463:
622:
4-chloro-2-2methylbenzenediazonium salt with 3-phospho-2-naphthoic acid-2'-4'-dimethyl aniline in Tris buffer. This stain is commercially sold as a kit for staining gels. If the protein is present, the mechanism of the reaction takes place in the following order: it starts with the de-phosphorylation of 3-phospho-2-naphthoic acid-2'-4'-dimethyl aniline by alkaline phosphatase (water is needed for the reaction). The phosphate group is released and replaced by an alcohol group from water. The electrophile 4- chloro-2-2 methylbenzenediazonium (Fast Red TR Diazonium salt) displaces the alcohol group forming the final product Red Azo dye. As its name implies, this is the final visible-red product of the reaction. In undergraduate academic experimentation of protein purification, the gel is usually run next to commercial purified samples to visualize the results and conclude whether or not purification was successful.
661:, which is rarely used, based on Pubmed citations (LB), isoelectric histidine, pK matched goods buffers, etc.; in most cases the purported rationale is lower current (less heat) matched ion mobilities, which leads to longer buffer life. Borate is problematic; Borate can polymerize, or interact with cis diols such as those found in RNA. TAE has the lowest buffering capacity but provides the best resolution for larger DNA. This means a lower voltage and more time, but a better product. LB is relatively new and is ineffective in resolving fragments larger than 5 kbp; However, with its low conductivity, a much higher voltage could be used (up to 35 V/cm), which means a shorter analysis time for routine electrophoresis. As low as one base pair size difference could be resolved in 3% agarose gel with an extremely low conductivity medium (1 mM Lithium borate).
311:
pH, but running for too long can exhaust the buffering capacity of the solution. There are also limitations in determining the molecular weight by SDS-PAGE, especially when trying to find the MW of an unknown protein. Certain biological variables are difficult or impossible to minimize and can affect electrophoretic migration. Such factors include protein structure, post-translational modifications, and amino acid composition. For example, tropomyosin is an acidic protein that migrates abnormally on SDS-PAGE gels. This is because the acidic residues are repelled by the negatively charged SDS, leading to an inaccurate mass-to-charge ratio and migration. Further, different preparations of genetic material may not migrate consistently with each other, for morphological or other reasons.
92:
559:
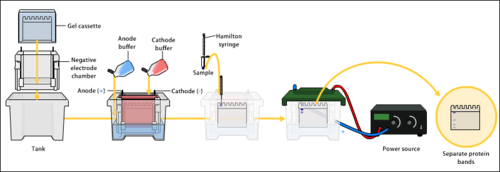
227:. The electric field consists of a negative charge at one end which pushes the molecules through the gel, and a positive charge at the other end that pulls the molecules through the gel. The molecules being sorted are dispensed into a well in the gel material. The gel is placed in an electrophoresis chamber, which is then connected to a power source. When the electric field is applied, the larger molecules move more slowly through the gel while the smaller molecules move faster. The different sized molecules form distinct bands on the gel.
286:(EMF) that is used to move the molecules through the gel matrix. By placing the molecules in wells in the gel and applying an electric field, the molecules will move through the matrix at different rates, determined largely by their mass when the charge-to-mass ratio (Z) of all species is uniform. However, when charges are not all uniform the electrical field generated by the electrophoresis procedure will cause the molecules to migrate differentially according to charge. Species that are net positively charged will migrate towards the
333:
955:(SDS) that coats the proteins with a negative charge. Generally, the amount of SDS bound is relative to the size of the protein (usually 1.4g SDS per gram of protein), so that the resulting denatured proteins have an overall negative charge, and all the proteins have a similar charge-to-mass ratio. Since denatured proteins act like long rods instead of having a complex tertiary shape, the rate at which the resulting SDS coated proteins migrate in the gel is relative only to their size and not their charge or shape.
29:
377:"Most agarose gels are made with between 0.7% (good separation or resolution of large 5–10kb DNA fragments) and 2% (good resolution for small 0.2–1kb fragments) agarose dissolved in electrophoresis buffer. Up to 3% can be used for separating very tiny fragments but a vertical polyacrylamide gel is more appropriate in this case. Low percentage gels are very weak and may break when you try to lift them. High percentage gels are often brittle and do not set evenly. 1% gels are common for many applications."
83:
3224:
209:
1013:
distribution), which then can be used in further products/processes (e.g. self-assembly processes). For the separation of nanoparticles within a gel, the key parameter is the ratio of the particle size to the mesh size, whereby two migration mechanisms were identified: the unrestricted mechanism, where the particle size << mesh size, and the restricted mechanism, where particle size is similar to mesh size.
3236:
829:. The results can be analyzed quantitatively by visualizing the gel with UV light and a gel imaging device. The image is recorded with a computer-operated camera, and the intensity of the band or spot of interest is measured and compared against standard or markers loaded on the same gel. The measurement and analysis are mostly done with specialized software.
927:
302:
separation of the components can lead to overlapping bands, or indistinguishable smears representing multiple unresolved components. Bands in different lanes that end up at the same distance from the top contain molecules that passed through the gel at the same speed, which usually means they are approximately the same size. There are
594:. Complexes remain—for the most part—associated and folded as they would be in the cell. One downside, however, is that complexes may not separate cleanly or predictably, as it is difficult to predict how the molecule's shape and size will affect its mobility. Addressing and solving this problem is a major aim of
877:, however, may show multiple bands, the speed of migration may depend on whether it is relaxed or supercoiled. Single-stranded DNA or RNA tends to fold up into molecules with complex shapes and migrate through the gel in a complicated manner based on their tertiary structure. Therefore, agents that disrupt the
911:
Electrophoresis of RNA samples can be used to check for genomic DNA contamination and also for RNA degradation. RNA from eukaryotic organisms shows distinct bands of 28s and 18s rRNA, the 28s band being approximately twice as intense as the 18s band. Degraded RNA has less sharply defined bands, has a
582:
Native gels are run in non-denaturing conditions so that the analyte's natural structure is maintained. This allows the physical size of the folded or assembled complex to affect the mobility, allowing for analysis of all four levels of the biomolecular structure. For biological samples, detergents
306:
available that contain a mixture of molecules of known sizes. If such a marker was run on one lane in the gel parallel to the unknown samples, the bands observed can be compared to those of the unknown to determine their size. The distance a band travels is approximately inversely proportional to the
617:
and intrinsic charge, but also the cross-sectional area, and thus experience different electrophoretic forces dependent on the shape of the overall structure. For proteins, since they remain in the native state they may be visualized not only by general protein staining reagents but also by specific
391:
Polyacrylamide gel electrophoresis (PAGE) is used for separating proteins ranging in size from 5 to 2,000 kDa due to the uniform pore size provided by the polyacrylamide gel. Pore size is controlled by modulating the concentrations of acrylamide and bis-acrylamide powder used in creating a gel. Care
365:
to several megabases (millions of bases), the largest of which require specialized apparatus. The distance between DNA bands of different lengths is influenced by the percent agarose in the gel, with higher percentages requiring longer run times, sometimes days. Instead high percentage agarose gels
310:
There are limits to electrophoretic techniques. Since passing a current through a gel causes heating, gels may melt during electrophoresis. Electrophoresis is performed in buffer solutions to reduce pH changes due to the electric field, which is important because the charge of DNA and RNA depends on
301:
If several samples have been loaded into adjacent wells in the gel, they will run parallel in individual lanes. Depending on the number of different molecules, each lane shows the separation of the components from the original mixture as one or more distinct bands, one band per component. Incomplete
648:
Buffers in gel electrophoresis are used to provide ions that carry a current and to maintain the pH at a relatively constant value. These buffers have plenty of ions in them, which is necessary for the passage of electricity through them. Something like distilled water or benzene contains few ions,
435:
are made in 6%, 8%, 10%, 12% or 15%. Stacking gel (5%) is poured on top of the resolving gel and a gel comb (which forms the wells and defines the lanes where proteins, sample buffer, and ladders will be placed) is inserted. The percentage chosen depends on the size of the protein that one wishes
319:
The types of gel most typically used are agarose and polyacrylamide gels. Each type of gel is well-suited to different types and sizes of the analyte. Polyacrylamide gels are usually used for proteins and have very high resolving power for small fragments of DNA (5-500 bp). Agarose gels, on the
1187:
A 1959 book on electrophoresis by Milan Bier cites references from the 1800s. However, Oliver
Smithies made significant contributions. Bier states: "The method of Smithies ... is finding wide application because of its unique separatory power." Taken in context, Bier clearly implies that Smithies'
161:
or other substances. Shorter molecules move faster and migrate farther than longer ones because shorter molecules migrate more easily through the pores of the gel. This phenomenon is called sieving. Proteins are separated by the charge in agarose because the pores of the gel are too large to sieve
37:
is placed in this buffer-filled box and an electric current is applied via the power supply to the rear. The negative terminal is at the far end (black wire), so DNA migrates toward the positively charged anode(red wire). This occurs because phosphate groups found in the DNA fragments possess a
169:
Gel electrophoresis uses a gel as an anticonvective medium or sieving medium during electrophoresis, the movement of a charged particle in an electric current. Gels suppress the thermal convection caused by the application of the electric field, and can also act as a sieving medium, slowing the
448:
potato starch makes for another non-toxic medium for protein electrophoresis. The gels are slightly more opaque than acrylamide or agarose. Non-denatured proteins can be separated according to charge and size. They are visualised using
Napthal Black or Amido Black staining. Typical starch gel
1012:
A novel application for gel electrophoresis is the separation or characterization of metal or metal oxide nanoparticles (e.g. Au, Ag, ZnO, SiO2) regarding the size, shape, or surface chemistry of the nanoparticles. The scope is to obtain a more homogeneous sample (e.g. narrower particle size
621:
A specific experiment example of an application of native gel electrophoresis is to check for enzymatic activity to verify the presence of the enzyme in the sample during protein purification. For example, for the protein alkaline phosphatase, the staining solution is a mixture of
407:
methods used polyacrylamide gels to separate DNA fragments differing by a single base-pair in length so the sequence could be read. Most modern DNA separation methods now use agarose gels, except for particularly small DNA fragments. It is currently most often used in the field of
324:(PFGE). Polyacrylamide gels are run in a vertical configuration while agarose gels are typically run horizontally in a submarine mode. They also differ in their casting methodology, as agarose sets thermally, while polyacrylamide forms in a chemical polymerization reaction.
357:. Agarose gels are easily cast and handled compared to other matrices because the gel setting is a physical rather than chemical change. Samples are also easily recovered. After the experiment is finished, the resulting gel can be stored in a plastic bag in a refrigerator.
668:
that significantly enhances the sharpness of the bands within the gel. During electrophoresis in a discontinuous gel system, an ion gradient is formed in the early stage of electrophoresis that causes all of the proteins to focus on a single sharp band in a process called
847:
86:
The image above shows how small DNA fragments will migrate through agarose quickly but large size DNA fragments move more slowly during electrophoresis. The graph to the right shows the nonlinear relationship between the size of the DNA fragment and the distance
274:
and must be handled using appropriate safety precautions to avoid poisoning. Agarose is composed of long unbranched chains of uncharged carbohydrates without cross-links resulting in a gel with large pores allowing for the separation of macromolecules and
673:. Separation of the proteins by size is achieved in the lower, "resolving" region of the gel. The resolving gel typically has a much smaller pore size, which leads to a sieving effect that now determines the electrophoretic mobility of the proteins.
298:), whereas species that are net negatively charged will migrate towards the positively charged anode. Mass remains a factor in the speed with which these non-uniformly charged molecules migrate through the matrix toward their respective electrodes.
170:
passage of molecules; gels can also simply serve to maintain the finished separation so that a post electrophoresis stain can be applied. DNA gel electrophoresis is usually performed for analytical purposes, often after amplification of DNA via
943:, unlike nucleic acids, can have varying charges and complex shapes, therefore they may not migrate into the polyacrylamide gel at similar rates, or all when placing a negative to positive EMF on the sample. Proteins, therefore, are usually
2094:
360:
Agarose gels do not have a uniform pore size, but are optimal for electrophoresis of proteins that are larger than 200 kDa. Agarose gel electrophoresis can also be used for the separation of DNA fragments ranging from 50
2356:
Barasinski, Matthäus; Garnweitner, Georg (12 February 2020). "Restricted and
Unrestricted Migration Mechanisms of Silica Nanoparticles in Agarose Gels and Their Utilization for the Separation of Binary Mixtures".
436:
to identify or probe in the sample. The smaller the known weight, the higher the percentage that should be used. Changes in the buffer system of the gel can help to further resolve proteins of very small sizes.
320:
other hand, have lower resolving power for DNA but have a greater range of separation, and are therefore used for DNA fragments of usually 50–20,000 bp in size, but the resolution of over 6 Mb is possible with
832:
Depending on the type of analysis being performed, other techniques are often implemented in conjunction with the results of gel electrophoresis, providing a wide range of field-specific applications.
523:
Denaturing conditions are necessary for proper estimation of molecular weight of RNA. RNA is able to form more intramolecular interactions than DNA which may result in change of its
1984:
1900:
371:
2035:
466:
TTGE profiles representing the bifidobacterial diversity of fecal samples from two healthy volunteers (A and B) before and after AMC (Oral
Amoxicillin-Clavulanic Acid) treatment
219:
is a process that enables the sorting of molecules based on charge, size, or shape. Using an electric field, molecules (such as DNA) can be made to move through a gel made of
904:" page for an example of a polyacrylamide DNA sequencing gel. Characterization through ligand interaction of nucleic acids or fragments may be performed by mobility shift
1541:
307:
logarithm of the size of the molecule (alternatively, this can be stated as the distance traveled is inversely proportional to the log of samples's molecular weight).
858:
In the case of nucleic acids, the direction of migration, from negative to positive electrodes, is due to the naturally occurring negative charge carried by their
2249:
2521:
2429:
2148:
95:
Gel electrophoresis is a process where an electric current is applied to DNA samples creating fragments that can be used for comparison between DNA samples.
869:
Double-stranded DNA fragments naturally behave as long rods, so their migration through the gel is relative to their size or, for cyclic fragments, their
2837:
755:. The gel will then be physically cut, and the protein complexes extracted from each portion separately. Each extract may then be analysed, such as by
473:
gels are run under conditions that disrupt the natural structure of the analyte, causing it to unfold into a linear chain. Thus, the mobility of each
2692:
462:
270:), the preferred matrix is purified agarose. In both cases, the gel forms a solid, yet porous matrix. Acrylamide, in contrast to polyacrylamide, is a
2687:
649:
which is not ideal for the use in electrophoresis. There are a number of buffers used for electrophoresis. The most common being, for nucleic acids
150:
to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.
432:
715:
dye. Other methods may also be used to visualize the separation of the mixture's components on the gel. If the molecules to be separated contain
2677:
2697:
563:
682:
2890:
3010:
2729:
238:
whose composition and porosity are chosen based on the specific weight and composition of the target to be analyzed. When separating
3131:
2977:
547:
179:
74:
1591:
Smisek, David L.; Hoagland, David A. (1989). "Agarose gel electrophoresis of high molecular weight, synthetic polyelectrolytes".
637:
665:
2972:
1530:
686:
266:, producing different sized mesh networks of polyacrylamide. When separating larger nucleic acids (greater than a few hundred
2284:
1795:
1461:
1428:
1395:
1362:
1324:
3162:
3091:
1203:
841:
2702:
1788:
Electrophoresis of
Proteins in Polyacrylamide and Starch Gels: Laboratory Techniques in Biochemistry and Molecular Biology
1764:
3157:
3136:
2682:
1705:
1228:
477:
depends only on its linear length and its mass-to-charge ratio. Thus, the secondary, tertiary, and quaternary levels of
234:" in this instance refers to the matrix used to contain, then separate the target molecules. In most cases, the gel is a
70:
735:
system. Gels are then commonly labelled for presentation and scientific records on the popular figure-creation website,
3289:
3121:
2204:
1675:
1575:
493:
386:
66:
38:
negative charge which is repelled by the negatively charged cathode and are attracted to the positively charged anode.
2562:
2179:
788:
392:
must be used when creating this type of gel, as acrylamide is a potent neurotoxin in its liquid and powdered forms.
3240:
3126:
2827:
2807:
1218:
1146:
367:
321:
404:
3167:
3116:
3056:
2788:
1027:
921:
1965:"Enhanced full-length transcription of Sindbis virus RNA by effective denaturation with methylmercury hydroxide"
3279:
3071:
3172:
3076:
3003:
2817:
2722:
1056:
543:
hydroxide was often used in denaturing RNA electrophoresis, but it may be method of choice for some samples.
2847:
1168:
1157:
2865:
400:
303:
2601:
Minde, David P.; Maurice, Madelon M.; Rüdiger, Stefan G. D. (3 October 2012). Uversky, Vladimir N. (ed.).
2453:"The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis"
3051:
2964:
2870:
2798:
1081:
944:
756:
602:
470:
341:
34:
3274:
3030:
1198:
966:
760:
3209:
3061:
2822:
2812:
2802:
2764:
1253:
1248:
1223:
1140:
1067:
989:
851:
784:
171:
62:
2544:
1824:"Zone electrophoresis in starch gels: group variations in the serum proteins of normal human adults"
3284:
3269:
3228:
3046:
2996:
2842:
2715:
2059:"Length-independent separation of DNA restriction fragments in two-dimensional gel electrophoresis"
985:
905:
901:
712:
558:
524:
276:
767:. This can provide a great deal of information about the identities of the proteins in a complex.
2006:
Fromin N; Hamelin J; Tarnawski S; Roesti D; Jourdain-Miserez K; Forestier N; et al. (2002).
1118:
517:
2309:
Hanauer, Matthias; Pierrat, Sebastien; Zins, Inga; Lotz, Alexander; Sönnichsen, Carsten (2007).
2913:
2875:
1276:"Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104"
1085:
952:
792:
508:, a method called reducing PAGE. Reducing conditions are usually maintained by the addition of
489:
478:
3101:
3086:
2832:
571:
539:
are the most often used denaturing agents to disrupt RNA structure. Originally, highly toxic
91:
3247:
3193:
2880:
2759:
2614:
2322:
1600:
1213:
1164:
1122:
748:
509:
505:
2603:"Determining Biophysical Protein Stability in Lysates by a Fast Proteolysis Assay, FASTpp"
142:
to separate proteins by charge or size (IEF agarose, essentially size independent) and in
8:
3188:
2793:
974:
496:
process. For full denaturation of proteins, it is also necessary to reduce the covalent
283:
235:
2618:
2326:
1604:
776:
Estimation of the size of DNA molecules following restriction enzyme digestion, e.g. in
546:
Denaturing gel electrophoresis is used in the DNA and RNA banding pattern-based methods
174:(PCR), but may be used as a preparative technique prior to use of other methods such as
2779:
2645:
2602:
2382:
2086:
1848:
1823:
1756:
1667:
1507:
1486:
1273:
1172:
870:
810:
777:
692:
532:
501:
139:
2469:
2452:
2008:"Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns"
1940:
1923:
1888:
889:, are used to denature the nucleic acids and cause them to behave as long rods again.
516:. For a general analysis of protein samples, reducing PAGE is the most common form of
3066:
2934:
2650:
2632:
2583:
2558:
2513:
2509:
2474:
2421:
2417:
2386:
2374:
2338:
2280:
2273:
2241:
2200:
2175:
2140:
2136:
2078:
2074:
2027:
2023:
1976:
1945:
1892:
1853:
1801:
1791:
1760:
1748:
1659:
1651:
1616:
1571:
1512:
1467:
1457:
1434:
1424:
1401:
1391:
1368:
1358:
1330:
1320:
1297:
1104:
814:
291:
175:
147:
2671:
1671:
936:– The indicated proteins are present in different concentrations in the two samples.
636:. However, native PAGE is also used to scan genes (DNA) for unknown mutations as in
3111:
2949:
2640:
2622:
2550:
2505:
2464:
2413:
2366:
2330:
2233:
2132:
2090:
2070:
2019:
1935:
1884:
1843:
1835:
1740:
1643:
1608:
1502:
1494:
1388:
Wilson and Walker's principles and techniques of biochemistry and molecular biology
1287:
1145:
1984 – pulsed-field gel electrophoresis enables separation of large DNA molecules (
1133:
993:
961:
are usually analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (
882:
764:
696:
670:
267:
2310:
332:
3081:
3019:
2918:
2742:
2707:
2627:
1647:
1176:
1045:
981:
932:
724:
255:
216:
203:
47:
2311:"Separation of Nanoparticles by Gel Electrophoresis According to Size and Shape"
28:
2857:
2774:
1208:
1150:
1129:
1060:
1037:
1950 – introduction of "zone electrophoresis" (Tiselius); paper electrophoresis
720:
614:
591:
576:
513:
497:
417:
396:
347:
224:
187:
154:
1697:
1570:. Vol. 1 (3rd ed.). Cold Spring Harbor Laboratory. p. 5.2–5.3.
1566:
Tom
Maniatis; E. F. Fritsch; Joseph Sambrook (1982). "Chapter 5, protocol 1".
424:
membrane to be probed with antibodies and corresponding markers, such as in a
3294:
3263:
3152:
3106:
2954:
2944:
2885:
2636:
2378:
2370:
2237:
2222:"History and principles of conductive media for standard DNA electrophoresis"
1655:
1620:
1405:
1031:
878:
802:
798:
716:
610:
601:
Unlike denaturing methods, native gel electrophoresis does not use a charged
587:
540:
474:
295:
191:
1635:
1471:
1438:
1372:
1334:
1319:(in Spanish). Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press.
747:
After separation, an additional separation method may then be used, such as
2939:
2908:
2769:
2654:
2587:
2342:
2245:
2221:
2031:
2007:
1924:"Synthesis of full length cDNAs from four partially purified oviduct mRNAs"
1857:
1752:
1728:
1663:
1516:
1301:
1292:
1275:
826:
822:
708:
700:
625:
425:
263:
243:
208:
163:
143:
138:, etc.) and their fragments, based on their size and charge. It is used in
2554:
2517:
2493:
2478:
2425:
2401:
2144:
2120:
1980:
1964:
1896:
1872:
2082:
2058:
1949:
1805:
1744:
1097:
1093:
704:
416:
of the same protein into separate bands. These can be transferred onto a
123:
2299:
Troubleshooting DNA agarose gel electrophoresis. Focus 19:3 p.66 (1997).
1612:
1565:
1059:); accurate control of parameters such as pore size and stability; and (
2751:
1243:
1052:
728:
691:
After the electrophoresis is complete, the molecules in the gel can be
654:
650:
629:
445:
409:
271:
259:
2334:
2121:"Detection of glucose-6-phosphate dehydrogenase in malarial parasites"
1839:
1390:. Cambridge, United Kingdom New York, NY: Cambridge University Press.
1274:
Kryndushkin DS; Alexandrov IM; Ter-Avanesyan MD; Kushnirov VV (2003).
162:
proteins. Gel electrophoresis can also be used for the separation of
82:
1238:
1180:
970:
948:
886:
863:
658:
633:
595:
362:
2402:"Electrophoretic separation of polyoma virus DNA from host cell DNA"
2197:
fundamental laboratory approaches for biochemistry and biotechnology
2005:
1921:
1108:
962:
818:
752:
606:
413:
412:
and protein analysis, often used to separate different proteins or
2988:
481:
are disrupted, leaving only the primary structure to be analyzed.
2738:
1873:"A thin-layer starch gel method for enzyme typing of bloodstains"
1498:
1487:"Agarose gel electrophoresis for the separation of DNA fragments"
1233:
1089:
1023:
958:
940:
874:
732:
566:
536:
354:
350:
287:
239:
220:
183:
158:
135:
980:
Characterization through ligand interaction may be performed by
336:
Inserting the gel comb in an agarose gel electrophoresis chamber
2674:, from the University of Utah's Genetic Science Learning Center
2195:
Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (2009).
1484:
1041:
1001:
997:
912:
smeared appearance, and the intensity ratio is less than 2:1.
2900:
859:
584:
258:) the gel is usually composed of different concentrations of
157:
to move the negatively charged molecules through a matrix of
2546:
1074:
550:(TGGE) and denaturing gradient gel electrophoresis (DGGE).
528:
485:
421:
2270:
926:
846:
1161:
900:
is usually done by agarose gel electrophoresis. See the "
897:
893:
683:
Gel electrophoresis of nucleic acids § Visualization
251:
247:
231:
131:
127:
2172:
Fundamental
Approaches to Biochemistry and Biotechnology
664:
Most SDS-PAGE protein separations are performed using a
2698:
Step by step photos of running a gel and extracting DNA
583:
are used only to the extent that they are necessary to
2672:
Biotechniques
Laboratory electrophoresis demonstration
2308:
1870:
2355:
2118:
736:
687:
Gel electrophoresis of proteins § Visualization
153:
Nucleic acid molecules are separated by applying an
2600:
2056:
1962:
1922:Buell GN; Wickens MP; Payvar F; Schimke RT (1978).
1531:"Molecular Weight Determination by SDS-PAGE, Rev B"
657:(TBE). Many other buffers have been proposed, e.g.
2737:
2580:Electrophoresis: theory, methods, and applications
2272:
1066:1965 – introduction of free-flow electrophoresis (
695:to make them visible. DNA may be visualized using
488:in the buffer, while proteins are denatured using
16:Method for separation and analysis of biomolecules
2365:(9). American Chemical Society (ACS): 5157–5166.
2194:
1599:(5). American Chemical Society (ACS): 2270–2277.
1454:Biochemical techniques : theory and practice
1423:(in Estonian). San Francisco: Benjamin Cummings.
1139:1981 – introduction of capillary electrophoresis
3261:
2678:Discontinuous native protein gel electrophoresis
2613:(10). Public Library of Science (PLoS): e46147.
290:which is negatively charged (because this is an
2450:
2219:
1817:
1815:
1114:1972 – agarose gels with ethidium bromide stain
484:Nucleic acids are often denatured by including
2349:
1590:
1485:Lee PY; Costumbrado J; Hsu CY; Kim YH (2012).
996:and determination of structural features like
797:Separation of restricted genomic DNA prior to
605:agent. The molecules being separated (usually
3004:
2723:
2491:
969:, by preparative native gel electrophoresis (
707:light, while protein may be visualised using
2693:Animation of gel analysis of DNA restriction
2213:
1999:
1812:
1720:
1698:"Agarose gel electrophoresis (basic method)"
1317:Molecular cloning : a laboratory manual
1308:
2485:
2444:
2393:
2169:
2112:
1956:
1864:
1478:
1267:
122:is a method for separation and analysis of
3011:
2997:
2730:
2716:
2542:
2279:(5th ed.). WH Freeman: New York, NY.
2174:. Bethesda, Md: Fitzgerald Science Press.
2050:
1915:
1821:
1779:
1726:
1584:
1379:
27:
2644:
2626:
2594:
2468:
2399:
1939:
1847:
1506:
1456:. Prospect Heights, Ill: Waveland Press.
1445:
1412:
1291:
628:gel electrophoresis is typically used in
3132:Temperature gradient gel electrophoresis
2571:
1348:
1346:
1344:
1314:
925:
845:
742:
557:
548:temperature gradient gel electrophoresis
461:
331:
207:
90:
81:
75:Temperature gradient gel electrophoresis
2271:Lodish H; Berk A; Matsudaira P (2004).
1568:Molecular Cloning - A Laboratory Manual
1357:(in Estonian). New York: W.H. Freeman.
666:"discontinuous" (or DISC) buffer system
638:single-strand conformation polymorphism
346:Agarose gels are made from the natural
3262:
2973:Photoactivated localization microscopy
2891:Protein–protein interaction prediction
2264:
1785:
1633:
1385:
1156:2004 – introduction of a standardized
2992:
2711:
1547:from the original on 17 November 2021
1451:
1418:
1341:
1055:gels; discontinuous electrophoresis (
107:Electric current applied to the gel.
101:Isolation and amplification of DNA.
3235:
3163:Gel electrophoresis of nucleic acids
3092:Electrophoretic mobility shift assay
2577:
1790:. Amsterdam: North-Holland Pub. Co.
1708:from the original on 11 October 2018
1678:from the original on 2 February 2022
1352:
1204:Electrophoretic mobility shift assay
1022:1930s – first reports of the use of
842:Gel electrophoresis of nucleic acids
731:can be taken of gels, often using a
3158:DNA separation by silica adsorption
3137:Two-dimensional gel electrophoresis
3018:
2848:Freeze-fracture electron microscopy
2359:The Journal of Physical Chemistry C
2199:. Hoboken, NJ: Wiley. p. 161.
1229:Two-dimensional gel electrophoresis
699:which, when intercalated into DNA,
71:Two-dimensional gel electrophoresis
33:Gel electrophoresis apparatus – an
13:
3122:Polyacrylamide gel electrophoresis
2549:. De Gruyter, ISBN 9783110761627.
387:Polyacrylamide gel electrophoresis
14:
3306:
2703:A typical method from wikiversity
2665:
2524:from the original on 11 June 2022
2432:from the original on 11 June 2022
2252:from the original on 11 June 2022
2100:from the original on 11 June 2022
2038:from the original on 11 June 2022
1987:from the original on 11 June 2022
1903:from the original on 11 June 2022
1871:Wraxall BG; Culliford BJ (1968).
1767:from the original on 11 June 2022
854:product compared to a DNA ladder.
564:Glucose-6-Phosphate Dehydrogenase
562:Specific enzyme-linked staining:
452:
380:
197:
110:DNA bands are separated by size.
3234:
3223:
3222:
3127:Pulsed-field gel electrophoresis
2828:Isothermal titration calorimetry
2808:Dual-polarization interferometry
2494:"The gel electrophoresis of DNA"
2151:from the original on 6 July 2023
2119:Hempelmann E; Wilson RJ (1981).
2024:10.1046/j.1462-2920.2002.00358.x
1421:Modern experimental biochemistry
1219:Pulsed field gel electrophoresis
1007:
835:
676:
322:pulsed field gel electrophoresis
212:Overview of gel electrophoresis.
3168:Gel electrophoresis of proteins
3117:Moving-boundary electrophoresis
3057:Capillary electrochromatography
2582:. Academic Press. p. 225.
2536:
2302:
2293:
2188:
2163:
1690:
1640:Current Protocols in Immunology
1627:
1028:moving-boundary electrophoresis
922:Gel electrophoresis of proteins
809:Gel electrophoresis is used in
770:
613:) therefore differ not only in
372:field inversion electrophoresis
314:
3072:Difference gel electrophoresis
2683:Drinking straw electrophoresis
2057:Fischer SG; Lerman LS (1979).
1559:
1523:
1125:, then SDS gel electrophoresis
449:concentrations are 5% to 10%.
282:Electrophoresis refers to the
194:for further characterization.
1:
3173:Serum protein electrophoresis
3077:Discontinuous electrophoresis
2818:Chromatin immunoprecipitation
2470:10.1016/S0021-9258(18)94333-4
1941:10.1016/S0021-9258(17)38097-3
1889:10.1016/s0015-7368(68)70449-7
1642:. Chapter 10: 10.4.1–10.4.8.
1636:"Agarose gel electrophoresis"
1260:
892:Gel electrophoresis of large
727:can be recorded of the gel.
457:
304:molecular weight size markers
104:DNA added to the gel wells.
2881:Protein structural alignment
2866:Protein structure prediction
2628:10.1371/journal.pone.0046147
2510:10.1016/0005-2787(72)90426-1
2418:10.1016/0042-6822(66)90029-8
2170:Ninfa AJ, Ballou DP (1998).
2137:10.1016/0166-6851(81)90100-6
2075:10.1016/0092-8674(79)90200-9
1963:Schelp C, Kaaden OR (1989).
1648:10.1002/0471142735.im1004s02
1111:using a stacking gel and SDS
787:products, e.g. in molecular
368:pulsed field electrophoresis
7:
3052:Agarose gel electrophoresis
2965:Super-resolution microscopy
2871:Protein function prediction
2799:Peptide mass fingerprinting
2794:Protein immunoprecipitation
2688:How to run a DNA or RNA gel
1634:Voytas, Daniel (May 2001).
1191:
1117:1975 – 2-dimensional gels (
1107:separated 28 components of
1044:gels, mediocre separation (
915:
757:peptide mass fingerprinting
342:Agarose gel electrophoresis
10:
3311:
3031:History of electrophoresis
2451:Weber K; Osborn M (1969).
2220:Brody JR; Kern SE (2004).
1199:History of electrophoresis
1188:method is an improvement.
1016:
967:native gel electrophoresis
919:
839:
761:de novo peptide sequencing
680:
643:
384:
339:
327:
201:
3290:Polymerase chain reaction
3218:
3210:Electrophoresis (journal)
3202:
3181:
3145:
3062:Capillary electrophoresis
3039:
3026:
2963:
2927:
2899:
2856:
2823:Surface plasmon resonance
2813:Microscale thermophoresis
2803:Protein mass spectrometry
2765:Green fluorescent protein
2750:
1315:Sambrook, Joseph (2001).
1254:Free-flow electrophoresis
1249:Fast parallel proteolysis
1224:Nonlinear frictiophoresis
1026:for gel electrophoresis;
990:capillary electrophoresis
553:
492:, usually as part of the
439:
172:polymerase chain reaction
63:Capillary electrophoresis
58:
53:
43:
26:
3047:Affinity electrophoresis
2843:Cryo-electron microscopy
2492:Aaij C; Borst P (1972).
2371:10.1021/acs.jpcc.9b10644
2238:10.1016/j.ab.2004.05.054
986:affinity electrophoresis
906:affinity electrophoresis
902:chain termination method
713:Coomassie brilliant blue
618:enzyme-linked staining.
525:electrophoretic mobility
277:macromolecular complexes
113:DNA bands are stained.
2876:Protein–protein docking
2789:Protein electrophoresis
1080:1969 – introduction of
1051:1959 – introduction of
1040:1955 – introduction of
873:. Circular DNA such as
596:preparative native PAGE
518:protein electrophoresis
2775:Protein immunostaining
2275:Molecular Cell Biology
1419:Boyer, Rodney (2000).
1386:Wilson, Keith (2018).
1293:10.1074/jbc.M307996200
1141:(Jorgenson and Lukacs)
953:sodium dodecyl sulfate
937:
855:
793:genetic fingerprinting
579:
490:sodium dodecyl sulfate
479:biomolecular structure
467:
337:
213:
116:
88:
3280:Laboratory techniques
3102:Immunoelectrophoresis
3087:Electrochromatography
2833:X-ray crystallography
2555:10.1515/9783110761641
2125:Mol Biochem Parasitol
1786:Gordon, A.H. (1969).
1353:Berg, Jeremy (2002).
1061:Raymond and Weintraub
992:as for estimation of
947:in the presence of a
929:
849:
801:, or of RNA prior to
743:Downstream processing
681:Further information:
572:Plasmodium falciparum
561:
500:that stabilize their
465:
366:should be run with a
335:
211:
94:
85:
3248:Analytical Chemistry
3194:Isoelectric focusing
2760:Protein purification
2578:Bier, Milan (1959).
2498:Biochim Biophys Acta
1745:10.1038/nprot.2006.4
1702:Biological Protocols
1452:Robyt, John (1990).
1214:Isoelectric focusing
1123:isoelectric focusing
1073:1966 – first use of
850:An agarose gel of a
749:isoelectric focusing
510:beta-mercaptoethanol
506:quaternary structure
3189:Electrical mobility
3097:Gel electrophoresis
2785:Gel electrophoresis
2619:2012PLoSO...746147M
2543:Michov, B. (2022).
2327:2007NanoL...7.2881H
1822:Smithies O (1955).
1727:Schägger H (2006).
1613:10.1021/ma00195a048
1605:1989MaMol..22.2270S
1158:polymerization time
975:2-D electrophoresis
778:restriction mapping
719:, for example in a
399:techniques such as
284:electromotive force
236:crosslinked polymer
120:Gel electrophoresis
23:
22:Gel electrophoresis
2928:Display techniques
2780:Protein sequencing
2400:Thorne HV (1966).
1877:J Forensic Sci Soc
1729:"Tricine-SDS-PAGE"
1084:agents especially
1057:Ornstein and Davis
938:
871:radius of gyration
856:
580:
468:
338:
214:
140:clinical chemistry
117:
98:DNA is extracted.
89:
21:
3275:Molecular biology
3257:
3256:
3067:Dielectrophoresis
2986:
2985:
2935:Bacterial display
2335:10.1021/nl071615y
2286:978-0-7167-4366-8
2012:Environ Microbiol
1840:10.1042/bj0610629
1797:978-0-7204-4202-1
1463:978-0-88133-556-9
1430:978-0-8053-3111-0
1397:978-1-316-61476-1
1364:978-0-7167-4955-4
1326:978-0-87969-576-7
994:binding constants
988:in agarose or by
815:molecular biology
803:Northern transfer
799:Southern transfer
789:genetic diagnosis
651:Tris/Acetate/EDTA
192:Southern blotting
176:mass spectrometry
148:molecular biology
124:biomacromolecules
80:
79:
3302:
3238:
3237:
3226:
3225:
3112:Isotachophoresis
3013:
3006:
2999:
2990:
2989:
2950:Ribosome display
2886:Protein ontology
2732:
2725:
2718:
2709:
2708:
2659:
2658:
2648:
2630:
2598:
2592:
2591:
2575:
2569:
2568:
2540:
2534:
2533:
2531:
2529:
2489:
2483:
2482:
2472:
2448:
2442:
2441:
2439:
2437:
2397:
2391:
2390:
2353:
2347:
2346:
2321:(9): 2881–2885.
2306:
2300:
2297:
2291:
2290:
2278:
2268:
2262:
2261:
2259:
2257:
2217:
2211:
2210:
2192:
2186:
2185:
2167:
2161:
2160:
2158:
2156:
2131:(3–4): 197–204.
2116:
2110:
2109:
2107:
2105:
2099:
2054:
2048:
2047:
2045:
2043:
2003:
1997:
1996:
1994:
1992:
1960:
1954:
1953:
1943:
1919:
1913:
1912:
1910:
1908:
1868:
1862:
1861:
1851:
1819:
1810:
1809:
1783:
1777:
1776:
1774:
1772:
1724:
1718:
1717:
1715:
1713:
1694:
1688:
1687:
1685:
1683:
1631:
1625:
1624:
1588:
1582:
1581:
1563:
1557:
1556:
1554:
1552:
1546:
1535:
1527:
1521:
1520:
1510:
1482:
1476:
1475:
1449:
1443:
1442:
1416:
1410:
1409:
1383:
1377:
1376:
1350:
1339:
1338:
1312:
1306:
1305:
1295:
1286:(49): 49636–43.
1271:
1171:, in particular
1000:content through
883:sodium hydroxide
765:in-gel digestion
697:ethidium bromide
671:isotachophoresis
655:Tris/Borate/EDTA
256:oligonucleotides
54:Other techniques
31:
24:
20:
3310:
3309:
3305:
3304:
3303:
3301:
3300:
3299:
3285:Electrophoresis
3270:Protein methods
3260:
3259:
3258:
3253:
3214:
3198:
3177:
3141:
3082:Electroblotting
3035:
3022:
3020:Electrophoresis
3017:
2987:
2982:
2959:
2923:
2919:Secretion assay
2895:
2852:
2746:
2736:
2668:
2663:
2662:
2599:
2595:
2576:
2572:
2565:
2541:
2537:
2527:
2525:
2490:
2486:
2463:(16): 4406–12.
2449:
2445:
2435:
2433:
2398:
2394:
2354:
2350:
2307:
2303:
2298:
2294:
2287:
2269:
2265:
2255:
2253:
2218:
2214:
2207:
2193:
2189:
2182:
2168:
2164:
2154:
2152:
2117:
2113:
2103:
2101:
2097:
2055:
2051:
2041:
2039:
2004:
2000:
1990:
1988:
1961:
1957:
1920:
1916:
1906:
1904:
1869:
1865:
1820:
1813:
1798:
1784:
1780:
1770:
1768:
1725:
1721:
1711:
1709:
1696:
1695:
1691:
1681:
1679:
1632:
1628:
1589:
1585:
1578:
1564:
1560:
1550:
1548:
1544:
1538:www.bio-rad.com
1533:
1529:
1528:
1524:
1483:
1479:
1464:
1450:
1446:
1431:
1417:
1413:
1398:
1384:
1380:
1365:
1351:
1342:
1327:
1313:
1309:
1272:
1268:
1263:
1258:
1194:
1177:electrophoresis
1160:for acrylamide
1019:
1010:
982:electroblotting
933:autoradiography
924:
918:
844:
838:
773:
745:
689:
679:
646:
588:lipid membranes
577:Red blood cells
556:
498:disulfide bonds
460:
455:
442:
389:
383:
353:extracted from
344:
330:
317:
217:Electrophoresis
206:
204:Electrophoresis
200:
73:
69:
65:
48:Electrophoresis
39:
17:
12:
11:
5:
3308:
3298:
3297:
3292:
3287:
3282:
3277:
3272:
3255:
3254:
3252:
3251:
3244:
3232:
3219:
3216:
3215:
3213:
3212:
3206:
3204:
3200:
3199:
3197:
3196:
3191:
3185:
3183:
3179:
3178:
3176:
3175:
3170:
3165:
3160:
3155:
3149:
3147:
3143:
3142:
3140:
3139:
3134:
3129:
3124:
3119:
3114:
3109:
3104:
3099:
3094:
3089:
3084:
3079:
3074:
3069:
3064:
3059:
3054:
3049:
3043:
3041:
3037:
3036:
3034:
3033:
3027:
3024:
3023:
3016:
3015:
3008:
3001:
2993:
2984:
2983:
2981:
2980:
2975:
2969:
2967:
2961:
2960:
2958:
2957:
2952:
2947:
2942:
2937:
2931:
2929:
2925:
2924:
2922:
2921:
2916:
2911:
2905:
2903:
2897:
2896:
2894:
2893:
2888:
2883:
2878:
2873:
2868:
2862:
2860:
2858:Bioinformatics
2854:
2853:
2851:
2850:
2845:
2840:
2835:
2830:
2825:
2820:
2815:
2810:
2805:
2796:
2791:
2782:
2777:
2772:
2767:
2762:
2756:
2754:
2748:
2747:
2735:
2734:
2727:
2720:
2712:
2706:
2705:
2700:
2695:
2690:
2685:
2680:
2675:
2667:
2666:External links
2664:
2661:
2660:
2593:
2570:
2563:
2535:
2504:(2): 192–200.
2484:
2443:
2392:
2348:
2301:
2292:
2285:
2263:
2212:
2206:978-0470087664
2205:
2187:
2180:
2162:
2111:
2069:(1): 191–200.
2049:
2018:(11): 634–43.
1998:
1975:(3): 297–302.
1955:
1934:(7): 2471–82.
1914:
1863:
1811:
1796:
1778:
1719:
1689:
1626:
1593:Macromolecules
1583:
1577:978-0879691363
1576:
1558:
1522:
1477:
1462:
1444:
1429:
1411:
1396:
1378:
1363:
1340:
1325:
1307:
1265:
1264:
1262:
1259:
1257:
1256:
1251:
1246:
1241:
1236:
1231:
1226:
1221:
1216:
1211:
1209:Gel extraction
1206:
1201:
1195:
1193:
1190:
1185:
1184:
1169:gel properties
1154:
1143:
1137:
1126:
1115:
1112:
1101:
1088:separation of
1078:
1071:
1064:
1049:
1038:
1035:
1018:
1015:
1009:
1006:
920:Main article:
917:
914:
879:hydrogen bonds
840:Main article:
837:
834:
807:
806:
795:
781:
780:of cloned DNA.
772:
769:
744:
741:
721:DNA sequencing
678:
675:
659:lithium borate
645:
642:
615:molecular mass
555:
552:
514:dithiothreitol
459:
456:
454:
453:Gel conditions
451:
441:
438:
433:resolving gels
418:nitrocellulose
397:DNA sequencing
385:Main article:
382:
381:Polyacrylamide
379:
348:polysaccharide
340:Main article:
329:
326:
316:
313:
225:polyacrylamide
199:
198:Physical basis
196:
188:DNA sequencing
155:electric field
115:
114:
111:
108:
105:
102:
99:
78:
77:
60:
56:
55:
51:
50:
45:
44:Classification
41:
40:
32:
15:
9:
6:
4:
3:
2:
3307:
3296:
3293:
3291:
3288:
3286:
3283:
3281:
3278:
3276:
3273:
3271:
3268:
3267:
3265:
3250:
3249:
3245:
3243:
3242:
3233:
3231:
3230:
3221:
3220:
3217:
3211:
3208:
3207:
3205:
3201:
3195:
3192:
3190:
3187:
3186:
3184:
3180:
3174:
3171:
3169:
3166:
3164:
3161:
3159:
3156:
3154:
3153:DNA laddering
3151:
3150:
3148:
3144:
3138:
3135:
3133:
3130:
3128:
3125:
3123:
3120:
3118:
3115:
3113:
3110:
3108:
3107:Iontophoresis
3105:
3103:
3100:
3098:
3095:
3093:
3090:
3088:
3085:
3083:
3080:
3078:
3075:
3073:
3070:
3068:
3065:
3063:
3060:
3058:
3055:
3053:
3050:
3048:
3045:
3044:
3042:
3038:
3032:
3029:
3028:
3025:
3021:
3014:
3009:
3007:
3002:
3000:
2995:
2994:
2991:
2979:
2976:
2974:
2971:
2970:
2968:
2966:
2962:
2956:
2955:Yeast display
2953:
2951:
2948:
2946:
2945:Phage display
2943:
2941:
2938:
2936:
2933:
2932:
2930:
2926:
2920:
2917:
2915:
2914:Protein assay
2912:
2910:
2907:
2906:
2904:
2902:
2898:
2892:
2889:
2887:
2884:
2882:
2879:
2877:
2874:
2872:
2869:
2867:
2864:
2863:
2861:
2859:
2855:
2849:
2846:
2844:
2841:
2839:
2836:
2834:
2831:
2829:
2826:
2824:
2821:
2819:
2816:
2814:
2811:
2809:
2806:
2804:
2800:
2797:
2795:
2792:
2790:
2786:
2783:
2781:
2778:
2776:
2773:
2771:
2768:
2766:
2763:
2761:
2758:
2757:
2755:
2753:
2749:
2744:
2740:
2733:
2728:
2726:
2721:
2719:
2714:
2713:
2710:
2704:
2701:
2699:
2696:
2694:
2691:
2689:
2686:
2684:
2681:
2679:
2676:
2673:
2670:
2669:
2656:
2652:
2647:
2642:
2638:
2634:
2629:
2624:
2620:
2616:
2612:
2608:
2604:
2597:
2589:
2585:
2581:
2574:
2566:
2564:9783110761641
2560:
2556:
2552:
2548:
2547:
2539:
2523:
2519:
2515:
2511:
2507:
2503:
2499:
2495:
2488:
2480:
2476:
2471:
2466:
2462:
2458:
2454:
2447:
2431:
2427:
2423:
2419:
2415:
2411:
2407:
2403:
2396:
2388:
2384:
2380:
2376:
2372:
2368:
2364:
2360:
2352:
2344:
2340:
2336:
2332:
2328:
2324:
2320:
2316:
2312:
2305:
2296:
2288:
2282:
2277:
2276:
2267:
2251:
2247:
2243:
2239:
2235:
2231:
2227:
2223:
2216:
2208:
2202:
2198:
2191:
2183:
2181:9781891786006
2177:
2173:
2166:
2150:
2146:
2142:
2138:
2134:
2130:
2126:
2122:
2115:
2096:
2092:
2088:
2084:
2080:
2076:
2072:
2068:
2064:
2060:
2053:
2037:
2033:
2029:
2025:
2021:
2017:
2013:
2009:
2002:
1986:
1982:
1978:
1974:
1970:
1966:
1959:
1951:
1947:
1942:
1937:
1933:
1929:
1925:
1918:
1902:
1898:
1894:
1890:
1886:
1882:
1878:
1874:
1867:
1859:
1855:
1850:
1845:
1841:
1837:
1834:(4): 629–41.
1833:
1829:
1825:
1818:
1816:
1807:
1803:
1799:
1793:
1789:
1782:
1766:
1762:
1758:
1754:
1750:
1746:
1742:
1738:
1734:
1730:
1723:
1707:
1703:
1699:
1693:
1677:
1673:
1669:
1665:
1661:
1657:
1653:
1649:
1645:
1641:
1637:
1630:
1622:
1618:
1614:
1610:
1606:
1602:
1598:
1594:
1587:
1579:
1573:
1569:
1562:
1543:
1539:
1532:
1526:
1518:
1514:
1509:
1504:
1500:
1496:
1492:
1488:
1481:
1473:
1469:
1465:
1459:
1455:
1448:
1440:
1436:
1432:
1426:
1422:
1415:
1407:
1403:
1399:
1393:
1389:
1382:
1374:
1370:
1366:
1360:
1356:
1349:
1347:
1345:
1336:
1332:
1328:
1322:
1318:
1311:
1303:
1299:
1294:
1289:
1285:
1281:
1277:
1270:
1266:
1255:
1252:
1250:
1247:
1245:
1242:
1240:
1237:
1235:
1232:
1230:
1227:
1225:
1222:
1220:
1217:
1215:
1212:
1210:
1207:
1205:
1202:
1200:
1197:
1196:
1189:
1182:
1178:
1174:
1173:gel stability
1170:
1166:
1163:
1159:
1155:
1152:
1148:
1144:
1142:
1138:
1135:
1131:
1127:
1124:
1120:
1116:
1113:
1110:
1106:
1102:
1099:
1095:
1091:
1087:
1083:
1079:
1076:
1072:
1069:
1065:
1062:
1058:
1054:
1050:
1047:
1043:
1039:
1036:
1033:
1029:
1025:
1021:
1020:
1014:
1008:Nanoparticles
1005:
1003:
999:
995:
991:
987:
983:
978:
976:
972:
968:
964:
960:
956:
954:
950:
946:
942:
935:
934:
928:
923:
913:
909:
907:
903:
899:
895:
890:
888:
884:
880:
876:
872:
867:
865:
861:
853:
848:
843:
836:Nucleic acids
833:
830:
828:
824:
820:
816:
812:
804:
800:
796:
794:
790:
786:
782:
779:
775:
774:
768:
766:
762:
758:
754:
750:
740:
738:
734:
730:
726:
725:autoradiogram
722:
718:
717:radioactivity
714:
710:
706:
702:
698:
694:
688:
684:
677:Visualization
674:
672:
667:
662:
660:
656:
652:
641:
639:
635:
631:
627:
623:
619:
616:
612:
611:nucleic acids
608:
604:
599:
597:
593:
589:
586:
578:
574:
573:
568:
565:
560:
551:
549:
544:
542:
541:methylmercury
538:
534:
530:
526:
521:
519:
515:
511:
507:
503:
499:
495:
491:
487:
482:
480:
476:
475:macromolecule
472:
464:
450:
447:
437:
434:
429:
427:
423:
419:
415:
411:
406:
402:
401:Maxam-Gilbert
398:
393:
388:
378:
375:
373:
369:
364:
358:
356:
352:
349:
343:
334:
325:
323:
312:
308:
305:
299:
297:
296:galvanic cell
293:
289:
285:
280:
278:
273:
269:
265:
261:
257:
253:
249:
245:
244:nucleic acids
241:
237:
233:
228:
226:
222:
218:
210:
205:
195:
193:
189:
185:
181:
177:
173:
167:
165:
164:nanoparticles
160:
156:
151:
149:
145:
141:
137:
133:
129:
125:
121:
112:
109:
106:
103:
100:
97:
96:
93:
84:
76:
72:
68:
64:
61:
57:
52:
49:
46:
42:
36:
30:
25:
19:
3246:
3239:
3227:
3146:Applications
3096:
2940:mRNA display
2909:Enzyme assay
2784:
2770:Western blot
2752:Experimental
2610:
2606:
2596:
2579:
2573:
2545:
2538:
2526:. Retrieved
2501:
2497:
2487:
2460:
2456:
2446:
2434:. Retrieved
2412:(2): 234–9.
2409:
2405:
2395:
2362:
2358:
2351:
2318:
2315:Nano Letters
2314:
2304:
2295:
2274:
2266:
2254:. Retrieved
2229:
2226:Anal Biochem
2225:
2215:
2196:
2190:
2171:
2165:
2153:. Retrieved
2128:
2124:
2114:
2102:. Retrieved
2066:
2062:
2052:
2040:. Retrieved
2015:
2011:
2001:
1989:. Retrieved
1972:
1968:
1958:
1931:
1927:
1917:
1905:. Retrieved
1880:
1876:
1866:
1831:
1827:
1787:
1781:
1769:. Retrieved
1739:(1): 16–22.
1736:
1732:
1722:
1710:. Retrieved
1701:
1692:
1680:. Retrieved
1639:
1629:
1596:
1592:
1586:
1567:
1561:
1549:. Retrieved
1537:
1525:
1499:10.3791/3923
1490:
1480:
1453:
1447:
1420:
1414:
1387:
1381:
1355:Biochemistry
1354:
1316:
1310:
1283:
1279:
1269:
1186:
1167:to optimize
1011:
979:
957:
939:
930:
910:
891:
868:
857:
831:
827:biochemistry
823:microbiology
808:
783:Analysis of
771:Applications
746:
709:silver stain
690:
663:
647:
624:
620:
600:
581:
570:
545:
522:
483:
469:
443:
430:
426:western blot
395:Traditional
394:
390:
376:
359:
345:
318:
315:Types of gel
309:
300:
294:rather than
292:electrolytic
281:
264:cross-linker
229:
215:
168:
152:
144:biochemistry
119:
118:
18:
2978:Vertico SMI
2838:Protein NMR
2457:J Biol Chem
2232:(1): 1–13.
1928:J Biol Chem
1883:(2): 81–2.
1280:J Biol Chem
729:Photographs
705:ultraviolet
634:metallomics
35:agarose gel
3264:Categories
3040:Techniques
1969:Acta Virol
1733:Nat Protoc
1261:References
1244:Zymography
1181:Kastenholz
1130:sequencing
1082:denaturing
1053:acrylamide
881:, such as
866:backbone.
630:proteomics
603:denaturing
567:isoenzymes
471:Denaturing
458:Denaturing
446:hydrolysed
444:Partially
431:Typically
410:immunology
370:(PFE), or
272:neurotoxin
260:acrylamide
230:The term "
202:See also:
2637:1932-6203
2387:213566317
2379:1932-7447
1828:Biochem J
1761:209529082
1656:1934-368X
1621:0024-9297
1491:J Vis Exp
1406:998750377
1239:QPNC-PAGE
1175:, during
1165:solutions
1119:O’Farrell
1092:subunit (
1004:binding.
973:), or by
971:QPNC-PAGE
949:detergent
945:denatured
931:SDS-PAGE
887:formamide
864:phosphate
811:forensics
701:fluoresce
575:infected
363:base pair
242:or small
87:migrated.
3229:Category
3203:Journals
2745:of study
2739:Proteins
2655:23056252
2607:PLOS ONE
2528:23 March
2522:Archived
2436:23 March
2430:Archived
2406:Virology
2343:17718532
2256:23 March
2250:Archived
2246:15351274
2155:23 March
2149:Archived
2104:23 March
2095:Archived
2042:23 March
2036:Archived
2032:12460271
1991:23 March
1985:Archived
1907:23 March
1901:Archived
1858:13276348
1771:23 March
1765:Archived
1753:17406207
1712:23 March
1706:Archived
1676:Archived
1672:39623776
1664:18432695
1551:23 March
1542:Archived
1517:22546956
1472:22549624
1439:44493241
1373:48055706
1335:45015638
1302:14507919
1192:See also
1147:Schwartz
1109:T4 phage
1046:Smithies
1032:Tiselius
963:SDS-PAGE
959:Proteins
951:such as
941:Proteins
916:Proteins
875:plasmids
819:genetics
753:SDS-PAGE
723:gel, an
607:proteins
502:tertiary
494:SDS-PAGE
414:isoforms
351:polymers
240:proteins
136:proteins
67:SDS-PAGE
3241:Commons
2743:methods
2646:3463568
2615:Bibcode
2588:1175404
2518:5063906
2479:5806584
2426:4287545
2323:Bibcode
2145:7012616
2091:9369012
1981:2570517
1897:5738223
1849:1215845
1682:1 March
1601:Bibcode
1508:4846332
1234:SDD-AGE
1128:1977 –
1103:1970 –
1090:protein
1024:sucrose
1017:History
733:Gel Doc
693:stained
653:(TAE),
644:Buffers
590:in the
537:glyoxal
355:seaweed
328:Agarose
288:cathode
221:agarose
184:cloning
182:, PCR,
159:agarose
59:Related
3182:Theory
2741:: key
2653:
2643:
2635:
2586:
2561:
2516:
2477:
2424:
2385:
2377:
2341:
2283:
2244:
2203:
2178:
2143:
2089:
2083:369706
2081:
2030:
1979:
1950:632280
1948:
1895:
1856:
1846:
1804:
1794:
1759:
1751:
1670:
1662:
1654:
1619:
1574:
1515:
1505:
1493:(62).
1470:
1460:
1437:
1427:
1404:
1394:
1371:
1361:
1333:
1323:
1300:
1151:Cantor
1134:Sanger
1132:gels (
1105:Lämmli
1098:Osborn
1068:Hannig
1042:starch
1002:lectin
998:glycan
984:or by
965:), by
763:after
737:SciUGo
703:under
685:, and
626:Native
554:Native
440:Starch
405:Sanger
262:and a
2901:Assay
2383:S2CID
2098:(PDF)
2087:S2CID
1806:21766
1757:S2CID
1668:S2CID
1545:(PDF)
1534:(PDF)
1094:Weber
860:sugar
268:bases
254:, or
190:, or
3295:Gels
2651:PMID
2633:ISSN
2584:OCLC
2559:ISBN
2530:2022
2514:PMID
2475:PMID
2438:2022
2422:PMID
2375:ISSN
2339:PMID
2281:ISBN
2258:2022
2242:PMID
2201:ISBN
2176:ISBN
2157:2022
2141:PMID
2106:2022
2079:PMID
2063:Cell
2044:2022
2028:PMID
1993:2022
1977:PMID
1946:PMID
1909:2022
1893:PMID
1854:PMID
1802:OCLC
1792:ISBN
1773:2022
1749:PMID
1714:2022
1684:2023
1660:PMID
1652:ISSN
1617:ISSN
1572:ISBN
1553:2022
1513:PMID
1468:OCLC
1458:ISBN
1435:OCLC
1425:ISBN
1402:OCLC
1392:ISBN
1369:OCLC
1359:ISBN
1331:OCLC
1321:ISBN
1298:PMID
1149:and
1096:and
1077:gels
1075:agar
825:and
632:and
592:cell
585:lyse
535:and
533:DMSO
529:Urea
504:and
486:urea
422:PVDF
180:RFLP
146:and
2641:PMC
2623:doi
2551:doi
2506:doi
2502:269
2465:doi
2461:244
2414:doi
2367:doi
2363:124
2331:doi
2234:doi
2230:333
2133:doi
2071:doi
2020:doi
1936:doi
1932:253
1885:doi
1844:PMC
1836:doi
1741:doi
1644:doi
1609:doi
1503:PMC
1495:doi
1288:doi
1284:278
1162:gel
1121:);
1086:SDS
898:RNA
896:or
894:DNA
885:or
852:PCR
791:or
785:PCR
759:or
751:or
711:or
609:or
569:in
512:or
420:or
403:or
252:RNA
248:DNA
232:gel
223:or
132:RNA
128:DNA
3266::
2649:.
2639:.
2631:.
2621:.
2609:.
2605:.
2557:.
2520:.
2512:.
2500:.
2496:.
2473:.
2459:.
2455:.
2428:.
2420:.
2410:29
2408:.
2404:.
2381:.
2373:.
2361:.
2337:.
2329:.
2317:.
2313:.
2248:.
2240:.
2228:.
2224:.
2147:.
2139:.
2127:.
2123:.
2093:.
2085:.
2077:.
2067:16
2065:.
2061:.
2034:.
2026:.
2014:.
2010:.
1983:.
1973:33
1971:.
1967:.
1944:.
1930:.
1926:.
1899:.
1891:.
1879:.
1875:.
1852:.
1842:.
1832:61
1830:.
1826:.
1814:^
1800:.
1763:.
1755:.
1747:.
1735:.
1731:.
1704:.
1700:.
1674:.
1666:.
1658:.
1650:.
1638:.
1615:.
1607:.
1597:22
1595:.
1540:.
1536:.
1511:.
1501:.
1489:.
1466:.
1433:.
1400:.
1367:.
1343:^
1329:.
1296:.
1282:.
1278:.
977:.
908:.
821:,
817:,
813:,
739:.
640:.
598:.
531:,
527:.
520:.
428:.
374:.
279:.
250:,
186:,
178:,
166:.
134:,
130:,
3012:e
3005:t
2998:v
2801:/
2787:/
2731:e
2724:t
2717:v
2657:.
2625::
2617::
2611:7
2590:.
2567:.
2553::
2532:.
2508::
2481:.
2467::
2440:.
2416::
2389:.
2369::
2345:.
2333::
2325::
2319:7
2289:.
2260:.
2236::
2209:.
2184:.
2159:.
2135::
2129:2
2108:.
2073::
2046:.
2022::
2016:4
1995:.
1952:.
1938::
1911:.
1887::
1881:8
1860:.
1838::
1808:.
1775:.
1743::
1737:1
1716:.
1686:.
1646::
1623:.
1611::
1603::
1580:.
1555:.
1519:.
1497::
1474:.
1441:.
1408:.
1375:.
1337:.
1304:.
1290::
1183:)
1179:(
1153:)
1136:)
1100:)
1070:)
1063:)
1048:)
1034:)
1030:(
862:-
805:.
246:(
126:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.