135:
subsititions (SNPs). When looking between distantly related species, recombination is less of a problem since recombination between branches from common ancestors is prevented after true speciation occurs. When examining closely related species, or branching within species, recombination creates a large number of 'irrelevant SNPs' for cladistic analysis. MtDNA, through the process of organelle division, became clonal over time; very little, or often none, of that paternal mtDNA is passed. While recombination may occur in mtDNA, there is little risk that it will be passed to the next generation. As a result, mtDNA become clonal copies of each other, except when a new mutation arises. As a result, mtDNA does not have pitfalls of autosomal loci when studied in interbreeding groups. Another advantage of mtDNA is that the hyper-variable regions evolve very quickly; this shows that certain regions of mitochondrial DNA approach neutrality. This allowed the use of mitochondrial DNA to determine that the relative age of the human population was small, having gone through a recent constriction at about 150,000 years ago (see
409:. Some of the mutations that did occur between the two have reverted, hiding an initially higher rate. Selection may play into this, a rare mutation may be selective at point X in time, but later climate may change or the species migrates and it is not longer selective, and pressure exerted on new mutations that revert the change, and sometimes the reversion of a nt can occur, the greater the distance between two species the more likely this is going to occur. In addition, from that ancestral species both species may randomly mutate a site to the same nucleotide. Many times this can be resolved by obtaining DNA samples from species in the branches, creating a parsimonious tree in which the order of mutation can be deduced, creating branch-length diagram. This diagram will then produce a more accurate estimate of mutations between two species. Statistically one can assign variance based on the problem of randomnicity, back mutations, and parallel mutations (homoplasies) in creating an error range.
265:, Fix, Il2rg, Plp, Gk, Ids, Alas2, Rrm2p4, AmeIX, Tnfsf5, Licam, and Msn. The time to most recent common ancestor (TMRCA) ranges from fixed to ~1.8 million years, with a median around 700ky. These studies roughly plot to the expected fixation distribution of alleles, given linkage disequilibrium between adjacent sites. For some alleles the point of origin is elusive, for others, the point of origin points toward Sub-Saharan Africa. There are some distinctions within SSA that suggest a smaller region, but there is not adequate enough sample size and coverage to define a place of most recent common ancestor. The TMRCA is consistent with and extends the bottleneck implied by mtDNA, confidently to about 500,000 years.
540:(180,000 years ago) in the human population. A year later Brown and Wilson were looking at RFLP fragments and determined the human population expanded more recently than other ape populations. In 1984 the first DNA sequence from an extinct animal was done. Sibley and Ahlquist apply DNA-DNA hybridization technology to anthropoid phylogeny, and see pan/human split closer than gorilla/pan or gorilla/human split, a highly controversial claim. However, in 1987 they were able to support their claim. In 1987, Cann, Stoneking and Wilson suggest, by RFLP analysis of human mitochondrial DNA, that humans evolved from a constrict in Africa of a single female in a small population, ~10,00 individuals, 200,000 years ago.
220:). Unlike mtDNA, it has mutations in the non-recombinant portion (NRY) of the chromosome spaced widely apart, so far apart that finding the mutations on new Y chromosomes is labor-intensive compared with mtDNA. Many studies rely on tandem repeats; however, tandem repeats can expand and retract rapidly and in some predictable patterns. The Y chromosome only tracks male lines, and is not found in females, whereas mtDNA can be traced in males even though they fail to pass on mtDNA. In addition, it has been estimated that effective male populations in the prehistoric period were typically two females per male, and recent studies show that
413:
branch (underestimating the age), lies in a third branch (underestimating the age) or in the case of being within the LCA species, may have been millions of years older than the branch. To date the only way to assess this variance is to apply molecular phylogenetics on species claimed to be branch points. This only, however identifies the 'outlying' anchor points. And since it is more likely the more abundant fossils are younger than the branch point the outlying fossil may simply be a rare older representative. These unknowns create uncertainty that is difficult to quantify, and often not attempted.
95:
405:
third nt may mutate 1 per million years. Unless scientist study the sequence of a great many animals, particularly those close to the branch being examined, they generally do not know what the rate of mutation for a given site. Mutations do occur at 1st and 2nd positions of codons, but in most cases these mutations are under negative selection and so are removed from the population over small periods of time. In defining the rate of evolution in the anchor one has the problem that random mutation creates. For example, a rate of .005 or .010 can also explain 24 mutations according to the
228:(TMRCA). The estimates for Y TMRCA range from 1/4 to less than 1/2 that of mtDNA TMRCA. It is unclear whether this is due to high male-to-female ratios in the past coupled with repeat migrations from Africa, as a result of mutational rate change, or as some have even proposed that females of the LCA between chimps and humans continued to pass DNA millions after males ceased to pass DNA. At present the best evidence suggests that in migration the male to female ratio in humans may have declined, causing a trimming of Y diversity on multiple occasions within and outside of Africa.
274:
127:
162:
316:
512:
370:
178:
417:
assigned by researchers to some major branch points have almost doubled in age over the last 30 years. An excellent example of this is the debate over LM3 (Mungo lake 3) in
Australia. Originally it was dated to around 30 ky by carbon dating, carbon dating has problems, however, for sampled over 20ky in age, and severe problems for samples around 30ky in age. Another study looked at the fossil and estimated the age to be 62 ky in age.
447:
170:
humans moved from Africa's south-eastern regions, that more mutations accumulated in the coding region than expected, and in passage to the new world some groups are believed to have passed from the Asian tropics to
Siberia to an ancient land region called Beringia and quickly migrated to South America. Many of the mtDNA have far more mutations and at rarely mutated coding sites relative to expectations of neutral mutations.
232:
206:
575:
592:. The result of that experiment is that the differences between humans living in Europe, many of which were derived from haplogroup H (CRS), Neandertals branched from humans more than 300,000 years before haplogroup H reached Europe. While the mtDNA and other studies continued to support a unique recent African origin, this new study basically answered critiques from the Neandertal side.
114:(mtDNA). MtDNA is almost always only passed to the next generation by females, but under highly exceptional circumstances mtDNA can be passed through males. The non-recombinant portion of the Y chromosome and the mtDNA, under normal circumstances, do not undergo productive recombination. Part of the Y chromosome can undergo recombination with the X chromosome and within
549:
566:
had actually written a review a year earlier, but this had gone unnoticed). A stream of papers would follow from both sides, many with highly flawed methods and sampling. One of the more interesting was Harris and Hey, 1998 which showed that the TMCRA (time to most recent common ancestor) for the PDHA1 gene was well in excess of 1 million years. Given a
174:
ancient DNA, in which the DNA is highly degraded, the number of copies of DNA is helpful in extending and bridging short fragments together, and decreases the amount of bone extracted from highly valuable fossil/ancient remains. Unlike Y chromosome, both male and female remains carry mtDNA in roughly equal quantities.
601:
sites. In 2007, Gonder et al. proposed that a core population of humans, with greatest level of diversity and lowest selection, once lived in the region of
Tanzania and proximal parts of southern Africa, since humans left this part of Africa, mitochondria have been selectively evolving to new regions.
169:
More recently, the mtDNA genome has been used to estimate branching patterns in peoples around the world, such as when the new world was settled and how. The problem with these studies have been that they rely heavily on mutations in the coding region. Researchers have increasingly discovered that as
260:
of 1.5. However, in humans the effective ploidy is somewhat higher, ~1.7, as females in the breeding population have tended to outnumber males by 2:1 during a large portion of human prehistory. Like mtDNA, X-linked DNA tends to over emphasize female population history much more than male. There have
404:
There are several problems not seen in the above. First, mutations occur as random events. Second, the chance that any site in the genome varies is different from the next site, a very good example is the codons for amino acids, the first two nt in a codon may mutate at 1 per billion years, but the
247:
by mtDNA appear to have migrated from
Eastern China or Taiwan, by Y chromosome from the Papua New Guinea region. When HLA haplotypes were used to evaluate the two hypotheses, it was uncovered that both were right, that the Maori were an admixed population. Such admixtures appear to be common in the
134:
Mitochondrial DNA became an area of research in phylogenetics in the late 1970s. Unlike genomic DNA, it offered advantages in that it did not undergo recombination. The process of recombination, if frequent enough, corrupts the ability to create parsimonious trees because of stretches of amino acid
626:
The ability to resolve population sizes based on the 2N rule, proposed by Kimura in the 1950s. To use that information to compare relative sizes of population and come to a conclusion about abundance that contrasted observations based on the paleontological record. While human fossils in the early
600:
Significant progress has been made in genomic sequencing since Ingman and colleague published their finding on mitochondrial genome. Several papers on genomic mtDNA have been published; there is considerable variability in the rate of evolution, and rate variation and selection are evident at many
570:
at this locus of 1.5 (3 fold higher than mtDNA) the TMRCA was more than double the expectation. While this falls into the 'fixation curve' of 1.5 ploidy (averaging 2 female and 1 male) the suggested age of 1.8 my is close a significantly deviant p-value for the population size, possibly indicating
565:
sequence. At the time, however Ayala was not aware of rapid evolution of HLA loci via recombinatory process. In 1996, Parham and Ohta published their finds on the rapid evolution of HLA by short-distance recombination ('gene conversion' or 'abortive recombination'), weakening Ayala's claim (Parham
382:
The molecular phylogenetics is based on quantification substitutions and then comparing sequence with other species, there are several points in the process which create errors. The first and greatest challenge is finding "anchors" that allow the research to calibrate the system. In this example,
395:
and humans, from about 14 million years ago. So that the researcher can use orangutan and human comparison and comes up with a difference of 24. Using this he can estimate (24/(14*2, the "2" is for the length of the branch to human (14my) and the branch to orangutan (14 my) from their last common
556:
In 1987, PCR-amplification of mtDNA was first used to determine sequences. In 1991 Vigilante et al. published the seminal work on mtDNA phylogeny implicating sub-saharan Africa as the place of humans most recent common ancestors for all mtDNAs. The war between out-of-Africa and multiregionalism,
412:
There is another problem in calibration however that has defied statistical analysis. There is a true/false designation of a fossil to a least common ancestor. In reality the odds of having the least common ancestor of two extant species as an anchor is low, often that fossil already lies in one
173:
Mitochondrial DNA offers another advantage over autosomal DNA. There are generally 2 to 4 copies of each chromosome in each cell (1 to 2 from each parent chromosome). For mtDNA there can be dozens to hundreds in each cell. This increases the amount of each mtDNA loci by at least a magnitude. For
85:
similarities between humans and chimpanzees, for example, certain studies also have concluded that there is roughly a 98 percent commonality between the DNA of both species. However, more recent studies have modified the commonality of 98 percent to a commonality of 94 percent, showing that the
416:
Recent papers have been able to estimate, roughly, variance. The general trend as new fossils are discovered, is that the older fossils underestimated the age of the branch point. In addition to this dating of fossils has had a history of errors and there have been many revised datings. The age
396:
ancestor (LCA). The mutation rate at 0.857 for a stretch of sequence. Mutation rates are given, however, as rate per nucleotide(nt)-site, so if the sequence were say 100 nt in length that rate would be 0.00857/nt per million years. Ten mutations*100nt/(0.00857*2) = 5.8 million years.
57:
in different populations, scientists can determine the closeness of relationships between populations (or within populations). Certain similarities in genetic makeup let molecular anthropologists determine whether or not different groups of people belong to the same
498:
estimate, but not as old as Leakey claimed, either. However, Leakey was correct in the divergence of old and new world monkeys, the value Sarich and wilson used was a significant underestimate. This error in prediction capability highlights a common theme. (See
354:. The MCR1 gene has also been sequenced but the results are controversial, with one study claiming that contamination issues cannot be resolved from human Neandertal similarities. Critically, however, no DNA sequence has been obtained from
420:
At the point one has an estimation of mutation rate, given the above there must be two sources of variance that need to be cross-multiplied to generate an overall variance. This is infrequently done in the literature.
587:
Ancient DNA sequencing had been conducted on a limited scale up to the late 1990s when the staff at the Max Planck
Institute shocked the anthropology world by sequencing DNA from an estimated 40,000-year-old
86:
genetic gap between humans and chimps is larger than originally thought. Such information is useful in searching for common ancestors and coming to a better understanding of how humans evolved.
623:
Realization that in DNA there are multiple independent comparisons. Two techniques, mtDNA and hybridization converge on a single answer, chimps as a species are most closely related to humans.
620:
branching from chimp to human to putative LCA, that there was an inequity in morphological evolution. Comparative morphology based on fossils could be biased by different rates of change.
536:), which was more affordable at the time compared to sequencing. In 1980, W.M. Brown, looking at the relative variation between human and other species, recognized there was a recent
557:
already simmering with the critiques of Allan
Templeton, soon escalated with the paleoanthropologist, like Milford Wolpoff, getting involved. In 1995, F. Ayala published his critical
454:
With DNA newly discovered as the genetic material, in the early 1960s protein sequencing was beginning to take off. Protein sequencing began on cytochrome C and
Hemoglobin.
765:
243:
For short-range molecular phylogenetics and molecular clocking, the Y chromosome is highly effective and creates a second perspective. One argument that arose was that the
571:
that the human population shrank or split off of another population. Oddly, the next X-linked loci they examined, Factor IX, showed a TMRCA of less than 300,000 years.
515:
Restriction fragment length polymorphisms studies the cutting of mtDNA into fragments, Later the focus of PCR would be on the D 'control'-loop, at the top of the circle
537:
350:
Krings
Neandertal mtDNA have been sequenced, and sequence similarity indicates an equally recent origin from a small population on the Neanderthal branch of late
474:
and A.C. Wilson found that albumin and hemoglobin has comparable rates of evolution, indicating chimps and humans split about 4 to 5 million years ago. In 1970,
165:
A population bottleneck, as illustrated was detected by intrahuman mtDNA phylogenetic studies; the length of the bottleneck itself is indeterminate per mtDNA.
789:
A.C.Wilson and N.O.Kaplan (1963) Enzymes and nucleic acids in systematics. Proceedings of the XVI International
Congress of Zoology Vol.4, pp.125-127.
110:, which is passed from father to son. Anatomical females carry a Y chromosome only rarely, as a result of genetic defect. The other linkage group is the
38:
links between ancient and modern human populations, as well as between contemporary species. Generally, comparisons are made between sequences, either
366:, or any of the other late hominids. Some of the ancient sequences obtained have highly probable errors, and proper control to avoid contamination.
627:
and middle stone age are far more abundant than chimpanzee or gorilla, there are few unambiguous chimpanzee or gorilla fossils from the same period
1434:
Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (September 1991). "African populations and the evolution of human mitochondrial DNA".
98:
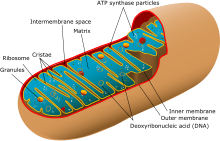
Image of mitochondrion. There are many mitochondria within a cell, and DNA in them replicates independently of the chromosomes in the nucleus.
387:
and humans, but the researcher has no known fossils that are agreeably ancestral to both but not ancestral to the next species in the tree,
1869:
Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S (July 1997). "Neandertal DNA sequences and the origin of modern humans".
256:
The X-chromosome is also a form of nuclear DNA. Since it is found as 1 copy in males and 2 non-identical chromosomes in females it has a
1140:
Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC (1984). "DNA sequences from the quagga, an extinct member of the horse family".
2065:
529:
769:
1922:
Ingman M, Kaessmann H, Pääbo S, Gyllensten U (December 2000). "Mitochondrial genome variation and the origin of modern humans".
261:
been several studies of loci on X chromosome, in total 20 sites have been examined. These include PDHA1, PDHA1, Xq21.3, Xq13.3,
130:
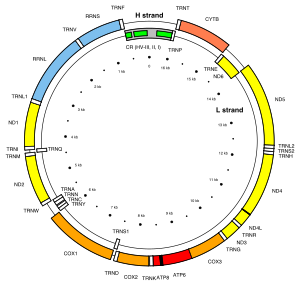
Illustration of the human mitochondrial DNA with the control region (CR, in grey) containing hypervariable sequences I and II.
682:
450:
Structure of human hemoglobin. Hemoglobins from dozens of animals and even plants were sequenced in the 1960s and early 1970s
262:
613:
That molecular phylogenetics could compete with comparative anthropology for determining the proximity of species to humans.
2016:"Process Leading to Quasi-Fixation of Genes in Natural Populations Due to Random Fluctuation of Selection Intensities"
1283:
Sibley CG, Ahlquist JE (1987). "DNA hybridization evidence of hominoid phylogeny: results from an expanded data set".
2070:
470:
began to promote the idea of a "molecular clock". By 1969 molecular clocking was applied to anthropoid evolution and
406:
118:
history the boundary has changed. Such recombinant changes in the non-recombinant region of Y are extremely rare.
552:
PCR could rapidly amplify DNA from one molecule to billions, allowing sequencing from human hairs or ancient DNA.
1191:
Sibley CG, Ahlquist JE (1984). "The phylogeny of the hominoid primates, as indicated by DNA-DNA hybridization".
326:
293:
77:
Molecular anthropology has been extremely useful in establishing the evolutionary tree of humans and other
578:
Cross-linked DNA extracted from the 4,000-year-old liver of an
Ancient Egyptian priest called Nekht-Ankh
2085:
224:
plays a large role in the passage of Y. This has created discordance between males and females for the
74:, which gives helpful insight as to how contemporary populations have formed and progressed over time.
1583:
Parham P, Ohta T (April 1996). "Population biology of antigen presentation by MHC class I molecules".
2015:
2090:
467:
1244:"The phylogeny of the hominoid primates: a statistical analysis of the DNA-DNA hybridization data"
1387:"Length mutations in human mitochondrial DNA: direct sequencing of enzymatically amplified DNA"
1024:"Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis"
478:
confronted this conclusion with arguing for improper calibration of molecular clocks. By 1975
282:
39:
1931:
1749:
1690:
1592:
1547:
1443:
1343:
1292:
1200:
1149:
1094:
1035:
976:
929:
871:
812:
491:
433:) combines the errors in calibration with errors in determining the age of a local branch.
273:
82:
81:, including closely related species like chimps and gorillas. While there are clearly many
1891:
1785:
Handt O, Höss M, Krings M, Pääbo S (June 1994). "Ancient DNA: methodological challenges".
1634:
Parham P, Adams EJ, Arnett KL (February 1995). "The origins of HLA-A, B, C polymorphism".
1260:
1243:
248:
human population and thus the use of a single haploid loci can give a biased perspective.
8:
617:
466:, in total more than hundreds of sequences from wide-ranging species were done. In 1967
94:
1935:
1753:
1694:
1596:
1551:
1447:
1347:
1296:
1204:
1153:
1098:
1039:
980:
933:
875:
816:
126:
2040:
1955:
1904:
1846:
1829:
1810:
1659:
1647:
1616:
1385:
Wrischnik LA, Higuchi RG, Stoneking M, Erlich HA, Arnheim N, Wilson AC (January 1987).
1367:
1316:
1224:
1173:
729:
704:
654:- Wilson group, 1980, 1981, 1984, 1987, 1989, 1991(posthumously) - TMRCA about 170 kya.
479:
455:
1882:
1762:
1737:
1411:
1386:
1117:
1082:
1058:
1023:
999:
964:
920:
King MC, Wilson AC (April 1975). "Evolution at two levels in humans and chimpanzees".
894:
859:
835:
800:
161:
2045:
1996:
1947:
1896:
1851:
1802:
1767:
1718:
1713:
1678:
1651:
1608:
1565:
1459:
1416:
1359:
1308:
1265:
1216:
1165:
1122:
1063:
1004:
945:
899:
840:
734:
720:
362:
221:
210:
111:
27:
1959:
1908:
1663:
1620:
1320:
2035:
2027:
1986:
1939:
1886:
1878:
1841:
1814:
1794:
1757:
1708:
1698:
1643:
1600:
1555:
1486:
1451:
1406:
1398:
1371:
1351:
1300:
1255:
1228:
1208:
1177:
1157:
1112:
1102:
1053:
1043:
994:
984:
937:
889:
879:
830:
820:
724:
716:
616:
Wilson and King realized in 1975, that while there was equity between the level of
66:
origin. This is significant because it allows anthropologists to trace patterns of
2071:
DNAanthro - Molecular Anthropology - Tech groups - Yahoo (a free discussion group)
1560:
1535:
1490:
236:
1334:
Cann RL, Stoneking M, Wilson AC (1987). "Mitochondrial DNA and human evolution".
278:
67:
2031:
1604:
1477:
Templeton AR (1993). "The 'Eve' Hypothesis: A genetic critique and reanalysis".
705:"Reconstructing phylogenies and phenotypes: A molecular view of human evolution"
244:
471:
289:
1738:"Human populations show reduced DNA sequence variation at the factor IX locus"
528:
in animals, and found they were evolving rapidly. The technique they used was
2079:
640:
194:
103:
54:
1991:
1974:
1455:
1107:
941:
2049:
2000:
1951:
1771:
1722:
1703:
1402:
1048:
989:
825:
738:
677:(X-linked) Harris and Hey - TMRCA for locus greater than 1.5 million years.
657:
635:
475:
356:
190:
107:
63:
35:
31:
1900:
1855:
1806:
1655:
1612:
1569:
1463:
1420:
1363:
1312:
1269:
1220:
1169:
1126:
1067:
949:
903:
884:
844:
685:, Fix, Il2rg, Plp, Gk, Ids, Alas2, Rrm2p4, AmeIX, Tnfsf5, Licam, and Msn
1008:
589:
369:
288:
chromosome pairs, both the female (XX) and male (XY) versions of the two
217:
1973:
Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA (March 2007).
486:
combined were used to propose that humans closest living relative (as a
315:
154:, and to verify the relationship of these three species relative to the
1798:
1304:
1212:
645:
511:
459:
384:
143:
59:
30:
has contributed to the understanding of human evolution. This field of
1943:
1355:
1161:
463:
392:
299:
285:
186:
182:
155:
483:
446:
78:
47:
1975:"Whole-mtDNA genome sequence analysis of ancient African lineages"
181:
Schematic of typical animal cell, showing subcellular components.
177:
1433:
487:
388:
351:
151:
43:
524:
In 1979, W.M.Brown and Wilson began looking at the evolution of
142:
Mitochondrial DNA has also been used to verify the proximity of
1384:
671:(Intron) - Tishkoff, 1996 - most of the diversity is in Africa.
662:
651:
567:
562:
257:
231:
205:
1921:
574:
860:"The relationship of African apes, man and old world monkeys"
674:
525:
373:
Comparison of differences between human and Neanderthal mtDNA
147:
106:
in humans that are carried by a single sex. The first is the
1868:
1139:
1081:
Ferris SD, Brown WM, Davidson WS, Wilson AC (October 1981).
1972:
71:
216:
The Y chromosome is found in the nucleus of normal cells (
1083:"Extensive polymorphism in the mitochondrial DNA of apes"
1080:
668:
391:. However, there are fossils believed to be ancestral to
115:
89:
2066:
Allan Wilson - Recent History of Science and Technology
494:(LCA) from humans and chimps appears to older than the
1536:"The myth of Eve: molecular biology and human origins"
548:
631:
Loci that have been used in molecular phylogenetics:
1827:
1784:
1828:Handt O, Krings M, Ward RH, Pääbo S (August 1996).
1679:"X chromosome evidence for ancient human histories"
1333:
665:- Ayala 1995 - TMRCA for locus is 60 million years.
609:Critical in the history of molecular anthropology:
46:sequences; however, early studies used comparative
1633:
962:
768:. Scientific American. 2006-12-19. Archived from
2077:
1520:Wolpoff M and Thorne A. The case against Eve.
424:
1830:"The retrieval of ancient human DNA sequences"
1282:
1190:
561:article "The Myth about Eve", which relied on
965:"Rapid evolution of animal mitochondrial DNA"
915:
913:
766:"Humans and Chimps: Close But Not That Close"
519:
2007:
1966:
1915:
1729:
1670:
1476:
1241:
963:Brown WM, George M, Wilson AC (April 1979).
801:"A molecular time scale for human evolution"
798:
1862:
1821:
1778:
1627:
1576:
1527:
1427:
1378:
1327:
1276:
1235:
1184:
1133:
1074:
1582:
1015:
919:
910:
851:
792:
2039:
1990:
1890:
1845:
1761:
1735:
1712:
1702:
1676:
1559:
1410:
1259:
1116:
1106:
1057:
1047:
998:
988:
956:
893:
883:
857:
834:
824:
728:
648:- Braunitizer, 1960s, Harding et al. 1997
399:
345:
2013:
1533:
1021:
573:
547:
530:restriction fragment length polymorphism
510:
490:) was the chimpanzee. In hindsight, the
445:
368:
272:
230:
204:
176:
160:
125:
93:
702:
226:Time to the Most Recent Common Ancestor
2078:
90:Haploid loci in molecular anthropology
1505:The multiregional evolution of Humans
1261:10.1093/oxfordjournals.molbev.a040363
681:Xlinked loci: PDHA1, Xq21.3, Xq13.3,
595:
500:
429:Time to most recent common ancestor (
136:
799:Wilson AC, Sarich VM (August 1969).
604:
310:
121:
377:
251:
13:
1648:10.1111/j.1600-065X.1995.tb00674.x
62:, and thus if they share a common
14:
2102:
2059:
758:
407:binomial probability distribution
306:
268:
721:10.1111/j.1469-7580.2007.00840.x
314:
1677:Harris EE, Hey J (March 1999).
1514:
1497:
1470:
1242:Templeton AR (September 1985).
383:there are 10 mutations between
200:
1892:11858/00-001M-0000-0025-0960-8
783:
755:. New York: McGraw-Hill, 2005.
745:
696:
582:
441:
1:
1883:10.1016/S0092-8674(00)80310-4
1763:10.1016/S0960-9822(01)00223-8
1736:Harris EE, Hey J (May 2001).
1561:10.1126/science.270.5244.1930
1491:10.1525/aa.1993.95.1.02a00030
690:
543:
1683:Proc. Natl. Acad. Sci. U.S.A
1087:Proc. Natl. Acad. Sci. U.S.A
1028:Proc. Natl. Acad. Sci. U.S.A
969:Proc. Natl. Acad. Sci. U.S.A
864:Proc. Natl. Acad. Sci. U.S.A
805:Proc. Natl. Acad. Sci. U.S.A
425:Problems in estimating TMRCA
7:
1605:10.1126/science.272.5258.67
703:Bradley, Brenda J. (2008).
10:
2107:
1534:Ayala FJ (December 1995).
858:Leakey LS (October 1970).
520:RLFP and DNA hybridization
506:
436:
297:
2032:10.1093/genetics/39.3.280
102:There are two continuous
1511:(April) pp. 28-33 (1992)
1503:Thorne A and Wolpoff M.
751:Kottak, Conrad Phillip.
1479:American Anthropologist
1456:10.1126/science.1840702
1108:10.1073/pnas.78.10.6319
942:10.1126/science.1090005
1704:10.1073/pnas.96.6.3320
1049:10.1073/pnas.77.6.3605
1022:Brown WM (June 1980).
990:10.1073/pnas.76.4.1967
826:10.1073/pnas.63.4.1088
579:
553:
516:
451:
400:Problem of calibration
374:
346:Ancient DNA sequencing
323:This section is empty.
303:
240:
213:
197:
166:
131:
99:
26:, is the study of how
20:Molecular anthropology
2014:Kimura M (May 1954).
1992:10.1093/molbev/msl209
885:10.1073/pnas.67.2.746
577:
551:
514:
449:
372:
298:Further information:
276:
234:
208:
180:
164:
129:
97:
16:Field of anthropology
1403:10.1093/nar/15.2.529
687:Autosomal:Numerous.
652:Mitochondrial D-loop
492:last common ancestor
294:mitochondrial genome
24:genetic anthropology
1936:2000Natur.408..708I
1754:2001CBio...11..774H
1695:1999PNAS...96.3320H
1597:1996Sci...272...67P
1552:1995Sci...270.1930A
1509:Scientific American
1448:1991Sci...253.1503V
1348:1987Natur.325...31C
1297:1987JMolE..26...99S
1205:1984JMolE..20....2S
1154:1984Natur.312..282H
1099:1981PNAS...78.6319F
1040:1980PNAS...77.3605B
981:1979PNAS...76.1967B
934:1975Sci...188..107K
876:1970PNAS...67..746L
817:1969PNAS...63.1088W
753:Windows on Humanity
618:molecular evolution
239:showing genetic map
1799:10.1007/BF01921720
1305:10.1007/BF02111285
1213:10.1007/BF02101980
709:Journal of Anatomy
596:Genomic sequencing
580:
554:
517:
480:protein sequencing
456:Gerhard Braunitzer
452:
375:
304:
296:(at bottom left).
241:
237:human X chromosome
214:
211:human Y chromosome
198:
167:
132:
100:
2086:Genetic genealogy
1834:Am. J. Hum. Genet
1524:(1991) pp. 37-41.
1391:Nucleic Acids Res
605:Critical progress
526:mitochondrial DNA
496:Sarich and Wilson
363:Homo floresiensis
343:
342:
292:, as well as the
222:cultural hegemony
137:#Causes of errors
122:Mitochondrial DNA
112:mitochondrial DNA
28:molecular biology
2098:
2054:
2053:
2043:
2011:
2005:
2004:
1994:
1970:
1964:
1963:
1944:10.1038/35047064
1930:(6813): 708–13.
1919:
1913:
1912:
1894:
1866:
1860:
1859:
1849:
1825:
1819:
1818:
1782:
1776:
1775:
1765:
1733:
1727:
1726:
1716:
1706:
1674:
1668:
1667:
1631:
1625:
1624:
1580:
1574:
1573:
1563:
1546:(5244): 1930–6.
1531:
1525:
1518:
1512:
1501:
1495:
1494:
1474:
1468:
1467:
1442:(5027): 1503–7.
1431:
1425:
1424:
1414:
1382:
1376:
1375:
1356:10.1038/325031a0
1331:
1325:
1324:
1280:
1274:
1273:
1263:
1239:
1233:
1232:
1188:
1182:
1181:
1162:10.1038/312282a0
1137:
1131:
1130:
1120:
1110:
1078:
1072:
1071:
1061:
1051:
1019:
1013:
1012:
1002:
992:
960:
954:
953:
928:(4184): 107–16.
917:
908:
907:
897:
887:
855:
849:
848:
838:
828:
796:
790:
787:
781:
780:
778:
777:
762:
756:
749:
743:
742:
732:
700:
482:and comparative
378:Causes of errors
338:
335:
325:You can help by
318:
311:
252:X-linked studies
209:Illustration of
22:, also known as
2106:
2105:
2101:
2100:
2099:
2097:
2096:
2095:
2091:Human evolution
2076:
2075:
2062:
2057:
2012:
2008:
1979:Mol. Biol. Evol
1971:
1967:
1920:
1916:
1867:
1863:
1826:
1822:
1783:
1779:
1734:
1730:
1675:
1671:
1632:
1628:
1591:(5258): 67–74.
1581:
1577:
1532:
1528:
1519:
1515:
1502:
1498:
1475:
1471:
1432:
1428:
1383:
1379:
1332:
1328:
1291:(1–2): 99–121.
1281:
1277:
1248:Mol. Biol. Evol
1240:
1236:
1189:
1185:
1148:(5991): 282–4.
1138:
1134:
1093:(10): 6319–23.
1079:
1075:
1020:
1016:
961:
957:
918:
911:
856:
852:
797:
793:
788:
784:
775:
773:
764:
763:
759:
750:
746:
701:
697:
693:
686:
607:
598:
585:
546:
522:
509:
501:Causes of Error
444:
439:
427:
402:
380:
348:
339:
333:
330:
309:
302:
290:sex chromosomes
279:human karyotype
271:
254:
203:
124:
92:
17:
12:
11:
5:
2104:
2094:
2093:
2088:
2074:
2073:
2068:
2061:
2060:External links
2058:
2056:
2055:
2006:
1965:
1914:
1861:
1820:
1777:
1728:
1669:
1626:
1575:
1526:
1513:
1496:
1469:
1426:
1377:
1342:(6099): 31–6.
1326:
1275:
1234:
1183:
1132:
1073:
1014:
975:(4): 1967–71.
955:
909:
850:
811:(4): 1088–93.
791:
782:
757:
744:
715:(4): 337–353.
694:
692:
689:
679:
678:
672:
666:
660:
655:
649:
643:
638:
629:
628:
624:
621:
614:
606:
603:
597:
594:
584:
581:
545:
542:
521:
518:
508:
505:
443:
440:
438:
435:
426:
423:
401:
398:
379:
376:
347:
344:
341:
340:
321:
319:
308:
307:Rate variation
305:
281:. It shows 22
270:
269:Autosomal loci
267:
253:
250:
202:
199:
123:
120:
104:linkage groups
91:
88:
15:
9:
6:
4:
3:
2:
2103:
2092:
2089:
2087:
2084:
2083:
2081:
2072:
2069:
2067:
2064:
2063:
2051:
2047:
2042:
2037:
2033:
2029:
2026:(3): 280–95.
2025:
2021:
2017:
2010:
2002:
1998:
1993:
1988:
1985:(3): 757–68.
1984:
1980:
1976:
1969:
1961:
1957:
1953:
1949:
1945:
1941:
1937:
1933:
1929:
1925:
1918:
1910:
1906:
1902:
1898:
1893:
1888:
1884:
1880:
1876:
1872:
1865:
1857:
1853:
1848:
1843:
1840:(2): 368–76.
1839:
1835:
1831:
1824:
1816:
1812:
1808:
1804:
1800:
1796:
1792:
1788:
1781:
1773:
1769:
1764:
1759:
1755:
1751:
1748:(10): 774–8.
1747:
1743:
1739:
1732:
1724:
1720:
1715:
1710:
1705:
1700:
1696:
1692:
1689:(6): 3320–4.
1688:
1684:
1680:
1673:
1665:
1661:
1657:
1653:
1649:
1645:
1641:
1637:
1630:
1622:
1618:
1614:
1610:
1606:
1602:
1598:
1594:
1590:
1586:
1579:
1571:
1567:
1562:
1557:
1553:
1549:
1545:
1541:
1537:
1530:
1523:
1522:New Scientist
1517:
1510:
1506:
1500:
1492:
1488:
1484:
1480:
1473:
1465:
1461:
1457:
1453:
1449:
1445:
1441:
1437:
1430:
1422:
1418:
1413:
1408:
1404:
1400:
1397:(2): 529–42.
1396:
1392:
1388:
1381:
1373:
1369:
1365:
1361:
1357:
1353:
1349:
1345:
1341:
1337:
1330:
1322:
1318:
1314:
1310:
1306:
1302:
1298:
1294:
1290:
1286:
1279:
1271:
1267:
1262:
1257:
1254:(5): 420–33.
1253:
1249:
1245:
1238:
1230:
1226:
1222:
1218:
1214:
1210:
1206:
1202:
1198:
1194:
1187:
1179:
1175:
1171:
1167:
1163:
1159:
1155:
1151:
1147:
1143:
1136:
1128:
1124:
1119:
1114:
1109:
1104:
1100:
1096:
1092:
1088:
1084:
1077:
1069:
1065:
1060:
1055:
1050:
1045:
1041:
1037:
1034:(6): 3605–9.
1033:
1029:
1025:
1018:
1010:
1006:
1001:
996:
991:
986:
982:
978:
974:
970:
966:
959:
951:
947:
943:
939:
935:
931:
927:
923:
916:
914:
905:
901:
896:
891:
886:
881:
877:
873:
869:
865:
861:
854:
846:
842:
837:
832:
827:
822:
818:
814:
810:
806:
802:
795:
786:
772:on 2007-10-11
771:
767:
761:
754:
748:
740:
736:
731:
726:
722:
718:
714:
710:
706:
699:
695:
688:
684:
676:
673:
670:
667:
664:
661:
659:
656:
653:
650:
647:
644:
642:
641:Serum albumin
639:
637:
634:
633:
632:
625:
622:
619:
615:
612:
611:
610:
602:
593:
591:
576:
572:
569:
564:
560:
550:
541:
539:
535:
531:
527:
513:
504:
502:
497:
493:
489:
485:
481:
477:
473:
469:
465:
461:
457:
448:
434:
432:
422:
418:
414:
410:
408:
397:
394:
390:
386:
371:
367:
365:
364:
359:
358:
353:
337:
328:
324:
320:
317:
313:
312:
301:
295:
291:
287:
284:
280:
275:
266:
264:
259:
249:
246:
238:
233:
229:
227:
223:
219:
212:
207:
196:
192:
188:
184:
179:
175:
171:
163:
159:
157:
153:
149:
145:
140:
138:
128:
119:
117:
113:
109:
105:
96:
87:
84:
83:morphological
80:
75:
73:
69:
65:
61:
56:
55:DNA sequences
53:By examining
51:
49:
45:
41:
37:
33:
29:
25:
21:
2023:
2019:
2009:
1982:
1978:
1968:
1927:
1923:
1917:
1877:(1): 19–30.
1874:
1870:
1864:
1837:
1833:
1823:
1793:(6): 524–9.
1790:
1786:
1780:
1745:
1741:
1731:
1686:
1682:
1672:
1639:
1636:Immunol. Rev
1635:
1629:
1588:
1584:
1578:
1543:
1539:
1529:
1521:
1516:
1508:
1504:
1499:
1482:
1478:
1472:
1439:
1435:
1429:
1394:
1390:
1380:
1339:
1335:
1329:
1288:
1285:J. Mol. Evol
1284:
1278:
1251:
1247:
1237:
1196:
1193:J. Mol. Evol
1192:
1186:
1145:
1141:
1135:
1090:
1086:
1076:
1031:
1027:
1017:
972:
968:
958:
925:
921:
870:(2): 746–8.
867:
863:
853:
808:
804:
794:
785:
774:. Retrieved
770:the original
760:
752:
747:
712:
708:
698:
680:
658:Y-chromosome
636:Cytochrome C
630:
608:
599:
586:
558:
555:
538:constriction
533:
523:
495:
476:Louis Leakey
453:
430:
428:
419:
415:
411:
403:
381:
361:
357:Homo erectus
355:
349:
331:
327:adding to it
322:
255:
242:
225:
215:
201:Y chromosome
195:mitochondria
172:
168:
150:relative to
141:
133:
108:Y chromosome
101:
76:
64:geographical
52:
36:evolutionary
32:anthropology
23:
19:
18:
1787:Experientia
1199:(1): 2–15.
590:Neanderthal
583:Ancient DNA
468:A.C. Wilson
442:Protein era
277:Diagram of
235:Diagram of
218:nuclear DNA
144:chimpanzees
2080:Categories
1742:Curr. Biol
1642:: 141–80.
776:2006-12-20
691:References
646:Hemoglobin
544:Era of PCR
460:hemoglobin
458:sequenced
393:orangutans
385:chimpanzee
283:homologous
183:Organelles
156:orangutans
72:settlement
60:haplogroup
34:examines
1485:: 51–72.
472:V. Sarich
464:myoglobin
334:July 2010
300:Karyotype
286:autosomal
187:nucleolus
68:migration
2050:17247483
2020:Genetics
2001:17194802
1960:52850476
1952:11130070
1909:13581775
1772:11378388
1723:10077682
1664:39486851
1621:22209086
1321:40231451
739:18380860
484:serology
352:hominids
152:gorillas
79:primates
48:serology
2041:1209652
1932:Bibcode
1901:9230299
1856:8755923
1847:1914746
1815:6742827
1807:8020612
1750:Bibcode
1691:Bibcode
1656:7558075
1613:8600539
1593:Bibcode
1585:Science
1570:8533083
1548:Bibcode
1540:Science
1464:1840702
1444:Bibcode
1436:Science
1421:2881260
1372:4285418
1364:3025745
1344:Bibcode
1313:3125341
1293:Bibcode
1270:3939706
1229:6658046
1221:6429338
1201:Bibcode
1178:4313241
1170:6504142
1150:Bibcode
1127:6273863
1095:Bibcode
1068:6251473
1036:Bibcode
977:Bibcode
950:1090005
930:Bibcode
922:Science
904:5002096
872:Bibcode
845:4982244
813:Bibcode
730:2409108
559:Science
507:DNA era
488:species
437:History
389:gorilla
191:nucleus
44:protein
2048:
2038:
1999:
1958:
1950:
1924:Nature
1907:
1899:
1854:
1844:
1813:
1805:
1770:
1721:
1711:
1662:
1654:
1619:
1611:
1568:
1462:
1419:
1412:340450
1409:
1370:
1362:
1336:Nature
1319:
1311:
1268:
1227:
1219:
1176:
1168:
1142:Nature
1125:
1118:349030
1115:
1066:
1059:349666
1056:
1009:109836
1007:
1000:383514
997:
948:
902:
895:283268
892:
843:
836:223432
833:
737:
727:
663:HLA-DR
568:ploidy
563:HLA-DR
258:ploidy
185:: (1)
148:humans
1956:S2CID
1905:S2CID
1811:S2CID
1714:15940
1660:S2CID
1617:S2CID
1368:S2CID
1317:S2CID
1225:S2CID
1174:S2CID
675:PDHA1
431:TMRCA
245:Maori
2046:PMID
1997:PMID
1948:PMID
1897:PMID
1871:Cell
1852:PMID
1803:PMID
1768:PMID
1719:PMID
1652:PMID
1609:PMID
1566:PMID
1460:PMID
1417:PMID
1360:PMID
1309:PMID
1266:PMID
1217:PMID
1166:PMID
1123:PMID
1064:PMID
1005:PMID
946:PMID
900:PMID
841:PMID
735:PMID
534:RFLP
462:and
193:(9)
189:(2)
70:and
2036:PMC
2028:doi
1987:doi
1940:doi
1928:408
1887:hdl
1879:doi
1842:PMC
1795:doi
1758:doi
1709:PMC
1699:doi
1644:doi
1640:143
1601:doi
1589:272
1556:doi
1544:270
1487:doi
1452:doi
1440:253
1407:PMC
1399:doi
1352:doi
1340:325
1301:doi
1256:doi
1209:doi
1158:doi
1146:312
1113:PMC
1103:doi
1054:PMC
1044:doi
995:PMC
985:doi
938:doi
926:188
890:PMC
880:doi
831:PMC
821:doi
725:PMC
717:doi
713:212
683:Zfx
669:CD4
329:.
263:Zfx
146:to
139:).
116:ape
42:or
40:DNA
2082::
2044:.
2034:.
2024:39
2022:.
2018:.
1995:.
1983:24
1981:.
1977:.
1954:.
1946:.
1938:.
1926:.
1903:.
1895:.
1885:.
1875:90
1873:.
1850:.
1838:59
1836:.
1832:.
1809:.
1801:.
1791:50
1789:.
1766:.
1756:.
1746:11
1744:.
1740:.
1717:.
1707:.
1697:.
1687:96
1685:.
1681:.
1658:.
1650:.
1638:.
1615:.
1607:.
1599:.
1587:.
1564:.
1554:.
1542:.
1538:.
1507:.
1483:95
1481:.
1458:.
1450:.
1438:.
1415:.
1405:.
1395:15
1393:.
1389:.
1366:.
1358:.
1350:.
1338:.
1315:.
1307:.
1299:.
1289:26
1287:.
1264:.
1250:.
1246:.
1223:.
1215:.
1207:.
1197:20
1195:.
1172:.
1164:.
1156:.
1144:.
1121:.
1111:.
1101:.
1091:78
1089:.
1085:.
1062:.
1052:.
1042:.
1032:77
1030:.
1026:.
1003:.
993:.
983:.
973:76
971:.
967:.
944:.
936:.
924:.
912:^
898:.
888:.
878:.
868:67
866:.
862:.
839:.
829:.
819:.
809:63
807:.
803:.
733:.
723:.
711:.
707:.
503:)
360:,
158:.
50:.
2052:.
2030::
2003:.
1989::
1962:.
1942::
1934::
1911:.
1889::
1881::
1858:.
1817:.
1797::
1774:.
1760::
1752::
1725:.
1701::
1693::
1666:.
1646::
1623:.
1603::
1595::
1572:.
1558::
1550::
1493:.
1489::
1466:.
1454::
1446::
1423:.
1401::
1374:.
1354::
1346::
1323:.
1303::
1295::
1272:.
1258::
1252:2
1231:.
1211::
1203::
1180:.
1160::
1152::
1129:.
1105::
1097::
1070:.
1046::
1038::
1011:.
987::
979::
952:.
940::
932::
906:.
882::
874::
847:.
823::
815::
779:.
741:.
719::
532:(
336:)
332:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.