699:PMID
648:PMID
597:PMID
548:PMID
499:PMID
448:PMID
398:PMID
380:ISSN
273:and
257:rice
1043:PMC
1035:doi
994:PMC
984:doi
937:doi
925:439
884:doi
845:doi
802:doi
790:282
730:doi
689:doi
638:PMC
628:doi
587:PMC
579:doi
538:PMC
530:doi
489:PMC
479:doi
440:doi
430:".
388:PMC
370:doi
114:syn
108:σύν
40:In
28:In
1122::
1051:.
1041:.
1031:40
1029:.
1025:.
1002:.
992:.
978:.
974:.
951:.
943:.
935:.
923:.
900:.
892:.
878:.
874:.
851:.
841:12
839:.
816:.
808:.
800:.
788:.
768:.
746:.
738:.
726:23
724:.
720:.
697:.
687:.
675:24
673:.
669:.
646:.
636:.
622:.
618:.
595:.
585:.
575:17
573:.
569:.
546:.
536:.
526:17
524:.
520:.
497:.
487:.
475:12
473:.
469:.
446:.
436:17
434:.
396:.
386:.
378:.
364:.
360:.
203:.
166:.
124:,
112:,
104::
44:,
1059:.
1037::
1010:.
986::
980:5
959:.
939::
931::
908:.
886::
880:5
859:.
847::
824:.
804::
796::
772:.
754:.
732::
705:.
691::
654:.
630::
624:8
603:.
581::
554:.
532::
505:.
481::
454:.
442::
404:.
372::
366:8
75:.
60:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.