3822:
3593:(SELEX). DNAzymes catalyze variety of chemical reactions including RNA-DNA cleavage, RNA-DNA ligation, amino acids phosphorylation-dephosphorylation, carbon-carbon bond formation, etc. DNAzymes can enhance catalytic rate of chemical reactions up to 100,000,000,000-fold over the uncatalyzed reaction. The most extensively studied class of DNAzymes is RNA-cleaving types which have been used to detect different metal ions and designing therapeutic agents. Several metal-specific DNAzymes have been reported including the GR-5 DNAzyme (lead-specific), the CA1-3 DNAzymes (copper-specific), the 39E DNAzyme (uranyl-specific) and the NaA43 DNAzyme (sodium-specific). The NaA43 DNAzyme, which is reported to be more than 10,000-fold selective for sodium over other metal ions, was used to make a real-time sodium sensor in cells.
3281:, in which a segment of a single strand in each helix is annealed to the complementary strand in the other helix. The Holliday junction is a tetrahedral junction structure that can be moved along the pair of chromosomes, swapping one strand for another. The recombination reaction is then halted by cleavage of the junction and re-ligation of the released DNA. Only strands of like polarity exchange DNA during recombination. There are two types of cleavage: east-west cleavage and north–south cleavage. The north–south cleavage nicks both strands of DNA, while the east–west cleavage has one strand of DNA intact. The formation of a Holliday junction during recombination makes it possible for genetic diversity, genes to exchange on chromosomes, and expression of wild-type viral genomes.
4089:
43:
592:
1507:
176:
124:
3975:
2907:, which are proteins that regulate transcription. Each transcription factor binds to one particular set of DNA sequences and activates or inhibits the transcription of genes that have these sequences close to their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription. Alternatively, transcription factors can bind
1656:. With DNA in its "relaxed" state, a strand usually circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands become more tightly or more loosely wound. If the DNA is twisted in the direction of the helix, this is positive supercoiling, and the bases are held more tightly together. If they are twisted in the opposite direction, this is negative supercoiling, and the bases come apart more easily. In nature, most DNA has slight negative supercoiling that is introduced by
343:
2052:
2229:
1855:
3196:
102:
880:
15668:
2892:
1295:
15580:
14752:
457:
1942:
2691:
3187:
1935:
152:
3682:
2746:. This enzyme makes the complementary strand by finding the correct base through complementary base pairing and bonding it onto the original strand. As DNA polymerases can only extend a DNA strand in a 5′ to 3′ direction, different mechanisms are used to copy the antiparallel strands of the double helix. In this way, the base on the old strand dictates which base appears on the new strand, and the cell ends up with a perfect copy of its DNA.
2386:
2095:
2569:
15606:
4109:, in which they said, "It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material." This letter was followed by a letter from Franklin and Gosling, which was the first publication of their own X-ray diffraction data and of their original analysis method. Then followed a letter by Wilkins and two of his colleagues, which contained an analysis of
15651:
1695:
14225:
15685:
2324:, due to normal cellular processes that produce reactive oxygen species, the hydrolytic activities of cellular water, etc., also occur frequently. Although most of these damages are repaired, in any cell some DNA damage may remain despite the action of repair processes. These remaining DNA damages accumulate with age in mammalian postmitotic tissues. This accumulation appears to be an important underlying cause of aging.
1356:
1343:
3320:
there is no direct evidence of ancient genetic systems, as recovery of DNA from most fossils is impossible because DNA survives in the environment for less than one million years, and slowly degrades into short fragments in solution. Claims for older DNA have been made, most notably a report of the isolation of a viable bacterium from a salt crystal 250 million years old, but these claims are controversial.
15634:
2827:
2938:
2088:
1310:. As the strands are not symmetrically located with respect to each other, the grooves are unequally sized. The major groove is 22 ångströms (2.2 nm) wide, while the minor groove is 12 Å (1.2 nm) in width. Due to the larger width of the major groove, the edges of the bases are more accessible in the major groove than in the minor groove. As a result, proteins such as
2102:
1998:. Dubbed S, B, P, and Z, these artificial bases are capable of bonding with each other in a predictable way (S–B and P–Z), maintain the double helix structure of DNA, and be transcribed to RNA. Their existence could be seen as an indication that there is nothing special about the four natural nucleobases that evolved on Earth. On the other hand, DNA is tightly related to
3219:
1418:. As a result of this base pair complementarity, all the information in the double-stranded sequence of a DNA helix is duplicated on each strand, which is vital in DNA replication. This reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in organisms.
3223:
recombination is thought to occur by the Double
Holliday Junction (DHJ) model, illustrated on the right, above. NCO recombinants are thought to occur primarily by the Synthesis Dependent Strand Annealing (SDSA) model, illustrated on the left, above. Most recombination events appear to be the SDSA type.
4056:
and was critical to their obtaining the correct structure of DNA. Franklin told Crick and Watson that the backbones had to be on the outside. Before then, Linus
Pauling, and Watson and Crick, had erroneous models with the chains inside and the bases pointing outwards. Franklin's identification of the
3705:
properties of DNA and other nucleic acids to create self-assembling branched DNA complexes with useful properties. DNA is thus used as a structural material rather than as a carrier of biological information. This has led to the creation of two-dimensional periodic lattices (both tile-based and using
2433:
of genetic information in genes is achieved via complementary base pairing. For example, in transcription, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence through the attraction between the DNA and the correct RNA nucleotides. Usually, this RNA
1397:
to pyrimidines, with adenine bonding only to thymine in two hydrogen bonds, and cytosine bonding only to guanine in three hydrogen bonds. This arrangement of two nucleotides binding together across the double helix (from six-carbon ring to six-carbon ring) is called a Watson-Crick base pair. DNA with
12559:
On p. 264, Kossel remarked presciently: Der
Erforschung der quantitativen Verhältnisse der vier stickstoffreichen Basen, der Abhängigkeit ihrer Menge von den physiologischen Zuständen der Zelle, verspricht wichtige Aufschlüsse über die elementaren physiologisch-chemischen Vorgänge. (The study of the
3552:
law in some places, this can allow cases to be reopened where prior trials have failed to produce sufficient evidence to convince a jury. People charged with serious crimes may be required to provide a sample of DNA for matching purposes. The most obvious defense to DNA matches obtained forensically
3547:
The development of forensic science and the ability to now obtain genetic matching on minute samples of blood, skin, saliva, or hair has led to re-examining many cases. Evidence can now be uncovered that was scientifically impossible at the time of the original examination. Combined with the removal
1000:
Therefore, any DNA strand normally has one end at which there is a phosphate group attached to the 5′ carbon of a ribose (the 5′ phosphoryl) and another end at which there is a free hydroxyl group attached to the 3′ carbon of a ribose (the 3′ hydroxyl). The orientation of the 3′ and 5′ carbons along
3319:
of the current genetic code based on four nucleotide bases. This would occur, since the number of different bases in such an organism is a trade-off between a small number of bases increasing replication accuracy and a large number of bases increasing the catalytic efficiency of ribozymes. However,
2754:
Naked extracellular DNA (eDNA), most of it released by cell death, is nearly ubiquitous in the environment. Its concentration in soil may be as high as 2 μg/L, and its concentration in natural aquatic environments may be as high at 88 μg/L. Various possible functions have been proposed for eDNA: it
2299:
produce multiple forms of damage, including base modifications, particularly of guanosine, and double-strand breaks. A typical human cell contains about 150,000 bases that have suffered oxidative damage. Of these oxidative lesions, the most dangerous are double-strand breaks, as these are difficult
1794:
spiral, with a shallow, wide minor groove and a narrower, deeper major groove. The A form occurs under non-physiological conditions in partly dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, and in enzyme-DNA complexes. Segments of DNA where
923:). The pair of chains have a radius of 10 Å (1.0 nm). According to another study, when measured in a different solution, the DNA chain measured 22–26 Å (2.2–2.6 nm) wide, and one nucleotide unit measured 3.3 Å (0.33 nm) long. The buoyant density of most DNA is 1.7g/cm.
8081:
A certain irreducible background incidence of cancer is to be expected regardless of circumstances: mutations can never be absolutely avoided, because they are an inescapable consequence of fundamental limitations on the accuracy of DNA replication, as discussed in
Chapter 5. If a human could live
3222:
A current model of meiotic recombination, initiated by a double-strand break or gap, followed by pairing with an homologous chromosome and strand invasion to initiate the recombinational repair process. Repair of the gap can lead to crossover (CO) or non-crossover (NCO) of the flanking regions. CO
1971:
occurs when non-complementary regions exist at the end of an otherwise complementary double-strand of DNA. However, branched DNA can occur if a third strand of DNA is introduced and contains adjoining regions able to hybridize with the frayed regions of the pre-existing double-strand. Although the
1902:
In addition to these stacked structures, telomeres also form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a long circle stabilized by telomere-binding proteins. At the very end of the T-loop, the single-stranded telomere DNA is held onto a
1497:
necessary to break half of the hydrogen bonds. When all the base pairs in a DNA double helix melt, the strands separate and exist in solution as two entirely independent molecules. These single-stranded DNA molecules have no single common shape, but some conformations are more stable than others.
3041:
in DNA. Some of these enzymes work by cutting the DNA helix and allowing one section to rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break. Other types of these enzymes are capable of cutting one DNA helix and then passing a second strand of DNA through this
3871:
identified the base, sugar, and phosphate nucleotide unit of RNA (then named "yeast nucleic acid"). In 1929, Levene identified deoxyribose sugar in "thymus nucleic acid" (DNA). Levene suggested that DNA consisted of a string of four nucleotide units linked together through the phosphate groups
2923:
and development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these base-interactions are made in the major groove, where the bases are most
2871:
and changing the rate of transcription. Other non-specific DNA-binding proteins in chromatin include the high-mobility group proteins, which bind to bent or distorted DNA. These proteins are important in bending arrays of nucleosomes and arranging them into the larger structures that make up
4166:
of the
University of Leicester to verify or disprove a suspect's rape-murder "confession". In this particular case, the suspect had confessed to two rape-murders, but had later retracted his confession. DNA testing at the university labs soon disproved the veracity of the suspect's original
1614:
copy that is translated into protein. The sequence on the opposite strand is called the "antisense" sequence. Both sense and antisense sequences can exist on different parts of the same strand of DNA (i.e. both strands can contain both sense and antisense sequences). In both prokaryotes and
1025:
1782:
is most common under the conditions found in cells, it is not a well-defined conformation but a family of related DNA conformations that occur at the high hydration levels present in cells. Their corresponding X-ray diffraction and scattering patterns are characteristic of molecular
1729:
forms, although only B-DNA and Z-DNA have been directly observed in functional organisms. The conformation that DNA adopts depends on the hydration level, DNA sequence, the amount and direction of supercoiling, chemical modifications of the bases, the type and concentration of metal
3536:, are compared between people. This method is usually an extremely reliable technique for identifying a matching DNA. However, identification can be complicated if the scene is contaminated with DNA from several people. DNA profiling was developed in 1984 by British geneticist Sir
2763:
of several bacterial species. It may act as a recognition factor to regulate the attachment and dispersal of specific cell types in the biofilm; it may contribute to biofilm formation; and it may contribute to the biofilm's physical strength and resistance to biological stress.
2002:
which does not only act as a transcript of DNA but also performs as molecular machines many tasks in cells. For this purpose it has to fold into a structure. It has been shown that to allow to create all possible structures at least four bases are required for the corresponding
1898:
of a metal ion in the centre of each four-base unit. Other structures can also be formed, with the central set of four bases coming from either a single strand folded around the bases, or several different parallel strands, each contributing one base to the central structure.
12384:(Therefore, in my experiments I subsequently limited myself to the whole nucleus, leaving to a more favorable material the separation of the substances, that for the present, without further prejudice, I will designate as soluble and insoluble nuclear material ("Nuclein"))
3666:
and their possible functions in an organism even before they have been isolated experimentally. Entire genomes may also be compared, which can shed light on the evolutionary history of particular organism and permit the examination of complex evolutionary events.
12827:. By this term I designate the axial thread of the chromosome, in which the geneticists locate the linear combination of genes; … In the normal chromosome there is usually only one genoneme; before cell-division this genoneme has become divided into two strands."
2353:. For an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. This inhibits both transcription and DNA replication, causing toxicity and mutations. As a result, DNA intercalators may be
1583:
forms closed circular molecules, each of which contains 16,569 DNA base pairs, with each such molecule normally containing a full set of the mitochondrial genes. Each human mitochondrion contains, on average, approximately 5 such mtDNA molecules. Each human
3658:, are difficult to use without the annotations that identify the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have the characteristic patterns associated with protein- or RNA-coding genes can be identified by
13521:"What Rosalind Franklin truly contributed to the discovery of DNA's structure – Franklin was no victim in how the DNA double helix was solved. An overlooked letter and an unpublished news article, both written in 1953, reveal that she was an equal player"
2729:
is essential for an organism to grow, but, when a cell divides, it must replicate the DNA in its genome so that the two daughter cells have the same genetic information as their parent. The double-stranded structure of DNA provides a simple mechanism for
2879:
is the best-understood member of this family and is used in processes where the double helix is separated, including DNA replication, recombination, and DNA repair. These binding proteins seem to stabilize single-stranded DNA and protect it from forming
3139:, which is required for the replication of telomeres. For example, HIV reverse transcriptase is an enzyme for AIDS virus replication. Telomerase is an unusual polymerase because it contains its own RNA template as part of its structure. It synthesizes
3091:
of these enzymes, the incoming nucleoside triphosphate base-pairs to the template: this allows polymerases to accurately synthesize the complementary strand of their template. Polymerases are classified according to the type of template that they use.
1177:
present in bacteria. This enzyme system acts at least in part as a molecular immune system protecting bacteria from infection by viruses. Modifications of the bases cytosine and adenine, the more common and modified DNA bases, play vital roles in the
1013:. The asymmetric ends of DNA strands are said to have a directionality of five prime end (5′ ), and three prime end (3′), with the 5′ end having a terminal phosphate group and the 3′ end a terminal hydroxyl group. One major difference between DNA and
4120:
In April 2023, scientists, based on new evidence, concluded that
Rosalind Franklin was a contributor and "equal player" in the discovery process of DNA, rather than otherwise, as may have been presented subsequently after the time of the discovery.
4073:
proposed a model for nucleic acids containing three intertwined chains, with the phosphates near the axis, and the bases on the outside. Watson and Crick completed their model, which is now accepted as the first correct model of the double helix of
1803:. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form. These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in the regulation of transcription.
12029:
Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CL, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J (May 2009). "Self-assembly of a nanoscale DNA box with a controllable lid".
1428:
Most DNA molecules are actually two polymer strands, bound together in a helical fashion by noncovalent bonds; this double-stranded (dsDNA) structure is maintained largely by the intrastrand base stacking interactions, which are strongest for
1459:
value), which is the temperature at which 50% of the double-strand molecules are converted to single-strand molecules; melting temperature is dependent on ionic strength and the concentration of DNA. As a result, it is both the percentage of
7795:
Gommers-Ampt JH, Van
Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P (December 1993). "beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei".
2838:
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called
2811:
can be non-specific, or the protein can bind specifically to a single DNA sequence. Enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
3099:
make copies of DNA polynucleotide chains. To preserve biological information, it is essential that the sequence of bases in each copy are precisely complementary to the sequence of bases in the template strand. Many DNA polymerases have a
4135:, which foretold the relationship between DNA, RNA, and proteins, and articulated the "adaptor hypothesis". Final confirmation of the replication mechanism that was implied by the double-helical structure followed in 1958 through the
1547:
per cell extends for 6.37 Gigabase pairs (Gbp), is 208.23 cm long and weighs 6.51 picograms (pg). Male values are 6.27 Gbp, 205.00 cm, 6.41 pg. Each DNA polymer can contain hundreds of millions of nucleotides, such as in
13488:"Untangling Rosalind Franklin's Role in DNA Discovery, 70 Years On – Historians have long debated the role that Dr. Franklin played in identifying the double helix. A new opinion essay argues that she was an "equal contributor.""
593:
12381:
Ich habe mich daher später mit meinen
Versuchen an die ganzen Kerne gehalten, die Trennung der Körper, die ich einstweilen ohne weiteres Präjudiz als lösliches und unlösliches Nuclein bezeichnen will, einem günstigeren Material
2676:, giving most amino acids more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region; these are the TAG, TAA, and TGA codons, (UAG, UAA, and UGA on the mRNA).
3634:. String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides. The DNA sequence may be
3556:
DNA profiling is also used successfully to positively identify victims of mass casualty incidents, bodies or body parts in serious accidents, and individual victims in mass war graves, via matching to family members.
1885:
These guanine-rich sequences may stabilize chromosome ends by forming structures of stacked sets of four-base units, rather than the usual base pairs found in other DNA molecules. Here, four guanine bases, known as a
3150:
that copies the sequence of a DNA strand into RNA. To begin transcribing a gene, the RNA polymerase binds to a sequence of DNA called a promoter and separates the DNA strands. It then copies the gene sequence into a
3917:. At the time, "yeast nucleic acid" (RNA) was thought to occur only in plants, while "thymus nucleic acid" (DNA) only in animals. The latter was thought to be a tetramer, with the function of buffering cellular pH.
12560:
quantitative relations of the four nitrogenous bases— of the dependence of their quantity on the physiological states of the cell—promises important insights into the fundamental physiological-chemical processes.)
2914:
As these DNA targets can occur throughout an organism's genome, changes in the activity of one type of transcription factor can affect thousands of genes. Consequently, these proteins are often the targets of the
3893:
could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form. This system provided the first clear suggestion that DNA carries genetic information.
3876:
proposed that inherited traits would be inherited via a "giant hereditary molecule" made up of "two mirror strands that would replicate in a semi-conservative fashion using each strand as a template". In 1928,
3585:, also called DNAzymes or catalytic DNA, were first discovered in 1994. They are mostly single stranded DNA sequences isolated from a large pool of random DNA sequences through a combinatorial approach called
7868:
Douki T, Reynaud-Angelin A, Cadet J, Sage E (August 2003). "Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation".
1681:
1314:
that can bind to specific sequences in double-stranded DNA usually make contact with the sides of the bases exposed in the major groove. This situation varies in unusual conformations of DNA within the cell
2983:, which cut DNA at specific sequences. For instance, the EcoRV enzyme shown to the left recognizes the 6-base sequence 5′-GATATC-3′ and makes a cut at the horizontal line. In nature, these enzymes protect
3754:
Because DNA collects mutations over time, which are then inherited, it contains historical information, and, by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their
3564:
to determine if someone is the biological parent or grandparent of a child with the probability of parentage is typically 99.99% when the alleged parent is biologically related to the child. Normal
13096:"Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types: Induction of Transformation by a Desoxyribonucleic Acid Fraction Isolated from Pneumococcus Type III"
1588:
contains approximately 100 mitochondria, giving a total number of mtDNA molecules per human cell of approximately 500. However, the amount of mitochondria per cell also varies by cell type, and an
3104:
activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3′ to 5′
1638:, this overlap may be involved in the regulation of gene transcription, while in viruses, overlapping genes increase the amount of information that can be encoded within the small viral genome.
3231:". This physical separation of different chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is in
1306:
Twin helical strands form the DNA backbone. Another double helix may be found tracing the spaces, or grooves, between the strands. These voids are adjacent to the base pairs and may provide a
3060:(ATP), to break hydrogen bonds between bases and unwind the DNA double helix into single strands. These enzymes are essential for most processes where enzymes need to access the DNA bases.
1448:
basepairs) but also on sequence (since stacking is sequence specific) and also length (longer molecules are more stable). The stability can be measured in various ways; a common way is the
907:. The structure of DNA is dynamic along its length, being capable of coiling into tight loops and other shapes. In all species it is composed of two helical chains, bound to each other by
1634:. In these cases, some DNA sequences do double duty, encoding one protein when read along one strand, and a second protein when read in the opposite direction along the other strand. In
6218:
Basu HS, Feuerstein BG, Zarling DA, Shafer RH, Marton LJ (October 1988). "Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: experimental and theoretical studies".
3299:
DNA has performed this function, as it has been proposed that the earliest forms of life may have used RNA as their genetic material. RNA may have acted as the central part of early
13436:
3396:
has been recovered from ancient organisms at a timescale where genome evolution can be directly observed, including from extinct organisms up to millions of years old, such as the
3250:
and can be important in the rapid evolution of new proteins. Genetic recombination can also be involved in DNA repair, particularly in the cell's response to double-strand breaks.
5531:"Single-stranded adenine-rich DNA and RNA retain structural characteristics of their respective double-stranded conformations and show directional differences in stacking pattern"
1840:
was announced, though the research was disputed, and evidence suggests the bacterium actively prevents the incorporation of arsenic into the DNA backbone and other biomolecules.
2851:, which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones, making
1972:
simplest example of branched DNA involves only three strands of DNA, complexes involving additional strands and multiple branches are also possible. Branched DNA can be used in
926:
DNA does not usually exist as a single strand, but instead as a pair of strands that are held tightly together. These two long strands coil around each other, in the shape of a
11560:
11318:
14215:
5486:
deHaseth PL, Helmann JD (June 1995). "Open complex formation by
Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA".
2759:; it may provide nutrients; and it may act as a buffer to recruit or titrate ions or antibiotics. Extracellular DNA acts as a functional extracellular matrix component in the
2446:. The details of these functions are covered in other articles; here the focus is on the interactions between DNA and other molecules that mediate the function of the genome.
14840:
11084:"Million-year-old mammoth genomes shatter record for oldest ancient DNA – Permafrost-preserved teeth, up to 1.6 million years old, identify a new kind of mammoth in Siberia"
2855:
to the acidic sugar-phosphate backbone of the DNA, and are thus largely independent of the base sequence. Chemical modifications of these basic amino acid residues include
1874:, as the enzymes that normally replicate DNA cannot copy the extreme 3′ ends of chromosomes. These specialized chromosome caps also help protect the DNA ends, and stop the
1433:
stacks. The two strands can come apart—a process known as melting—to form two single-stranded DNA (ssDNA) molecules. Melting occurs at high temperatures, low salt and high
14213:
2369:
form DNA adducts that induce errors in replication. Nevertheless, due to their ability to inhibit DNA transcription and replication, other similar toxins are also used in
1742:
1414:, they can be broken and rejoined relatively easily. The two strands of DNA in a double helix can thus be pulled apart like a zipper, either by a mechanical force or high
11032:
1615:
eukaryotes, antisense RNA sequences are produced, but the functions of these RNAs are not entirely clear. One proposal is that antisense RNAs are involved in regulating
871:, compact and organize DNA. These compacting structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
4872:"RCSB PDB – 1D65: Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution"
4102:
published a series of five articles giving the Watson and Crick double-helix structure DNA and evidence supporting it. The structure was reported in a letter titled "
3590:
1464:
base pairs and the overall length of a DNA double helix that determines the strength of the association between the two strands of DNA. Long DNA helices with a high
13027:
1829:
of Earth that uses radically different biochemical and molecular processes than currently known life. One of the proposals was the existence of lifeforms that use
3227:
A DNA helix usually does not interact with other segments of DNA, and in human cells, the different chromosomes even occupy separate areas in the nucleus called "
6705:
Rothenburg S, Koch-Nolte F, Haag F (December 2001). "DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles".
5681:
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. (April 1981). "Sequence and organization of the human mitochondrial genome".
11141:
Goff SP, Berg P (December 1976). "Construction of hybrid viruses containing SV40 and lambda phage DNA segments and their propagation in cultured monkey cells".
9768:"RCSB PDB – 1RVA: Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Å resolution"
9442:
14590:
4484:
Irobalieva RN, Fogg JM, Catanese DJ, Catanese DJ, Sutthibutpong T, Chen M, Barker AK, Ludtke SJ, Harris SA, Schmid MF, Chiu W, Zechiedrich L (October 2015).
13062:
1862:
repeats. The looped conformation of the DNA backbone is very different from the typical DNA helix. The green spheres in the center represent potassium ions.
10022:
6850:
6070:
3553:
is to claim that cross-contamination of evidence has occurred. This has resulted in meticulous strict handling procedures with new cases of serious crime.
2672:, which carries amino acids. Since there are 4 bases in 3-letter combinations, there are 64 possible codons (4 combinations). These encode the twenty
14214:
3295:
DNA contains the genetic information that allows all forms of life to function, grow and reproduce. However, it is unclear how long in the 4-billion-year
1579:(mtDNA) which encodes certain proteins used by the mitochondria. The mtDNA is usually relatively small in comparison to the nuclear DNA. For example, the
1526:(to scale at bottom left). The blue scale to the left of each chromosome pair (and the mitochondrial genome) shows its length in terms of millions of DNA
11499:
1879:
2442:, which depends on the same interaction between RNA nucleotides. In an alternative fashion, a cell may copy its genetic information in a process called
1319:, but the major and minor grooves are always named to reflect the differences in width that would be seen if the DNA was twisted back into the ordinary
13938:
First published in
October 1974 by MacMillan, with foreword by Francis Crick; the definitive DNA textbook, revised in 1994 with a nine-page postscript.
8113:
5054:
Johnson TB, Coghill RD (1925). "Pyrimidines. CIII. The discovery of 5-methylcytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus".
2320:. Because of inherent limits in the DNA repair mechanisms, if humans lived long enough, they would all eventually develop cancer. DNA damages that are
1592:
can contain 100,000 mitochondria, corresponding to up to 1,500,000 copies of the mitochondrial genome (constituting up to 90% of the DNA of the cell).
737:
9893:
Schoeffler AJ, Berger JM (December 2005). "Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism".
9231:
8082:
long enough, it is inevitable that at least one of his or her cells would eventually accumulate a set of mutations sufficient for cancer to develop.
5561:
9351:
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (September 1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution".
14363:
10966:
14262:
10761:
Vreeland RH, Rosenzweig WD, Powers DW (October 2000). "Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal".
993:(five prime end) carbons, the prime symbol being used to distinguish these carbon atoms from those of the base to which the deoxyribose forms a
6334:
6289:
5232:
Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE (October 1980). "Crystal structure analysis of a complete turn of B-DNA".
13459:
13646:
13497:
7201:
Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (May 1999). "Mammalian telomeres end in a large duplex loop".
3821:
2591:
typically contain few genes but are important for the function and stability of chromosomes. An abundant form of noncoding DNA in humans are
2153:
protein core around which DNA is wrapped in the chromatin structure or else by remodeling carried out by chromatin remodeling complexes (see
10999:
6889:
6579:
5429:"A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques"
3042:
break, before rejoining the helix. Topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.
3246:
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of
2867:. These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to
13432:
5197:
Carell T, Kurz MQ, Müller M, Rossa M, Spada F (April 2018). "Non-canonical Bases in the Genome: The Regulatory Information Layer in DNA".
1749:
that provided only a limited amount of structural information for oriented fibers of DNA. An alternative analysis was proposed by Wilkins
9535:
Grosschedl R, Giese K, Pagel J (March 1994). "HMG domain proteins: architectural elements in the assembly of nucleoprotein structures".
2875:
A distinct group of DNA-binding proteins is the DNA-binding proteins that specifically bind single-stranded DNA. In humans, replication
1903:
region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This
1386:
In a DNA double helix, each type of nucleobase on one strand bonds with just one type of nucleobase on the other strand. This is called
14615:
12526:
5841:
488:
11544:
11310:
4326:
3277:
or damage to the DNA. A series of steps catalyzed in part by the recombinase then leads to joining of the two helices by at least one
2141:. Base modifications can be involved in packaging, with regions that have low or no gene expression usually containing high levels of
799:, these chromosomes are duplicated in the process of DNA replication, providing a complete set of chromosomes for each daughter cell.
14788:
9029:
Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS (February 2002). "Extracellular DNA required for bacterial biofilm formation".
7904:
Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S (March 1999). "Hydroxyl radicals and DNA base damage".
4983:
3143:
at the ends of chromosomes. Telomeres prevent fusion of the ends of neighboring chromosomes and protect chromosome ends from damage.
10847:
Nickle DC, Learn GH, Rain MW, Mullins JI, Mittler JE (January 2002). "Curiously modern DNA for a "250 million-year-old" bacterium".
8040:
6816:
6648:
6404:
3123:
RNA-dependent DNA polymerases are a specialized class of polymerases that copy the sequence of an RNA strand into DNA. They include
2903:
In contrast, other proteins have evolved to bind to particular DNA sequences. The most intensively studied of these are the various
5732:
3944:
2496:. The genetic information in a genome is held within genes, and the complete set of this information in an organism is called its
1472:
content have more weakly interacting strands. In biology, parts of the DNA double helix that need to separate easily, such as the
15175:
14737:
3654:
relationships and protein function. Data sets representing entire genomes' worth of DNA sequences, such as those produced by the
8718:
Harrison PM, Gerstein M (May 2002). "Studying genomes through the aeons: protein families, pseudogenes and proteome evolution".
7852:
4612:
3736:, synthetic oligonucleotide ligands for specific target molecules used in a range of biotechnology and biomedical applications.
3108:
activity is activated and the incorrect base removed. In most organisms, DNA polymerases function in a large complex called the
13664:
2770:
is found in the blood of the mother, and can be sequenced to determine a great deal of information about the developing fetus.
2641:
sequence, which then defines one or more protein sequences. The relationship between the nucleotide sequences of genes and the
2182:
have higher levels, with up to 1% of their DNA containing 5-methylcytosine. Despite the importance of 5-methylcytosine, it can
4756:
1185:
A number of noncanonical bases are known to occur in DNA. Most of these are modifications of the canonical bases plus uracil.
990:
986:
892:
888:
14956:
14125:
14080:
14057:
14016:
13997:
13410:
13394:
13306:
12823:
From page 313: "I think that the size of the chromosomes in the salivary glands is determined through the multiplication of
12775:
Koltzoff NK (1928). "Physikalisch-chemische Grundlagen der Morphologie" [The physical-chemical basis of morphology].
11818:
11781:
11201:
11022:
9476:
8821:
8107:
5763:
Satoh M, Kuroiwa T (September 1991). "Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell".
4962:
4320:
4125:
3087:
group at the end of the growing polynucleotide chain. As a consequence, all polymerases work in a 5′ to 3′ direction. In the
6472:
4128:. Nobel Prizes are awarded only to living recipients. A debate continues about who should receive credit for the discovery.
3724:
have also been demonstrated, and these DNA structures have been used to template the arrangement of other molecules such as
14595:
11055:
9066:"DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides"
4139:. Further work by Crick and co-workers showed that the genetic code was based on non-overlapping triplets of bases, called
4088:
4075:
2240:
772:, where DNA bases are exchanged for their corresponding bases except in the case of thymine (T), for which RNA substitutes
4470:
2421:
has approximately 3 billion base pairs of DNA arranged into 46 chromosomes. The information carried by DNA is held in the
15622:
14439:
14367:
8076:
4349:
1994:
Several artificial nucleobases have been synthesized, and successfully incorporated in the eight-base DNA analogue named
1410:(hydrogen bonding the 6-carbon ring to the 5-carbon ring) is a rare variation of base-pairing. As hydrogen bonds are not
1387:
13050:
10234:
Cremer T, Cremer C (April 2001). "Chromosome territories, nuclear architecture and gene regulation in mammalian cells".
1882:, telomeres are usually lengths of single-stranded DNA containing several thousand repeats of a simple TTAGGG sequence.
14820:
14610:
14404:
11525:
8596:"RCSB PDB – 1MSW: Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase"
4672:
Arrighi, Frances E.; Mandel, Manley; Bergendahl, Janet; Hsu, T. C. (June 1970). "Buoyant densities of DNA of mammals".
4132:
1812:
1002:
14373:
Seven-page, handwritten letter that Crick sent to his 12-year-old son Michael in 1953 describing the structure of DNA.
12982:
Brachet J (1933). "Recherches sur la synthese de l'acide thymonucleique pendant le developpement de l'oeuf d'Oursin".
12616:
5893:
Munroe SH (November 2004). "Diversity of antisense regulation in eukaryotes: multiple mechanisms, emerging patterns".
1664:. These enzymes are also needed to relieve the twisting stresses introduced into DNA strands during processes such as
15261:
14928:
14147:
14106:
14038:
13931:
13909:
13887:
13866:
13847:
11888:
8070:
7665:
4739:
4456:
4426:
4401:
3872:("tetranucleotide hypothesis"). Levene thought the chain was short and the bases repeated in a fixed order. In 1927,
1009:, the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are
2781:, monitoring the movements and presence of species in water, air, or on land, and assessing an area's biodiversity.
2287:. The type of DNA damage produced depends on the type of mutagen. For example, UV light can damage DNA by producing
15548:
14372:
9403:
8310:
Braña MF, Cacho M, Gradillas A, de Pascual-Teresa B, Ramos A (November 2001). "Intercalators as anticancer drugs".
7150:
Parkinson GN, Lee MP, Neidle S (June 2002). "Crystal structure of parallel quadruplexes from human telomeric DNA".
3375:
2795:
Neutrophil extracellular traps (NETs) are networks of extracellular fibers, primarily composed of DNA, which allow
2321:
2215:
14318:
14257:
12755:] (Speech). 3rd All-Union Meeting of Zoologist, Anatomists, and Histologists (in Russian). Leningrad, U.S.S.R.
12546:
8669:
Harrison PM, Hegyi H, Balasubramanian S, Luscombe NM, Bertone P, Echols N, Johnson T, Gerstein M (February 2002).
2035:
which leave behind negative charges on the phosphate groups. These negative charges protect DNA from breakdown by
15553:
15082:
15026:
14164:
13268:
13078:
2992:
2847:, while in prokaryotes multiple types of proteins are involved. The histones form a disk-shaped complex called a
2161:
between DNA methylation and histone modification, so they can coordinately affect chromatin and gene expression.
114:
encode genes, which provide functions. A human DNA can have up to 500 million base pairs with thousands of genes.
13323:
10320:
O'Driscoll M, Jeggo PA (January 2006). "The role of double-strand break repair – insights from human genetics".
10061:
8671:"Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22"
6842:
6109:
2911:
that modify the histones at the promoter. This changes the accessibility of the DNA template to the polymerase.
725:
to make double-stranded DNA. The complementary nitrogenous bases are divided into two groups, the single-ringed
15021:
2790:
2406:
481:
13143:
Chargaff E (June 1950). "Chemical specificity of nucleic acids and mechanism of their enzymatic degradation".
12761:
Koltsov HK (1928). "Физико-химические основы морфологии" [The physical-chemical basis of morphology].
11503:
361:
15450:
15336:
15109:
15040:
14781:
11834:
Sjölander K (January 2004). "Phylogenomic inference of protein molecular function: advances and challenges".
11399:
Weir BS, Triggs CM, Starling L, Stowell LI, Walsh KA, Buckleton J (March 1997). "Interpreting DNA mixtures".
8804:
Tani K, Nasu M (2010). "Roles of Extracellular DNA in Bacterial Ecosystems". In Kikuchi Y, Rykova EY (eds.).
6934:
4136:
2562:
8095:
2595:, which are copies of genes that have been disabled by mutation. These sequences are usually just molecular
14845:
14696:
12396:
Dahm R (January 2008). "Discovering DNA: Friedrich Miescher and the early years of nucleic acid research".
10078:
Johnson A, O'Donnell M (2005). "Cellular DNA replicases: components and dynamics at the replication fork".
4757:"Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents. Recommendations 1970"
4022:
3816:
3428:
3258:
2673:
2329:
1580:
1523:
1381:
base pair with two hydrogen bonds. Non-covalent hydrogen bonds between the pairs are shown as dashed lines.
42:
13269:"Pictures and Illustrations: Crystallographic photo of Sodium Thymonucleate, Type B. "Photo 51." May 1952"
10363:
Vispé S, Defais M (October 1997). "Mammalian Rad51 protein: a RecA homologue with pleiotropic functions".
8539:"Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project"
3568:
methods happen after birth, but there are new methods to test paternity while a mother is still pregnant.
2149:
bases. DNA packaging and its influence on gene expression can also occur by covalent modifications of the
15563:
15558:
15543:
15168:
14376:
8931:"Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms"
5530:
4258:
4162:
In 1986, DNA analysis was first used for criminal investigative purposes when police in the UK requested
4062:
3647:
2799:, a type of white blood cell, to kill extracellular pathogens while minimizing damage to the host cells.
1029:
1010:
753:
13487:
7477:
6489:
Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL (October 1980). "Polymorphism of DNA double helices".
1870:. The main function of these regions is to allow the cell to replicate chromosome ends using the enzyme
15596:
15060:
14444:
12787:
In 1934, Koltsov contended that the proteins that contain a cell's genetic information replicate. See:
12323:
Akram F, Haq IU, Ali H, Laghari AT (October 2018). "Trends to store digital data in DNA: an overview".
11810:
10898:
Callahan MP, Smith KE, Cleaves HJ, Ruzicka J, Stern JC, Glavin DP, House CH, Dworkin JP (August 2011).
10455:
Dickman MJ, Ingleston SM, Sedelnikova SE, Rafferty JB, Lloyd RG, Grasby JA, Hornby DP (November 2002).
6670:
Lu XJ, Shakked Z, Olson WK (July 2000). "A-form conformational motifs in ligand-bound DNA structures".
3856:
of discarded surgical bandages. As it resided in the nuclei of cells, he called it "nuclein". In 1878,
3623:
2223:
2137:
The expression of genes is influenced by how the DNA is packaged in chromosomes, in a structure called
1490:
In the laboratory, the strength of this interaction can be measured by finding the melting temperature
1165:
250:
10958:
9174:"Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals"
1080:). These four bases are attached to the sugar-phosphate to form the complete nucleotide, as shown for
14600:
14585:
14360:
Another DNA Learning Center site on DNA, genes, and heredity from Mendel to the human genome project.
14249:
4289:
4234: – Universal notation using the Roman characters A, C, G, and T to call the four DNA nucleotides
4152:
3452:
3436:
3262:
3079:. The sequence of their products is created based on existing polynucleotide chains—which are called
2313:
2276:
2008:
1601:
1006:
938:(which interacts with the other DNA strand in the helix). A nucleobase linked to a sugar is called a
749:
612:
474:
12789:
Koltzoff N (October 1934). "The structure of the chromosomes in the salivary glands of Drosophila".
12743:
Koltsov proposed that a cell's genetic information was encoded in a long chain of amino acids. See:
10569:
9304:"The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin"
8240:
Stephens TD, Bunde CJ, Fillmore BJ (June 2000). "Mechanism of action in thalidomide teratogenesis".
7215:
6302:"The structure of sodium thymonucleate fibres. II. The cylindrically symmetrical Patterson function"
6301:
6256:
3520:
to identify a matching DNA of an individual, such as a perpetrator. This process is formally termed
2504:
and is a region of DNA that influences a particular characteristic in an organism. Genes contain an
1437:(low pH also melts DNA, but since DNA is unstable due to acid depurination, low pH is rarely used).
717:. The nitrogenous bases of the two separate polynucleotide strands are bound together, according to
15104:
14892:
14805:
14774:
14605:
14424:
11848:
10812:
Hebsgaard MB, Phillips MJ, Willerslev E (May 2005). "Geologically ancient DNA: fact or artefact?".
3940:
3882:
3860:
isolated the non-protein component of "nuclein", nucleic acid, and later isolated its five primary
3784:
3254:
2920:
2756:
2624:
1714:
1665:
769:
268:
22:
13597:
1894:
structure. These structures are stabilized by hydrogen bonding between the edges of the bases and
15065:
14882:
14867:
13386:
12134:
Aldaye FA, Palmer AL, Sleiman HF (September 2008). "Assembling materials with DNA as the guide".
11972:
10989:
8534:
8162:
Freitas AA, de Magalhães JP (2011). "A review and appraisal of the DNA damage theory of ageing".
7499:
6873:
6610:
4629:
Mandelkern M, Elias JG, Eden D, Crothers DM (October 1981). "The dimensions of DNA in solution".
4030:
4006:
3686:
3163:, the enzyme that transcribes most of the genes in the human genome, operates as part of a large
3076:
3053:
2843:. In eukaryotes, this structure involves DNA binding to a complex of small basic proteins called
2536:
2430:
2191:
1890:, form a flat plate. These flat four-base units then stack on top of each other to form a stable
1775:
analysis of the DNA X-ray diffraction patterns to suggest that the structure was a double helix.
1772:
1759:
B-DNA X-ray diffraction-scattering patterns of highly hydrated DNA fibers in terms of squares of
1685:
1081:
713:) between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating
9064:
Hu W, Li L, Sharma S, Wang J, McHardy I, Lux R, Yang Z, He X, Gimzewski JK, Li Y, Shi W (2012).
3685:
The DNA structure at left (schematic shown) will self-assemble into the structure visualized by
3315:
where nucleic acid would have been used for both catalysis and genetics may have influenced the
2492:. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the
15725:
15715:
15583:
15276:
15161:
15070:
14995:
14887:
14397:
14345:
13897:
13003:"Jean Brachet's Cytochemical Embryology: Connections with the Renovation of Biology in France?"
11843:
10564:
8486:"The C-value enigma in plants and animals: a review of parallels and an appeal for partnership"
7210:
5936:
Makalowska I, Lin CF, Makalowski W (February 2005). "Overlapping genes in vertebrate genomes".
3836:, co-originators of the double-helix model based on the X-ray diffraction data and insights of
3228:
3101:
3057:
3000:
2174:
785:
13749:
13578:
13012:. Cahiers pour I'histoire de la recherche. Vol. 2. Paris: CNRS Editions. pp. 207–20.
12841:
Soyfer VN (September 2001). "The consequences of political dictatorship for Russian science".
12507:
12477:
12460:
12443:
12367:
11712:"In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing"
8813:
2991:
infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the
15460:
15304:
14985:
14970:
14850:
14732:
14566:
14531:
14188:
10091:
10041:
6964:"Identification of a specific telomere terminal transferase activity in Tetrahymena extracts"
4898:"Base-stacking and base-pairing contributions into thermal stability of the DNA double helix"
4237:
4231:
3702:
3694:
3615:
3561:
3296:
3243:
occurs. Chromosomal crossover is when two DNA helices break, swap a section and then rejoin.
3240:
3232:
3210:
3176:
3156:
3124:
3023:
2646:
2531:
encodes protein. For example, only about 1.5% of the human genome consists of protein-coding
2439:
1989:
1515:
1144:
1116:
761:
441:
421:
386:
298:
293:
12523:
9570:
Iftode C, Daniely Y, Borowiec JA (1999). "Replication protein A (RPA): the eukaryotic SSB".
8805:
5838:
3924:
produced the first X-ray diffraction patterns that showed that DNA had a regular structure.
15495:
15478:
15309:
15092:
14990:
14908:
14631:
14449:
14245:
13958:
13703:
13612:
13534:
13335:
12798:
12143:
12100:
12039:
11987:
11723:
11455:
11353:
11097:
10911:
10856:
10770:
10719:
10664:
10513:
10411:
9720:
9607:"RCSB PDB – 1LMB: Refined 1.8 Å crystal structure of the lambda repressor-operator complex"
9360:
9185:
9077:
8550:
8448:
8358:
8171:
7913:
7702:
7691:"The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain"
7434:
7367:
7316:
7159:
6761:
6594:
6440:
6372:
6316:
6271:
6089:
5690:
5645:
5440:
5383:
5241:
4979:
4828:
4572:
4497:
4310:
4079:
4002:
3983:
3760:
3655:
3418:
3312:
3290:
2904:
2868:
2808:
2628:
2154:
2077:
1506:
1311:
1041:
107:
26:
12060:
8982:"A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm"
8030:
7572:
Klose RJ, Bird AP (February 2006). "Genomic DNA methylation: the mark and its mediators".
6808:
4085:
in Cambridge, England to announce that he and Watson had "discovered the secret of life".
3767:
can learn the history of particular populations. This can be used in studies ranging from
8:
15490:
15473:
15331:
15214:
14918:
14711:
14701:
14661:
14561:
14541:
14434:
14429:
14178:
7352:
Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, Kim HJ, et al. (22 February 2019).
6357:
3952:
3932:
3768:
3764:
3529:
3464:
3451:
is a man-made DNA sequence that has been assembled from other DNA sequences. They can be
3422:
3236:
3159:, where it halts and detaches from the DNA. As with human DNA-dependent DNA polymerases,
2964:
2916:
2821:
2767:
2608:
2517:
2509:
2309:
2292:
2158:
1904:
1407:
1147:
have been created to study the properties of nucleic acids, or for use in biotechnology.
978:
860:
733:. In DNA, the pyrimidines are thymine and cytosine; the purines are adenine and guanine.
710:
416:
396:
331:
288:
282:
273:
245:
123:
82:
59:
13962:
13707:
13616:
13538:
13339:
13002:
12802:
12147:
12104:
12043:
11991:
11727:
11459:
11357:
11101:
10915:
10860:
10774:
10723:
10668:
10517:
10415:
10165:
Martinez E (December 2002). "Multi-protein complexes in eukaryotic gene transcription".
9724:
9364:
9271:
9246:
9189:
9172:
Foote AD, Thomsen PF, Sveegaard S, Wahlberg M, Kielgast J, Kyhn LA, et al. (2012).
9081:
8626:
Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences
8554:
8452:
8362:
8345:
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. (February 2001).
8175:
7917:
7706:
7438:
7371:
7320:
7163:
6765:
6598:
6444:
6376:
6320:
6275:
5736:
5694:
5649:
5632:
Gregory SG, Barlow KF, McLay KE, Kaul R, Swarbreck D, Dunham A, et al. (May 2006).
5444:
5387:
5299:
5245:
4832:
4576:
4501:
3037:
are enzymes with both nuclease and ligase activity. These proteins change the amount of
2834:(in blue). These proteins' basic amino acids bind to the acidic phosphate groups on DNA.
1017:
is the sugar, with the 2-deoxyribose in DNA being replaced by the related pentose sugar
175:
15384:
15379:
15254:
15053:
14936:
14671:
14556:
14551:
14526:
14350:
13804:
13787:
13638:
13560:
13492:
13358:
13245:
13220:
13168:
13120:
13095:
12910:
12885:
12866:
12592:
12575:
12421:
12348:
12300:
12275:
12167:
12073:
12011:
11953:
11807:
Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology
11746:
11711:
11646:
11621:
11481:
11424:
11251:
11226:
11166:
10934:
10899:
10880:
10794:
10743:
10633:
10590:
10537:
10486:
10432:
10399:
10345:
10302:
10259:
10190:
10147:
10053:
9434:
9384:
9333:
9284:
9208:
9173:
9149:
9124:
9100:
9065:
9006:
8981:
8957:
8930:
8862:
8646:
8621:
8571:
8538:
8510:
8485:
8035:
7831:
7772:
7747:
7723:
7690:
7549:
7522:
7458:
7424:
7388:
7353:
7280:
7255:
7236:
7183:
7127:
7102:
6916:
6730:
6718:
6545:
6464:
6396:
6200:
6154:
6101:
6046:
6021:
5949:
5918:
5816:
5799:
5714:
5609:
5582:
5511:
5499:
5404:
5371:
5347:
5322:
5265:
5174:
5139:
4930:
4897:
4789:
4705:
4604:
4526:
4485:
4144:
3878:
3849:
3690:
3676:
3635:
3432:
3265:
and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as
2980:
2942:
2513:
2505:
2480:, to fit the small available volumes of the cell. In eukaryotes, DNA is located in the
1924:
1746:
1480:
1174:
1169:
in 1925. The reason for the presence of these noncanonical bases in bacterial viruses (
411:
406:
381:
303:
14095:
13726:
13691:
13204:
13187:
12959:
12934:
12695:
12678:
12251:
12224:
11940:
11913:
11687:
11670:
11287:
11242:
11056:"World's oldest DNA sequenced from a mammoth that lived more than a million years ago"
10376:
9998:
9973:
9870:
9845:
9818:
9793:
9743:
9708:
8906:
8881:
8781:
8754:
8731:
8695:
8670:
8253:
7925:
7412:
7224:
7075:
7050:
6784:
6749:
6175:
Wang JC (June 2002). "Cellular roles of DNA topoisomerases: a molecular perspective".
5997:
5972:
5395:
4754:
2291:, which are cross-links between pyrimidine bases. On the other hand, oxidants such as
15468:
15430:
15423:
15374:
15326:
14755:
14686:
14676:
14666:
14500:
14390:
14334:
14143:
14121:
14102:
14076:
14069:
14053:
14034:
14012:
13993:
13976:
13927:
13905:
13883:
13862:
13843:
13809:
13731:
13630:
13564:
13552:
13390:
13363:
13302:
13295:
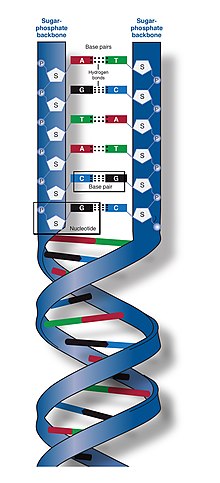
13250:
13160:
13125:
13070:
12964:
12915:
12858:
12814:
12726:
12597:
12413:
12340:
12305:
12256:
12159:
12116:
12065:
12003:
11945:
11894:
11884:
11861:
11814:
11787:
11777:
11751:
11692:
11651:
11602:
11598:
11552:
11473:
11416:
11381:
11376:
11341:
11291:
11256:
11207:
11197:
11158:
11154:
11123:
11115:
10939:
10872:
10829:
10786:
10735:
10692:
10687:
10652:
10625:
10582:
10529:
10478:
10473:
10456:
10437:
10380:
10337:
10294:
10251:
10182:
10139:
10095:
10045:
10003:
9989:
9954:
9949:
9910:
9875:
9823:
9748:
9689:
9648:
9644:
9587:
9552:
9548:
9517:
9482:
9472:
9426:
9376:
9325:
9320:
9303:
9276:
9213:
9154:
9105:
9046:
9011:
8997:
8962:
8911:
8897:
8854:
8817:
8806:
8786:
8735:
8700:
8651:
8576:
8515:
8466:
8417:
8376:
8327:
8292:
8288:
8257:
8222:
8218:
8187:
8144:
8103:
8066:
8011:
7970:
7929:
7886:
7849:
7823:
7809:
7777:
7728:
7671:
7661:
7630:
7589:
7554:
7450:
7393:
7358:
7334:
7285:
7228:
7175:
7132:
7080:
7031:
6985:
6980:
6963:
6903:
Cressey D (3 October 2012). "'Arsenic-life' Bacterium Prefers Phosphorus after all".
6789:
6722:
6687:
6652:
6549:
6506:
6502:
6456:
6388:
6235:
6204:
6192:
6146:
6142:
6093:
6051:
6037:
6002:
5953:
5910:
5875:
5821:
5780:
5776:
5706:
5663:
5614:
5553:
5503:
5468:
5463:
5428:
5409:
5352:
5303:
5257:
5214:
5179:
5161:
5120:
5112:
5071:
5036:
5028:
4958:
4935:
4917:
4852:
4844:
4794:
4776:
4735:
4697:
4689:
4654:
4646:
4642:
4596:
4588:
4557:
4531:
4513:
4462:
4452:
4422:
4397:
4390:
4316:
4267:
4213:
4156:
4045:
4041:
3837:
3749:
3639:
3414:
3332:
3278:
3247:
3206:
3160:
2996:
2976:
2774:
2735:
2572:
2305:
2296:
1576:
832:
461:
346:
225:
15137:
14354:, June 1953. First American newspaper coverage of the discovery of the DNA structure
14303:
13837:
13673:
13221:"Independent functions of viral protein and nucleic acid in growth of bacteriophage"
12870:
12711:
12352:
12241:
11957:
11857:
11428:
11170:
10884:
10637:
10490:
10306:
10194:
10151:
10057:
9337:
9288:
8866:
8771:
7835:
7462:
6920:
6734:
6158:
5922:
5858:
Hüttenhofer A, Schattner P, Polacek N (May 2005). "Non-coding RNAs: hope or hype?".
5515:
5023:
5006:
4709:
3362:, have also been formed in the laboratory under conditions mimicking those found in
2633:
A gene is a sequence of DNA that contains genetic information and can influence the
2557:". However, some DNA sequences that do not code protein may still encode functional
2190:. Other base modifications include adenine methylation in bacteria, the presence of
15720:
15294:
14716:
14691:
14313:
14283:
14268:
13966:
13799:
13721:
13711:
13642:
13620:
13542:
13525:
13353:
13343:
13240:
13232:
13199:
13172:
13152:
13115:
13107:
13042:
12954:
12950:
12946:
12905:
12897:
12850:
12806:
12690:
12659:
12628:
12587:
12405:
12332:
12295:
12287:
12246:
12236:
12198:
12171:
12151:
12108:
12077:
12055:
12047:
12015:
11995:
11935:
11925:
11853:
11741:
11731:
11682:
11641:
11633:
11594:
11485:
11463:
11408:
11371:
11361:
11283:
11246:
11238:
11189:
11150:
11105:
11088:
10929:
10919:
10864:
10821:
10798:
10778:
10747:
10727:
10682:
10672:
10621:
10617:
10594:
10574:
10541:
10521:
10468:
10427:
10419:
10372:
10349:
10329:
10286:
10263:
10243:
10174:
10129:
10087:
10037:
9993:
9985:
9944:
9902:
9865:
9857:
9813:
9809:
9805:
9738:
9728:
9679:
9640:
9579:
9544:
9509:
9464:
9438:
9418:
9388:
9368:
9315:
9266:
9258:
9203:
9193:
9144:
9136:
9095:
9085:
9038:
9001:
8993:
8952:
8942:
8901:
8893:
8846:
8776:
8766:
8727:
8690:
8682:
8641:
8633:
8566:
8558:
8505:
8497:
8456:
8407:
8366:
8319:
8284:
8249:
8214:
8179:
8136:
8001:
7960:
7921:
7878:
7813:
7805:
7767:
7759:
7718:
7710:
7653:
7620:
7581:
7544:
7534:
7442:
7383:
7375:
7324:
7275:
7267:
7220:
7187:
7167:
7122:
7114:
7070:
7062:
7021:
6975:
6908:
6881:
6779:
6769:
6714:
6679:
6644:
6602:
6537:
6498:
6468:
6448:
6400:
6380:
6324:
6279:
6231:
6227:
6184:
6138:
6105:
6085:
6041:
6033:
5992:
5984:
5945:
5902:
5867:
5811:
5772:
5718:
5698:
5653:
5604:
5594:
5545:
5495:
5458:
5448:
5399:
5391:
5342:
5334:
5295:
5269:
5249:
5206:
5169:
5151:
5102:
5063:
5018:
4925:
4909:
4836:
4802:
4784:
4768:
4681:
4638:
4608:
4580:
4521:
4505:
4207:
4148:
4098:
4026:
3974:
3627:
3619:
3493:
3472:
3343:
3015:
3011:
2896:
2715:
2711:
2707:
2604:
2477:
2435:
2338:
2172:
of chromosomes. The average level of methylation varies between organisms—the worm
2165:
2114:
1822:
1631:
1156:
1139:), usually takes the place of thymine in RNA and differs from thymine by lacking a
671:
632:
507:
353:
140:
14278:
13418:
12425:
12186:
10210:"RCSB PDB – 1M6G: Structural Characterisation of the Holliday Junction TCGGTACCGA"
9042:
7652:. Current Topics in Microbiology and Immunology. Vol. 301. pp. 283–315.
7240:
4363:
4140:
2637:
of an organism. Within a gene, the sequence of bases along a DNA strand defines a
1791:
15418:
15389:
15114:
14951:
14797:
14576:
14135:
13877:
12530:
11930:
10710:
Lindahl T (April 1993). "Instability and decay of the primary structure of DNA".
10277:
Pál C, Papp B, Lercher MJ (May 2006). "An integrated view of protein evolution".
10118:"The reverse transcriptase of HIV-1: from enzymology to therapeutic intervention"
10116:
Tarrago-Litvak L, Andréola ML, Nevinsky GA, Sarih-Cottin L, Litvak S (May 1994).
9929:
9198:
9090:
8947:
7856:
6425:
5845:
4294:
4183:
4124:
In 1962, after Franklin's death, Watson, Crick, and Wilkins jointly received the
4053:
4037:
3921:
3886:
3873:
3857:
3841:
3802:
3794:
3659:
3631:
3549:
3541:
3448:
3300:
3273:. The first step in recombination is a double-stranded break caused by either an
3164:
3049:
2860:
2731:
2685:
2443:
2327:
Many mutagens fit into the space between two adjacent base pairs, this is called
2268:
2252:
2073:
2024:
1784:
1760:
1669:
1616:
994:
840:
744:
when the two strands separate. A large part of DNA (more than 98% for humans) is
741:
702:
662:
The two DNA strands are known as polynucleotides as they are composed of simpler
13046:
13028:"Some recent developments in the X-ray study of proteins and related structures"
12810:
12480:[On the distribution of hypoxanthins in the animal and plant kingdoms].
11710:
Torabi SF, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y (May 2015).
11193:
9709:"A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells"
9468:
9247:"Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome"
8837:
Vlassov VV, Laktionov PP, Rykova EY (July 2007). "Extracellular nucleic acids".
8183:
6257:"The Structure of Sodium Thymonucleate Fibres I. The Influence of Water Content"
5581:
Piovesan A, Pelleri MC, Antonaros F, Strippoli P, Caracausi M, Vitale L (2019).
5107:
5090:
4558:"Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid"
4052:", at high hydration levels of DNA. This photo was given to Watson and Crick by
3797:
compared to electronic devices. However, high costs, slow read and write times (
3528:. In DNA profiling, the lengths of variable sections of repetitive DNA, such as
1028:
A section of DNA. The bases lie horizontally between the two spiraling strands (
15610:
15510:
15413:
15230:
14941:
14913:
14706:
14646:
14546:
13696:
Proceedings of the National Academy of Sciences of the United States of America
13547:
13520:
13383:
What Is Life?: investigating the nature of life in the age of synthetic biology
13328:
Proceedings of the National Academy of Sciences of the United States of America
11716:
Proceedings of the National Academy of Sciences of the United States of America
11346:
Proceedings of the National Academy of Sciences of the United States of America
11110:
11083:
10904:
Proceedings of the National Academy of Sciences of the United States of America
10657:
Proceedings of the National Academy of Sciences of the United States of America
10134:
10117:
9713:
Proceedings of the National Academy of Sciences of the United States of America
9684:
9667:
9463:. Current Topics in Microbiology and Immunology. Vol. 274. pp. 1–22.
8205:
Ferguson LR, Denny WA (September 1991). "The genetic toxicology of acridines".
8058:
7585:
7446:
7329:
7304:
6754:
Proceedings of the National Academy of Sciences of the United States of America
6750:"Z-DNA-binding proteins can act as potent effectors of gene expression in vivo"
5433:
Proceedings of the National Academy of Sciences of the United States of America
4446:
4201:
4195:
4014:
3948:
3936:
3868:
3825:
3798:
3725:
3607:
3602:
3565:
3397:
3147:
3096:
2967:. Nucleases that hydrolyse nucleotides from the ends of DNA strands are called
2743:
2703:
2661:
2558:
2554:
2362:
2301:
2264:
2244:
2203:
2169:
2007:, while a higher number is also possible but this would be against the natural
1973:
1887:
1607:
1585:
1544:
1519:
1468:-content have more strongly interacting strands, while short helices with high
951:
942:, and a base linked to a sugar and to one or more phosphate groups is called a
931:
908:
764:
of these four nucleobases along the backbone that encodes genetic information.
752:. The two strands of DNA run in opposite directions to each other and are thus
745:
714:
694:
648:
608:
401:
326:
12935:"Bacterial gene transfer by natural genetic transformation in the environment"
12901:
12663:
12632:
12409:
12336:
12291:
10900:"Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases"
10868:
10825:
10578:
10178:
9583:
9140:
7354:"Hachimoji DNA and RNA: A genetic system with eight building blocks (paywall)"
7271:
6912:
6606:
6329:
6284:
6129:
Champoux JJ (2001). "DNA topoisomerases: structure, function, and mechanism".
5871:
5599:
5529:
Isaksson J, Acharya S, Barman J, Cheruku P, Chattopadhyaya J (December 2004).
4840:
2228:
1854:
1682:
Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid
15704:
15445:
15435:
15316:
15271:
15249:
15142:
14829:
14097:
The Double Helix: A Personal Account of the Discovery of the Structure of DNA
14090:
12763:Успехи экспериментальной биологии (Advances in Experimental Biology) series B
12091:
Ishitsuka Y, Ha T (May 2009). "DNA nanotechnology: a nanomachine goes live".
11883:(2nd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
11556:
11119:
9861:
8323:
7965:
7948:
7657:
7539:
5321:
Nikolova EN, Zhou H, Gottardo FL, Alvey HS, Kimsey IJ, Al-Hashimi HM (2013).
5165:
5156:
5116:
5075:
5032:
4921:
4848:
4780:
4693:
4650:
4592:
4517:
4466:
4341:
4252:
4243:
4163:
4104:
MOLECULAR STRUCTURE OF NUCLEIC ACIDS A Structure for Deoxyribose Nucleic Acid
4066:
3994:
3829:
3756:
3745:
3721:
3651:
3582:
3577:
3537:
3533:
3521:
3488:
3195:
3152:
3038:
3034:
2988:
2726:
2699:
2638:
2576:
2540:
2485:
2471:
2288:
2199:
2028:
1995:
1968:
1768:
1689:
1661:
1653:
1647:
1611:
1411:
1394:
1372:
1299:
1170:
1037:
916:
912:
884:
828:
796:
722:
706:
652:
14233:
was created from a revision of this article dated 12 February 2007
13767:
Pray L (2008). "Discovery of DNA structure and function: Watson and Crick".
12202:
12155:
11898:
11791:
11736:
11529:
11366:
10924:
9733:
9422:
8371:
8346:
7714:
7379:
5453:
4819:
Ghosh A, Bansal M (April 2003). "A glossary of DNA structures from A to Z".
3540:, and first used in forensic science to convict Colin Pitchfork in the 1988
3010:
can rejoin cut or broken DNA strands. Ligases are particularly important in
1866:
At the ends of the linear chromosomes are specialized regions of DNA called
15710:
15528:
15348:
15321:
15266:
15185:
15016:
14946:
14815:
14656:
14536:
14293:
14230:
13980:
13833:
13813:
13735:
13634:
13556:
13367:
13254:
13164:
13129:
13074:
12919:
12862:
12818:
12601:
12417:
12344:
12309:
12260:
12163:
12120:
12112:
12069:
12007:
11949:
11865:
11769:
11755:
11655:
11295:
11260:
11211:
11127:
10994:
10943:
10876:
10833:
10790:
10677:
10629:
10586:
10533:
10482:
10441:
10341:
10298:
10255:
10186:
10099:
10049:
9958:
9914:
9879:
9752:
9693:
9652:
9591:
9521:
9500:
Thomas JO (August 2001). "HMG1 and 2: architectural DNA-binding proteins".
9486:
9430:
9329:
9217:
9158:
9109:
9050:
9015:
8966:
8915:
8858:
8790:
8739:
8704:
8655:
8637:
8580:
8519:
8470:
8421:
8380:
8331:
8261:
8191:
8148:
8015:
8006:
7989:
7933:
7890:
7781:
7732:
7675:
7634:
7593:
7558:
7454:
7397:
7338:
7289:
7232:
7179:
7136:
7066:
6793:
6774:
6726:
6691:
6683:
6460:
6392:
6196:
6150:
6097:
6006:
5957:
5914:
5879:
5825:
5667:
5618:
5557:
5472:
5413:
5356:
5218:
5210:
5183:
5124:
5091:"Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses"
4939:
4856:
4600:
4535:
4167:"confession", and the suspect was exonerated from the murder-rape charges.
4070:
4018:
3998:
3928:
3906:
3902:
3833:
3790:
3772:
3729:
3663:
3460:
3444:
3274:
3218:
2972:
2669:
2650:
2620:
2481:
2455:
2422:
2418:
2370:
2346:
2186:
to leave a thymine base, so methylated cytosines are particularly prone to
2051:
1959:
1920:
1891:
1849:
1818:
1764:
1549:
1307:
1140:
962:
927:
820:
777:
636:
620:
391:
128:
13716:
12968:
12730:
11696:
11606:
11477:
11420:
11385:
10739:
10696:
10400:"Clarifying the mechanics of DNA strand exchange in meiotic recombination"
10384:
10143:
10115:
10007:
9827:
9556:
9380:
9280:
9262:
8296:
8226:
8140:
7974:
7827:
7794:
7084:
7051:"Normal human chromosomes have long G-rich telomeric overhangs at one end"
7035:
7026:
7009:
6989:
6885:
6656:
6510:
6239:
6055:
5784:
5710:
5507:
5307:
5261:
5040:
4798:
4701:
4658:
4315:(6th ed.). Garland. p. Chapter 4: DNA, Chromosomes and Genomes.
3662:
algorithms, which allow researchers to predict the presence of particular
15440:
15286:
15153:
15119:
15048:
14636:
14300:
14290:
14026:
13942:
13919:
13348:
13111:
11637:
11162:
9125:"Recent advances in the prenatal interrogation of the human fetal genome"
8501:
7118:
6635:
Wahl MC, Sundaralingam M (1997). "Crystal structures of A-DNA duplexes".
4913:
4058:
3979:
3717:
3708:
3611:
3517:
3476:
3393:
3387:
3363:
3336:
3266:
3213:. The four separate DNA strands are coloured red, blue, green and yellow.
3105:
3088:
2968:
2864:
2856:
2796:
2777:
eDNA has seen increased use in the natural sciences as a survey tool for
2544:
2489:
2390:
2350:
2334:
2280:
2183:
2142:
2129:
2040:
1830:
1796:
1572:
1476:
1449:
1415:
1179:
966:
879:
836:
824:
698:
15241:
14183:
13971:
13946:
13625:
13236:
12051:
11999:
11184:
Houdebine LM (2007). "Transgenic animal models in biomedical research".
10555:
Orgel LE (2004). "Prebiotic chemistry and the origin of the RNA world".
10423:
9906:
9631:
Myers LC, Kornberg RD (2000). "Mediator of transcriptional regulation".
8562:
8537:, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. (June 2007).
7429:
5973:"Properties of overlapping genes are conserved across microbial genomes"
5658:
5633:
5067:
4755:
IUPAC-IUB Commission on Biochemical Nomenclature (CBN) (December 1970).
4210: – Health problem caused by one or more abnormalities in the genome
3014:
DNA replication, as they join the short segments of DNA produced at the
2891:
2660:
In transcription, the codons of a gene are copied into messenger RNA by
1294:
768:
strands are created using DNA strands as a template in a process called
15500:
15209:
15204:
15199:
15077:
14641:
14510:
14490:
14465:
13156:
11585:
Breaker RR, Joyce GF (December 1994). "A DNA enzyme that cleaves RNA".
9513:
8850:
7763:
7625:
7608:
6541:
5988:
4685:
4509:
4198: – Collection of microscopic DNA spots attached to a solid surface
3898:
3861:
3713:
3467:
organisms produced can be used to produce products such as recombinant
3367:
3136:
3132:
3068:
3019:
3007:
2960:
2852:
2848:
2719:
2642:
2592:
2588:
2463:
2410:
2398:
2354:
2272:
2179:
2036:
1875:
1871:
1553:
1399:
1120:
947:
943:
939:
935:
904:
844:
792:
781:
726:
674:
667:
321:
235:
215:
192:
111:
51:
14140:
The third man of the double helix the autobiography of Maurice Wilkins
12549:[Further contributions to the chemistry of the cell nucleus].
11412:
10608:
Davenport RJ (May 2001). "Ribozymes. Making copies in the RNA world".
10209:
9767:
9606:
8882:"DNA as a nutrient: novel role for bacterial competence gene homologs"
8686:
8595:
8412:
8395:
7882:
7171:
7049:
Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW (November 1997).
5906:
5549:
5338:
4871:
4772:
3955:, stating that in DNA from any species of any organism, the amount of
3447:
make intensive use of these techniques in recombinant DNA technology.
1941:
1084:. Adenine pairs with thymine and guanine pairs with cytosine, forming
791:
Within eukaryotic cells, DNA is organized into long structures called
15364:
12854:
11468:
11443:
11227:"Multigene engineering: dawn of an exciting new era in biotechnology"
10782:
10731:
10247:
8461:
7818:
6649:
10.1002/(SICI)1097-0282(1997)44:1<45::AID-BIP4>3.0.CO;2-#
6452:
6384:
5702:
5253:
4895:
4584:
4219:
3914:
3681:
3383:
3371:
3316:
3304:
3140:
3113:
3109:
3083:. These enzymes function by repeatedly adding a nucleotide to the 3′
2881:
2876:
2840:
2807:
All the functions of DNA depend on interactions with proteins. These
2690:
2634:
2459:
2402:
2366:
2358:
2138:
1895:
1878:
systems in the cell from treating them as damage to be corrected. In
1834:
1826:
1735:
1630:, blur the distinction between sense and antisense strands by having
1568:
1557:
1533:
1527:
1511:
1390:
1332:
1092:
958:
920:
864:
856:
800:
718:
278:
151:
144:
90:
12647:
12478:"Ueber die Verbreitung des Hypoxanthins im Thier- und Pflanzenreich"
11311:"From the crime scene to the courtroom: the journey of a DNA sample"
10525:
10333:
10290:
9930:"Unraveling DNA helicases. Motif, structure, mechanism and function"
9028:
8668:
8436:
8309:
4155:
to decipher the genetic code. These findings represent the birth of
3186:
2568:
1934:
15640:
15605:
15518:
15369:
15299:
15097:
15087:
15011:
14681:
14651:
14495:
14485:
14413:
10454:
9707:
Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B (July 2003).
9668:"Biological control through regulated transcriptional coactivators"
7990:"Regulation and mechanisms of mammalian double-strand break repair"
6188:
4309:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2014).
4177:
4049:
4010:
3960:
3733:
3643:
3586:
3379:
3355:
3308:
3257:, where the two chromosomes involved share very similar sequences.
3117:
3084:
3045:
2984:
2952:
2885:
2695:
2665:
2584:
2583:
Some noncoding DNA sequences play structural roles in chromosomes.
2501:
2497:
2493:
2385:
2342:
2233:
2219:
2187:
2146:
2109:
2094:
2062:
1867:
1859:
1694:
1635:
1589:
1057:
1045:
887:
shown as dotted lines. Each end of the double helix has an exposed
848:
678:
624:
616:
337:
220:
210:
167:
15691:
14766:
14339:
12274:
Panda D, Molla KA, Baig MJ, Swain A, Behera D, Dash M (May 2018).
10990:"DNA Building Blocks Can Be Made in Space, NASA Evidence Suggests"
9372:
9232:"Researchers Detect Land Animals Using DNA in Nearby Water Bodies"
8532:
8396:"The bacterial nucleoid: a highly organized and dynamic structure"
5634:"The DNA sequence and biological annotation of human chromosome 1"
5369:
4445:
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Peter W (2002).
3427:
Methods have been developed to purify DNA from organisms, such as
2657:
formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT).
1298:
DNA major and minor grooves. The latter is a binding site for the
15523:
14833:
14324:
13464:
12501:] (in German). Strassburg, Germany: K.J. Trübner. p. 19.
11188:. Methods in Molecular Biology. Vol. 360. pp. 163–202.
8045:
If we lived long enough, sooner or later we all would get cancer.
7010:"The telomerase reverse transcriptase: components and regulation"
6488:
5580:
4225:
4189:
4061:
for DNA crystals revealed to Crick that the two DNA strands were
3968:
3964:
3956:
3693:
is the field that seeks to design nanoscale structures using the
3468:
3456:
3440:
3359:
3328:
3324:
2844:
2831:
2778:
2760:
2668:
that reads the RNA sequence by base-pairing the messenger RNA to
2549:
2524:
2260:
2248:
2150:
2119:
1755:
1623:
1541:
1073:
1065:
1049:
970:
911:. Both chains are coiled around the same axis, and have the same
900:
868:
852:
816:
756:. Attached to each sugar is one of four types of nucleobases (or
690:
686:
682:
663:
640:
604:
75:Gene
68:DNA
52:Chromosome
15674:
14309:
11023:"NASA Ames Reproduces the Building Blocks of Life in Laboratory"
8127:
Hoeijmakers JH (October 2009). "DNA damage, aging, and cancer".
8094:
Bernstein H, Payne CM, Bernstein C, Garewal H, Dvorak K (2008).
6843:"Arsenic-Eating Bacteria Opens New Possibilities for Alien Life"
5370:
Clausen-Schaumann H, Rief M, Tolksdorf C, Gaub HE (April 2000).
5140:"Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond"
4246: – specific region of DNA that that codes for ribosomal RNA
3793:
for information has enormous potential since it has much higher
3018:
into a complete copy of the DNA template. They are also used in
2553:, among species, represent a long-standing puzzle known as the "
2476:
Genomic DNA is tightly and orderly packed in the process called
1024:
101:
15538:
15533:
15483:
14505:
14480:
13902:
The Eighth Day of Creation: Makers of the Revolution in Biology
11528:. National Institute of Justice. September 2006. Archived from
8275:
Jeffrey AM (1985). "DNA modification by chemical carcinogens".
7648:
Walsh CP, Xu GL (2006). "Cytosine methylation and DNA repair".
7200:
5680:
3509:
3351:
3072:
2995:. In technology, these sequence-specific nucleases are used in
2956:
2908:
2739:
2600:
2596:
2528:
2414:
2374:
2317:
2236:
2032:
1908:
1837:
1657:
1622:
A few DNA sequences in prokaryotes and eukaryotes, and more in
1355:
1342:
1160:
1155:
Modified bases occur in DNA. The first of these recognized was
1132:
1104:
1018:
974:
950:
comprising multiple linked nucleotides (as in DNA) is called a
812:
804:
773:
730:
644:
230:
205:
15657:
14364:
The Register of Francis Crick Personal Papers 1938 – 2007
10504:
Joyce GF (July 2002). "The antiquity of RNA-based evolution".
8093:
4483:
4092:
Pencil sketch of the DNA double helix by Francis Crick in 1953
4078:. On 28 February 1953 Crick interrupted patrons' lunchtime at
2826:
2031:. It will be fully ionized at a normal cellular pH, releasing
561:
513:
15616:
15341:
14855:
11274:
Job D (November 2002). "Plant biotechnology in agriculture".
9171:
8928:
7867:
6526:"Structural Order and Partial Disorder in Biological systems"
6525:
5007:"Modified oligonucleotides: synthesis and strategy for users"
3501:
3497:
3270:
3128:
2945:
2937:
2919:
processes that control responses to environmental changes or
2734:. Here, the two strands are separated and then each strand's
2284:
2195:
1800:
1726:
1722:
1718:
1707:
1703:
1699:
1627:
1320:
977:) sugar. The sugars are joined by phosphate groups that form
808:
628:
549:
132:
13460:"Rosalind Franklin's role in DNA discovery gets a new twist"
12276:"DNA as a digital information storage device: hope or hype?"
10959:"NASA Researchers: DNA Building Blocks Can Be Made in Space"
9974:"Polymerase structures and function: variations on a theme?"
8096:"Cancer and aging as consequences of un-repaired DNA damage"
7903:
5528:
5426:
5323:"A historical account of Hoogsteen base-pairs in duplex DNA"
4957:(4th ed.). Sudbury, Mass.: Jones and Barlett Learning.
1610:
is called a "sense" sequence if it is the same as that of a
1036:
The DNA double helix is stabilized primarily by two forces:
14382:
12495:
Untersuchungen über die Nucleine und ihre Spaltungsprodukte
11027:
10962:
10897:
10811:
6580:"X-ray scattering by partially disordered membrane systems"
6217:
5857:
5427:
Chalikian TV, Völker J, Plum GE, Breslauer KJ (July 1999).
4628:
4131:
In an influential presentation in 1957, Crick laid out the
3513:
3505:
3347:
3181:
2653:. The genetic code consists of three-letter 'words' called
2543:
in eukaryotic genomes and the extraordinary differences in
2532:
2467:
2426:
2128:
Structure of cytosine with and without the 5-methyl group.
2082:
2059:
Pure DNA extracted from cells forms white, stringy clumps.
2020:
1929:
1350:
1337:
982:
748:, meaning that these sections do not serve as patterns for
656:
582:
543:
537:
136:
74:
7256:"DNA enables nanoscale control of the structure of matter"
7101:
Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006).
5583:"On the length, weight and GC content of the human genome"
4671:
3712:
method) and three-dimensional structures in the shapes of
3303:
as it can both transmit genetic information and carry out
2975:
cut within strands. The most frequently used nucleases in
2087:
1799:
may undergo a larger change in conformation and adopt the
705:. The nucleotides are joined to one another in a chain by
619:
instructions for the development, functioning, growth and
546:
519:
15224:
14475:
14357:
14050:
DNA: A Graphic Guide to the Molecule that Shook the World
13856:
13010:
Les sciences biologiques et médicales en France 1920–1950
11398:
11060:
9350:
8929:
Mulcahy H, Charron-Mazenod L, Lewenza S (November 2008).
7048:
6809:"Arsenic-loving bacteria may help in hunt for alien life"
5320:
4896:
Yakovchuk P, Protozanova E, Frank-Kamenetskii MD (2006).
4451:(Fourth ed.). New York and London: Garland Science.
4082:
3986:
3910:
3853:
3591:
systematic evolution of ligands by exponential enrichment
3340:
2520:, which control transcription of the open reading frame.
2101:
2004:
1999:
1731:
1143:
on its ring. In addition to RNA and DNA, many artificial
1014:
765:
564:
528:
200:
14273:
11619:
10760:
9846:"Structural and mechanistic conservation in DNA ligases"
8434:
6704:
5137:
4204: – Process of determining the nucleic acid sequence
3852:
who, in 1869, discovered a microscopic substance in the
1440:
The stability of the dsDNA form depends not only on the
957:
The backbone of the DNA strand is made from alternating
13992:. Plainview, N.Y: Cold Spring Harbor Laboratory Press.
12028:
7100:
6874:"Arsenic-eating microbe may redefine chemistry of life"
5935:
4444:
4308:
4261: – family of non-destructive analytical techniques
4113:
B-DNA X-ray patterns, and which supported the presence
3732:
proteins. DNA and other nucleic acids are the basis of
3614:, search and manipulate biological data, including DNA
3155:
transcript until it reaches a region of DNA called the
3112:
that contains multiple accessory subunits, such as the
2067:
1434:
934:
of the molecule (which holds the chain together) and a
14071:
The Stuff of Life: A Graphic Guide to Genetics and DNA
10846:
10557:
Critical Reviews in Biochemistry and Molecular Biology
10111:
10109:
9572:
Critical Reviews in Biochemistry and Molecular Biology
8836:
8435:
Wolfsberg TG, McEntyre J, Schuler GD (February 2001).
8393:
7745:
6567:. Amsterdam – New York: North-Holland Publishers.
5631:
4040:, a graduate student working under the supervision of
3646:
that make them distinct. These techniques, especially
2535:, with over 50% of human DNA consisting of non-coding
15594:
13904:(2nd ed.). Cold Spring Harbor Laboratory Press.
13785:
12499:
Investigations into nuclein and its cleavage products
12368:"Ueber die chemische Zusammensetzung der Eiterzellen"
11973:"Folding DNA to create nanoscale shapes and patterns"
11186:
Target Discovery and Validation Reviews and Protocols
9706:
9569:
8239:
7523:"Epigenetic regulation of human embryonic stem cells"
6022:"Diversity of coding strategies in influenza viruses"
5800:"Mitochondria in oocyte aging: current understanding"
5797:
5286:
Pabo CO, Sauer RT (1984). "Protein-DNA recognition".
5196:
2603:
for the creation of new genes through the process of
2527:, only a small fraction of the total sequence of the
1518:, both the female (XX) and male (XY) versions of the
1005:(sometimes called polarity) to each DNA strand. In a
576:
573:
567:
555:
552:
531:
525:
522:
13857:
Calladine CR, Drew HR, Luisi BF, Travers AA (2003).
12370:[On the chemical composition of pus cells].
11620:
Chandra M, Sachdeva A, Silverman SK (October 2009).
10653:"What is the optimum size for the genetic alphabet?"
10207:
10077:
9534:
9459:
Ito T (2003). "Nucleosome Assembly and Remodeling".
8622:"The role of heterochromatin in centromere function"
8102:. New York: Nova Science Publishers. pp. 1–47.
7748:"N6-methyladenine: the other methylated base of DNA"
7746:
Ratel D, Ravanat JL, Berger F, Wion D (March 2006).
6800:
5798:
Zhang D, Keilty D, Zhang ZF, Chian RC (March 2017).
4870:
Edwards KJ, Brown DG, Spink N, Skelly JV, Neidle S.
4263:
Pages displaying wikidata descriptions as a fallback
4248:
Pages displaying wikidata descriptions as a fallback
3989:
in Cambridge, England commemorating Crick and Watson
3759:. This field of phylogenetics is a powerful tool in
2645:
sequences of proteins is determined by the rules of
651:), nucleic acids are one of the four major types of
579:
558:
540:
534:
510:
33:
13093:
13035:
Cold Spring Harbor Symposia on Quantitative Biology
12273:
12133:
11622:"DNA-catalyzed sequence-specific hydrolysis of DNA"
11580:
11578:
11441:
10319:
10106:
10020:
9244:
8161:
8057:Alberts B, Johnson A, Lewis J, et al. (2002).
8056:
6426:"Molecular structure of deoxypentose nucleic acids"
6423:
5231:
4869:
4240: – Succession of nucleotides in a nucleic acid
3618:data. These have led to widely applied advances in
3378:(PAHs), the most carbon-rich chemical found in the
2714:. This enzyme makes discontinuous segments (called
1652:DNA can be twisted like a rope in a process called
1103:The nucleobases are classified into two types: the
736:Both strands of double-stranded DNA store the same
516:
21:For a non-technical introduction to the topic, see
14094:
14068:
13924:The path to the double helix: the discovery of DNA
13859:Understanding DNA: the molecule & how it works
13294:
12322:
11709:
11668:
10987:
9665:
8394:Thanbichler M, Wang SC, Shapiro L (October 2005).
8344:
7149:
7103:"Quadruplex DNA: sequence, topology and structure"
4729:
4389:
4180: – Any chromosome other than a sex chromosome
3951:developed and published observations now known as
3763:. If DNA sequences within a species are compared,
3431:, and to manipulate it in the laboratory, such as
2694:DNA replication: The double helix is unwound by a
1976:to construct geometric shapes, see the section on
1487:content, making the strands easier to pull apart.
1182:control of gene expression in plants and animals.
13094:Avery OT, Macleod CM, McCarty M (February 1944).
12932:
12184:
11444:"Individual-specific 'fingerprints' of human DNA"
9245:Sandman K, Pereira SL, Reeve JN (December 1998).
7688:
7410:
7351:
6634:
6358:"Molecular configuration in sodium thymonucleate"
5848:JCBN/NC-IUB Newsletter 1989. Retrieved 7 May 2008
3610:involves the development of techniques to store,
3253:The most common form of chromosomal crossover is
3167:with multiple regulatory and accessory subunits.
2784:
15702:
13750:"The Nobel Prize in Physiology or Medicine 1968"
13579:"The Nobel Prize in Physiology or Medicine 1962"
13185:
13067:Symposia of the Society for Experimental Biology
12567:
11669:Carmi N, Shultz LA, Breaker RR (December 1996).
11575:
9892:
9022:
8717:
7609:"DNA methylation patterns and epigenetic memory"
6961:
6220:Journal of Biomolecular Structure & Dynamics
3146:Transcription is carried out by a DNA-dependent
2614:
2413:. The set of chromosomes in a cell makes up its
2063:Chemical modifications and altered DNA packaging
985:of adjacent sugar rings. These are known as the
930:. The nucleotide contains both a segment of the
12225:"Dating branches on the tree of life using DNA"
11526:"DNA Identification in Mass Fatality Incidents"
8979:
6424:Wilkins MH, Stokes AR, Wilson HR (April 1953).
6355:
6299:
6254:
5970:
5576:
5574:
5485:
4228: – Cell division producing haploid gametes
3571:
3029:
2164:For one example, cytosine methylation produces
1962:can form networks containing multiple branches.
13512:
13301:. Cambridge, Mass.: Harvard University Press.
13188:"Chargaff's Rules: the Work of Erwin Chargaff"
13136:
12712:"The search for the chemical structure of DNA"
12652:Berichte der Deutschen Chemischen Gesellschaft
12621:Berichte der Deutschen Chemischen Gesellschaft
12187:"Analysis of aptamer discovery and technology"
11308:
11224:
11014:
10276:
9063:
8619:
7007:
6935:"Structure and packing of human telomeric DNA"
6669:
6562:
5372:"Mechanical stability of single DNA molecules"
5088:
5053:
4186: – Scientific study of crystal structures
3848:DNA was first isolated by the Swiss physician
2749:
2599:, although they can occasionally serve as raw
15169:
14782:
14398:
14066:
14031:Francis Crick: discoverer of the genetic code
13875:
13689:
13479:
13315:
12709:
12316:
12267:
11774:Bioinformatics: The Machine Learning Approach
11339:
11075:
11047:
9630:
9401:
8922:
8873:
8830:
8204:
7987:
7305:"Four new DNA letters double life's alphabet"
6351:
6349:
5674:
5004:
4889:
4551:
4549:
4547:
4545:
3335:) may have been formed extraterrestrially in
3131:enzyme involved in the infection of cells by
2802:
1833:. A report in 2010 of the possibility in the
1790:Compared to B-DNA, the A-DNA form is a wider
1675:
1048:nucleobases. The four bases found in DNA are
670:. Each nucleotide is composed of one of four
611:chains that coil around each other to form a
482:
13692:"The Replication of DNA in Escherichia Coli"
13672:(Speech). Cambridge, England. Archived from
13598:"The double helix and the 'wronged heroine'"
13451:
13403:
13324:"A Proposed Structure For The Nucleic Acids"
13321:
13218:
13179:
12645:
12614:
11881:Bioinformatics: Sequence and Genome Analysis
11584:
10950:
10233:
10208:Thorpe JH, Gale BC, Teixeira SC, Cardin CJ.
9927:
9791:
8879:
8526:
6068:
5791:
5762:
5758:
5756:
5754:
5625:
5571:
5314:
5190:
5131:
5082:
3261:can be damaging to cells, as it can produce
2899:transcription factor bound to its DNA target
2019:The phosphate groups of DNA give it similar
1806:
1098:
780:, these RNA strands specify the sequence of
14366:at Mandeville Special Collections Library,
14321:National Centre for Biotechnology Education
14089:
14033:. Ashland, OH: Eminent Lives, Atlas Books.
13831:
13518:
13286:
13025:
12933:Lorenz MG, Wackernagel W (September 1994).
12547:"Weitere Beiträge zur Chemie des Zellkerns"
12090:
11768:
10956:
10497:
10397:
10362:
9971:
9839:
9837:
9765:
8980:Berne C, Kysela DT, Brun YV (August 2010).
8973:
8338:
8126:
8065:(4th ed.). New York: Garland Science.
8031:"Unearthing Prehistoric Tumors, and Debate"
7946:
7520:
6124:
6122:
6019:
5281:
5279:
5138:Kumar S, Chinnusamy V, Mohapatra T (2018).
5047:
4818:
4555:
3739:
3075:that synthesize polynucleotide chains from
2948:(green) in a complex with its substrate DNA
1907:structure is called a displacement loop or
1795:the bases have been chemically modified by
15183:
15176:
15162:
14937:Precursor mRNA (pre-mRNA / hnRNA)
14789:
14775:
14405:
14391:
13297:In pursuit of the gene: from Darwin to DNA
13186:Kresge N, Simoni RD, Hill RL (June 2005).
11671:"In vitro selection of self-cleaving DNAs"
11342:"Likelihood ratios for DNA identification"
9843:
9666:Spiegelman BM, Heinrich R (October 2004).
9122:
8797:
7411:Burghardt B, Hartmann AK (February 2007).
6962:Greider CW, Blackburn EH (December 1985).
6957:
6955:
6871:
6419:
6417:
6346:
4542:
4216: – DNA testing to infer relationships
2710:copy. Another DNA polymerase binds to the
2539:. The reasons for the presence of so much
655:that are essential for all known forms of
489:
475:
14047:
13970:
13803:
13792:The Malaysian Journal of Medical Sciences
13725:
13715:
13624:
13546:
13357:
13347:
13244:
13203:
13119:
13008:. In Debru C, Gayon J, Picard JF (eds.).
12958:
12909:
12753:The physical-chemical basis of morphology
12694:
12591:
12299:
12250:
12240:
12059:
11970:
11939:
11929:
11847:
11833:
11745:
11735:
11686:
11645:
11467:
11375:
11365:
11250:
11183:
11109:
10933:
10923:
10891:
10686:
10676:
10650:
10607:
10568:
10472:
10431:
10133:
9997:
9948:
9869:
9817:
9742:
9732:
9683:
9319:
9270:
9207:
9197:
9148:
9116:
9099:
9089:
9005:
8956:
8946:
8905:
8780:
8770:
8694:
8645:
8570:
8509:
8460:
8411:
8370:
8005:
7964:
7817:
7771:
7722:
7624:
7571:
7548:
7538:
7500:"How To Extract DNA From Anything Living"
7428:
7387:
7328:
7279:
7214:
7126:
7074:
7025:
7003:
7001:
6999:
6979:
6783:
6773:
6747:
6328:
6283:
6045:
5996:
5971:Johnson ZI, Chisholm SW (November 2004).
5815:
5751:
5657:
5608:
5598:
5462:
5452:
5403:
5346:
5285:
5173:
5155:
5106:
5022:
4929:
4788:
4525:
4486:"Structural diversity of supercoiled DNA"
4440:
4438:
3459:or in the appropriate format, by using a
2046:
1115:, which are fused five- and six-membered
16:Molecule that carries genetic information
14241:, and does not reflect subsequent edits.
14224:
14142:. Cambridge, England: University Press.
13457:
13292:
13142:
12886:"The Significance of Pneumococcal Types"
12883:
12788:
12774:
12580:The Yale Journal of Biology and Medicine
12576:"Albrecht Kossel, a biographical sketch"
12533:(On the chemistry of the cell nucleus),
12365:
11804:
11442:Jeffreys AJ, Wilson V, Thein SL (1985).
11140:
11081:
11020:
10164:
10092:10.1146/annurev.biochem.73.011303.073859
10042:10.1146/annurev.biochem.71.090501.150041
9834:
9057:
8803:
8022:
7096:
7094:
6836:
6834:
6565:Direct analysis of diffraction by matter
6255:Franklin RE, Gosling RG (6 March 1953).
6128:
6119:
5276:
5056:Journal of the American Chemical Society
4814:
4812:
4270: – Synthetic nucleic acid analogues
4087:
3973:
3820:
3680:
3217:
3170:
2936:
2932:
2927:
2890:
2825:
2689:
2567:
2384:
2227:
2050:
1853:
1843:
1693:
1505:
1293:
1023:
878:
150:
122:
110:long strand of DNA unraveled. The DNA's
14738:List of genetics research organizations
14134:
14006:
13060:
12981:
12834:
12760:
12746:
12463:[On nuclein in yeast, Part 2].
12185:Dunn MR, Jimenez RM, Chaput JC (2017).
11542:
11309:Curtis C, Hereward J (29 August 2017).
10709:
9461:Protein Complexes that Modify Chromatin
8483:
8274:
8028:
7988:Valerie K, Povirk LF (September 2003).
7647:
6952:
6902:
6840:
6748:Oh DB, Kim YG, Rich A (December 2002).
6414:
6170:
6168:
6078:Annual Review of Biomedical Engineering
4982:. Memorial University of Newfoundland.
4725:
4723:
4721:
4719:
4416:
4387:
4096:The 25 April 1953 issue of the journal
2959:that cut DNA strands by catalyzing the
2815:
2380:
2337:and planar molecules; examples include
2132:converts 5-methylcytosine into thymine.
1977:
1787:with a significant degree of disorder.
1745:—and also B-DNA—used analyses based on
891:phosphate on one strand and an exposed
15703:
14325:Genetic Education Modules for Teachers
14265:From the official Nobel Prize web site
14258:DNA binding site prediction on protein
14115:
14025:
13896:
13861:. Amsterdam: Elsevier Academic Press.
13595:
13485:
13000:
12840:
12676:
12544:
12510:[On xanthin and hypoxanthin].
12505:
12492:
12475:
12458:
12441:
11911:
11872:
10021:Hubscher U, Maga G, Spadari S (2002).
9965:
9604:
9499:
7469:
7302:
7253:
6996:
6896:
6892:from the original on 12 February 2012.
6806:
6577:
6523:
6356:Franklin RE, Gosling RG (April 1953).
6090:10.1146/annurev.bioeng.6.062403.132016
5892:
5089:Weigele P, Raleigh EA (October 2016).
4435:
4222: – Group of genes from one parent
4192: – Holiday celebrated on April 25
3778:
3408:
1698:From left to right, the structures of
1595:
1150:
88:
79:
72:
15157:
14957:Histone acetylation and deacetylation
14770:
14386:
14346:"Clue to chemistry of heredity found"
13788:"Forensic DNA Profiling and Database"
13786:Panneerchelvam S, Norazmi MN (2003).
13662:
13652:from the original on 17 October 2016.
13380:
13322:Pauling L, Corey RB (February 1953).
12679:"The structure of yeast nucleic acid"
12573:
12551:Zeitschrift für Physiologische Chemie
12535:Zeitschrift für physiologische Chemie
12512:Zeitschrift für physiologische Chemie
12482:Zeitschrift für physiologische Chemie
12465:Zeitschrift für physiologische Chemie
12448:Zeitschrift für physiologische Chemie
11878:
11002:from the original on 5 September 2011
10554:
10503:
9972:Joyce CM, Steitz TA (November 1995).
8880:Finkel SE, Kolter R (November 2001).
8620:Pidoux AL, Allshire RC (March 2005).
8593:
8116:from the original on 25 October 2014.
8079:from the original on 2 January 2016.
7091:
6831:
6177:Nature Reviews Molecular Cell Biology
5839:Designation of the two strands of DNA
4952:
4809:
4748:
4730:Berg J, Tymoczko J, Stryer L (2002).
4618:from the original on 4 February 2007.
4473:from the original on 1 November 2016.
4126:Nobel Prize in Physiology or Medicine
3670:
3638:with other DNA sequences to identify
3403:
2664:. This RNA copy is then decoded by a
2561:molecules, which are involved in the
2434:copy is then used to make a matching
2202:of uracil to produce the "J-base" in
1741:The first published reports of A-DNA
1001:the sugar-phosphate backbone confers
819:) store most of their DNA inside the
139:in the structure are colour-coded by
65:
56:
49:
15623:
15022:Ribosome-nascent chain complex (RNC)
13987:
13941:
13918:
13766:
13439:from the original on 30 January 2009
13100:The Journal of Experimental Medicine
12395:
12372:Medicinisch-chemische Untersuchungen
12222:
11321:from the original on 22 October 2017
11053:
10988:ScienceDaily Staff (9 August 2011).
10981:
9844:Doherty AJ, Suh SW (November 2000).
9402:Jenuwein T, Allis CD (August 2001).
9301:
9251:Cellular and Molecular Life Sciences
8752:
7689:Kriaucionis S, Heintz N (May 2009).
7606:
7008:Nugent CI, Lundblad V (April 1998).
6853:from the original on 4 December 2010
6819:from the original on 3 December 2010
6410:from the original on 3 January 2011.
6295:from the original on 9 January 2016.
6174:
6165:
4977:
4716:
4419:Principles of Nucleic Acid Structure
4352:from the original on 5 January 2017.
3943:, supporting Griffith's suggestion (
3366:, using starting chemicals, such as
2830:Interaction of DNA (in orange) with
2579:(green) from a DNA template (orange)
2449:
2357:, and in the case of thalidomide, a
2259:DNA can be damaged by many sorts of
2068:Base modifications and DNA packaging
1831:arsenic instead of phosphorus in DNA
1552:. Chromosome 1 is the largest human
784:within proteins in a process called
721:rules (A with T and C with G), with
14796:
14377:Crick's medal goes under the hammer
14368:University of California, San Diego
13882:. Basingstoke: Palgrave Macmillan.
13519:Cobb M, Comfort N (25 April 2023).
12749:Физико-химические основы морфологии
11273:
11225:Daniell H, Dhingra A (April 2002).
9458:
9448:from the original on 8 August 2017.
9123:Hui L, Bianchi DW (February 2013).
8129:The New England Journal of Medicine
8043:from the original on 24 June 2017.
7953:The Journal of Biological Chemistry
7947:Beckman KB, Ames BN (August 1997).
6020:Lamb RA, Horvath CM (August 1991).
5938:Computational Biology and Chemistry
5300:10.1146/annurev.bi.53.070184.001453
4339:
4117:of the Watson and Crick structure.
3905:suggested that DNA is found in the
3701:DNA nanotechnology uses the unique
2055:Impure DNA extracted from an orange
1983:
981:between the third and fifth carbon
143:and the detailed structures of two
91:Function
13:
14211:
13947:"Quiet debut for the double helix"
13824:
13690:Meselson M, Stahl FW (July 1958).
13500:from the original on 25 April 2023
13433:"Original X-ray diffraction image"
11502:. 14 December 2006. Archived from
11340:Collins A, Morton NE (June 1994).
9792:Bickle TA, Krüger DH (June 1993).
8347:"The sequence of the human genome"
8059:"The Preventable Causes of Cancer"
7475:
6719:10.1034/j.1600-065x.2001.1840125.x
6340:from the original on 29 June 2017.
5950:10.1016/j.compbiolchem.2004.12.006
5804:Facts, Views & Vision in ObGyn
5567:from the original on 10 June 2007.
5500:10.1111/j.1365-2958.1995.tb02309.x
4556:Watson JD, Crick FH (April 1953).
4329:from the original on 14 July 2014.
4133:central dogma of molecular biology
3095:In DNA replication, DNA-dependent
3052:. They use the chemical energy in
2178:lacks cytosine methylation, while
1813:hypothetical types of biochemistry
1423:
1283:2,6-Diaminopurine (2-Aminoadenine)
895:hydroxyl group (—OH) on the other.
14:
15737:
15300:Micro
14158:
13225:The Journal of General Physiology
11563:from the original on 24 June 2017
11035:from the original on 5 March 2015
10969:from the original on 23 June 2015
8437:"Guide to the draft human genome"
7650:DNA Methylation: Basic Mechanisms
6478:from the original on 13 May 2011.
4986:from the original on 19 July 2016
3805:has prevented its practical use.
3596:
1821:have proposed the existence of a
1402:is more stable than DNA with low
1131:. A fifth pyrimidine nucleobase,
903:made from repeating units called
863:. Within eukaryotic chromosomes,
15683:
15666:
15649:
15632:
15604:
15579:
15578:
14751:
14750:
14223:
13926:. New York: Dover Publications.
13779:
13760:
13742:
13683:
13656:
13589:
13571:
13425:
13374:
13261:
13219:Hershey AD, Chase M (May 1952).
13212:
13087:
13063:"X-ray studies of nucleic acids"
13016:
12994:
12975:
12926:
12877:
12737:
12703:
12670:
12639:
12608:
12432:
12389:
12359:
12216:
12178:
12127:
12084:
12022:
11964:
11905:
11827:
11798:
11762:
11703:
11662:
11613:
11536:
11518:
11492:
11435:
11392:
11333:
11302:
11267:
11231:Current Opinion in Biotechnology
11218:
11177:
11134:
10840:
10805:
10754:
10703:
10644:
10601:
10548:
10474:10.1046/j.1432-1033.2002.03250.x
10461:European Journal of Biochemistry
10448:
10398:Neale MJ, Keeney S (July 2006).
10391:
10356:
10313:
10270:
10227:
10201:
10158:
10071:
10014:
9990:10.1128/jb.177.22.6321-6329.1995
9950:10.1111/j.1432-1033.2004.04094.x
9937:European Journal of Biochemistry
9921:
9895:Biochemical Society Transactions
9886:
9785:
9759:
9700:
9659:
9645:10.1146/annurev.biochem.69.1.729
9624:
9598:
9563:
9528:
9502:Biochemical Society Transactions
9493:
9452:
9395:
9344:
9321:10.1111/j.1365-2958.2005.04598.x
9295:
9238:
9224:
9165:
8998:10.1111/j.1365-2958.2010.07267.x
8898:10.1128/JB.183.21.6288-6293.2001
8746:
8711:
8662:
8613:
8587:
8400:Journal of Cellular Biochemistry
8098:. In Kimura H, Suzuki A (eds.).
7413:"RNA secondary structure design"
6872:Katsnelson A (2 December 2010).
6300:Franklin RE, Gosling RG (1953).
6143:10.1146/annurev.biochem.70.1.369
5895:Journal of Cellular Biochemistry
4821:Acta Crystallographica Section D
3945:Avery–MacLeod–McCarty experiment
3482:
3376:polycyclic aromatic hydrocarbons
3194:
3185:
3048:are proteins that are a type of
2216:DNA damage (naturally occurring)
2100:
2093:
2086:
1940:
1933:
1354:
1341:
506:
456:
455:
342:
341:
174:
100:
41:
15255:precursor, heterogenous nuclear
15027:Post-translational modification
13411:"Double Helix of DNA: 50 Years"
13192:Journal of Biological Chemistry
12747:Koltsov HK (12 December 1927).
12508:"Ueber Xanthin und Hypoxanthin"
12446:[On nuclein in yeast].
12242:10.1186/gb-2001-3-1-reviews0001
11082:Callaway E (17 February 2021).
10957:Steigerwald J (8 August 2011).
9928:Tuteja N, Tuteja R (May 2004).
8772:10.1186/gb-2001-2-1-reviews3002
8477:
8428:
8387:
8303:
8277:Pharmacology & Therapeutics
8268:
8233:
8198:
8155:
8120:
8087:
8050:
7981:
7940:
7897:
7861:
7842:
7788:
7739:
7682:
7641:
7600:
7565:
7514:
7492:
7404:
7345:
7296:
7260:Quarterly Reviews of Biophysics
7247:
7194:
7143:
7042:
6927:
6865:
6741:
6698:
6663:
6628:
6571:
6556:
6517:
6482:
6246:
6211:
6062:
6013:
5964:
5929:
5886:
5851:
5832:
5725:
5522:
5479:
5420:
5363:
5225:
5024:10.1146/annurev.biochem.67.1.99
4998:
4980:"Watson-Crick Structure of DNA"
4971:
4946:
4863:
4665:
4622:
4396:. New York: Benjamin Cummings.
3897:In 1933, while studying virgin
2993:restriction modification system
1914:
1641:
1556:with approximately 220 million
1522:(bottom right), as well as the
1326:
1232:5-Glycosylhydroxymethylcytosine
15385:Trans-acting small interfering
15349:Enhancer RNAs
15267:Transfer
12951:10.1128/MMBR.58.3.563-602.1994
12710:Cohen JS, Portugal FH (1974).
12061:11858/00-001M-0000-0010-9362-B
11805:Gusfield D (15 January 1997).
10849:Journal of Molecular Evolution
10622:10.1126/science.292.5520.1278a
9810:10.1128/MMBR.57.2.434-450.1993
9404:"Translating the histone code"
8029:Johnson G (28 December 2010).
7574:Trends in Biochemical Sciences
6563:Hosemann R, Bagchi RN (1962).
6232:10.1080/07391102.1988.10507714
4477:
4410:
4381:
4356:
4333:
4302:
4295:Merriam-Webster.com Dictionary
4282:
4255: – DNA analysis technique
3913:is present exclusively in the
3560:DNA profiling is also used in
3455:into organisms in the form of
3063:
2791:Neutrophil extracellular traps
2785:Neutrophil extracellular traps
2679:
2027:and it can be considered as a
1619:through RNA-RNA base pairing.
855:) store their DNA only in the
147:are shown in the bottom right.
1:
15272:Ribosomal
15250:Messenger
14279:Double Helix: 50 years of DNA
14269:DNA under electron microscope
14052:. Columbia University Press.
13842:. New York: Alfred A. Knopf.
13273:scarc.library.oregonstate.edu
13205:10.1016/S0021-9258(20)61522-8
12696:10.1016/S0021-9258(18)87254-4
12646:Levene PA, Jacobs WA (1909).
12615:Levene PA, Jacobs WA (1909).
11858:10.1093/bioinformatics/bth021
11688:10.1016/S1074-5521(96)90170-2
11288:10.1016/S0300-9084(02)00013-5
11243:10.1016/S0958-1669(02)00297-5
10377:10.1016/S0300-9084(97)82007-X
10080:Annual Review of Biochemistry
10030:Annual Review of Biochemistry
9633:Annual Review of Biochemistry
9043:10.1126/science.295.5559.1487
8755:"Replicative DNA polymerases"
8732:10.1016/S0022-2836(02)00109-2
8312:Current Pharmaceutical Design
8254:10.1016/S0006-2952(99)00388-3
8063:Molecular biology of the cell
7926:10.1016/S0027-5107(99)00004-4
7303:Warren M (21 February 2019).
7225:10.1016/S0092-8674(00)80760-6
6841:Bortman H (2 December 2010).
6131:Annual Review of Biochemistry
6069:Benham CJ, Mielke SP (2005).
5396:10.1016/S0006-3495(00)76747-6
5288:Annual Review of Biochemistry
5011:Annual Review of Biochemistry
4448:Molecular Biology of the Cell
4421:. New York: Springer-Verlag.
4312:Molecular Biology of the Cell
4275:
2615:Transcription and translation
2563:regulation of gene expression
2508:that can be transcribed, and
2397:DNA usually occurs as linear
874:
14697:Missing heritability problem
14412:
14189:Resources in other libraries
14067:Schultz M, Cannon Z (2009).
14011:. Cold Spring Harbor Press.
13876:Carina D, Clayton J (2003).
13458:Burakoff M (25 April 2023).
13026:Astbury WT, Bell FO (1938).
12648:"Über die Hefe-Nucleinsäure"
11931:10.1371/journal.pbio.0020073
11599:10.1016/1074-5521(94)90014-0
11401:Journal of Forensic Sciences
11155:10.1016/0092-8674(76)90133-1
10023:"Eukaryotic DNA polymerases"
9794:"Biology of DNA restriction"
9549:10.1016/0168-9525(94)90232-1
9199:10.1371/journal.pone.0041781
9091:10.1371/journal.pone.0051905
8948:10.1371/journal.ppat.1000213
8720:Journal of Molecular Biology
8289:10.1016/0163-7258(85)90013-0
8219:10.1016/0165-1110(91)90006-H
7810:10.1016/0092-8674(93)90322-H
6981:10.1016/0092-8674(85)90170-9
6807:Palmer J (2 December 2010).
6672:Journal of Molecular Biology
6503:10.1016/0022-2836(80)90124-2
6491:Journal of Molecular Biology
6038:10.1016/0168-9525(91)90326-L
5777:10.1016/0014-4827(91)90467-9
5005:Verma S, Eckstein F (1998).
4734:. W.H. Freeman and Company.
4643:10.1016/0022-2836(81)90099-1
4631:Journal of Molecular Biology
3817:History of molecular biology
3697:properties of DNA molecules.
3572:DNA enzymes or catalytic DNA
3429:phenol-chloroform extraction
3284:
3259:Non-homologous recombination
3030:Topoisomerases and helicases
2738:sequence is recreated by an
2649:, known collectively as the
2316:. These mutations can cause
2118:
2113:
2108:
1951:
1948:
1713:DNA exists in many possible
1540:In humans, the total female
965:groups. The sugar in DNA is
7:
14009:DNA Science: A First Course
13666:A Note for the RNA Tie Club
13047:10.1101/sqb.1938.006.01.013
12884:Griffith F (January 1928).
12811:10.1126/science.80.2075.312
12574:Jones ME (September 1953).
12461:"Ueber Nucleïn der Hefe II"
11971:Rothemund PW (March 2006).
11054:Hunt K (17 February 2021).
11021:Marlaire R (3 March 2015).
9469:10.1007/978-3-642-55747-7_1
8808:Extracellular Nucleic Acids
8484:Gregory TR (January 2005).
8184:10.1016/j.mrrev.2011.05.001
7521:Hu Q, Rosenfeld MG (2012).
7480:. Michigan State University
7254:Seeman NC (November 2005).
5108:10.1021/acs.chemrev.6b00114
4259:X-ray scattering techniques
4170:
4013:was confirmed in 1952 when
3648:multiple sequence alignment
3624:string searching algorithms
2750:Extracellular nucleic acids
2373:to inhibit rapidly growing
2312:from the DNA sequence, and
883:Chemical structure of DNA;
647:and complex carbohydrates (
10:
15742:
15451:Multicopy single-stranded
15295:Interferential
13990:Francis Crick: A Biography
13548:10.1038/d41586-023-01313-5
13486:Anthes E (25 April 2023).
12524:"Zur Chemie des Zellkerns"
11811:Cambridge University Press
11543:Pollack A (19 June 2012).
11111:10.1038/d41586-021-00436-x
10651:Szathmáry E (April 1992).
10135:10.1096/fasebj.8.8.7514143
9685:10.1016/j.cell.2004.09.037
8100:New Research on DNA Damage
7586:10.1016/j.tibs.2005.12.008
7447:10.1103/PhysRevE.75.021920
7330:10.1038/d41586-019-00650-8
5765:Experimental Cell Research
3814:
3808:
3782:
3743:
3674:
3600:
3575:
3486:
3412:
3382:, may have been formed in
3288:
3263:chromosomal translocations
3174:
2819:
2803:Interactions with proteins
2788:
2683:
2618:
2453:
2314:chromosomal translocations
2300:to repair and can produce
2224:DNA damage theory of aging
2213:
2071:
2043:which could hydrolyze it.
2014:
1987:
1918:
1847:
1810:
1743:X-ray diffraction patterns
1679:
1676:Alternative DNA structures
1645:
1599:
1531:
1330:
1289:
1272:5-Hydroxymethyldeoxyuracil
1195:N6-carbamoyl-methyladenine
1166:Mycobacterium tuberculosis
20:
15574:
15509:
15459:
15402:
15365:Guide
15357:
15285:
15240:
15223:
15192:
15130:
15039:
15004:
14978:
14969:
14927:
14901:
14875:
14866:
14804:
14746:
14725:
14624:
14575:
14519:
14458:
14420:
14274:Dolan DNA Learning Center
14263:DNA the Double Helix Game
14184:Resources in your library
13596:Maddox B (January 2003).
12902:10.1017/S0022172400031879
12777:Biologisches Zentralblatt
12664:10.1002/cber.190904202148
12633:10.1002/cber.190904201196
12410:10.1007/s00439-007-0433-0
12337:10.1007/s11033-018-4280-y
12325:Molecular Biology Reports
12292:10.1007/s13205-018-1246-7
11194:10.1385/1-59745-165-7:163
10869:10.1007/s00239-001-0025-x
10826:10.1016/j.tim.2005.03.010
10579:10.1080/10409230490460765
9584:10.1080/10409239991209255
9141:10.1016/j.tig.2012.10.013
7855:22 September 2008 at the
7272:10.1017/S0033583505004087
6913:10.1038/nature.2012.11520
6607:10.1107/S0567739478001540
6330:10.1107/S0365110X53001940
6285:10.1107/S0365110X53001939
5872:10.1016/j.tig.2005.03.007
5600:10.1186/s13104-019-4137-z
4841:10.1107/S0907444903003251
4153:Marshall Warren Nirenberg
4137:Meselson–Stahl experiment
3828:(left) shakes hands with
3722:algorithmic self-assembly
3437:polymerase chain reaction
2981:restriction endonucleases
2700:topoisomerase
2333:. Most intercalators are
2277:electromagnetic radiation
2209:
2168:, which is important for
2009:principle of least effort
1858:DNA quadruplex formed by
1825:, a postulated microbial
1807:Alternative DNA chemistry
1602:Sense (molecular biology)
1501:
1159:, which was found in the
1123:, the six-membered rings
1099:Nucleobase classification
1007:nucleic acid double helix
127:The structure of the DNA
15327:Small nuclear
15088:sequestration (P-bodies)
13435:. Oregon State Library.
12773:Reprinted in German as:
12529:17 November 2017 at the
12444:"Ueber Nucleïn der Hefe"
12191:Nature Reviews Chemistry
11545:"Before Birth, Dad's ID"
10457:"The RuvABC resolvasome"
9766:Kostrewa D, Winkler FK.
8324:10.2174/1381612013397113
8242:Biochemical Pharmacology
7966:10.1074/jbc.272.32.19633
7949:"Oxidative decay of DNA"
7658:10.1007/3-540-31390-7_11
7540:10.3389/fgene.2012.00238
5157:10.3389/fgene.2018.00640
4023:Hershey–Chase experiment
3939:, identified DNA as the
3931:, along with co-workers
3889:of the "smooth" form of
3785:DNA digital data storage
3740:History and anthropology
3642:and locate the specific
3323:Building blocks of DNA (
3255:homologous recombination
3077:nucleoside triphosphates
3054:nucleoside triphosphates
2921:cellular differentiation
2757:horizontal gene transfer
2625:Transcription (genetics)
2484:, with small amounts in
2425:of pieces of DNA called
2363:benzopyrene diol epoxide
1514:of a human. It shows 22
1040:between nucleotides and
715:sugar-phosphate backbone
23:Introduction to genetics
15441:Genomic
15066:Gene regulatory network
14379:, Nature, 5 April 2013.
14310:ENCODE threads explorer
14118:DNA: The Secret of Life
13839:DNA: the secret of life
13387:Oxford University Press
12939:Microbiological Reviews
12843:Nature Reviews Genetics
12203:10.1038/s41570-017-0076
12156:10.1126/science.1154533
11912:Strong M (March 2004).
11737:10.1073/pnas.1420361112
11675:Chemistry & Biology
11626:Nature Chemical Biology
11587:Chemistry & Biology
11367:10.1073/pnas.91.13.6007
10925:10.1073/pnas.1106493108
10322:Nature Reviews Genetics
10279:Nature Reviews Genetics
10236:Nature Reviews Genetics
10179:10.1023/A:1021258713850
10167:Plant Molecular Biology
9978:Journal of Bacteriology
9798:Microbiological Reviews
9734:10.1073/pnas.1332764100
9423:10.1126/science.1063127
8886:Journal of Bacteriology
8372:10.1126/science.1058040
7715:10.1126/science.1169786
7613:Genes & Development
7607:Bird A (January 2002).
7380:10.1126/science.aat0971
7055:Genes & Development
7014:Genes & Development
5454:10.1073/pnas.96.14.7853
4761:The Biochemical Journal
4290:"deoxyribonucleic acid"
4031:enterobacteria phage T2
4025:showed that DNA is the
4007:University of Cambridge
3687:atomic force microscopy
3650:, are used in studying
2241:metabolically activated
2192:5-hydroxymethylcytosine
1763:. In the same journal,
1686:Molecular models of DNA
1581:human mitochondrial DNA
1082:adenosine monophosphate
989:(three prime end), and
15544:Artificial chromosomes
15332:Small nucleolar
15071:cis-regulatory element
14358:DNA from the Beginning
14337:Molecule of the Month
14329:DNA from the Beginning
14319:Double Helix 1953–2003
14219:
14199:Listen to this article
12890:The Journal of Hygiene
12725:(10): 551–52, 554–57.
12522:Albrect Kossel (1883)
12113:10.1038/nnano.2009.101
11914:"Protein nanomachines"
10814:Trends in Microbiology
10678:10.1073/pnas.89.7.2614
9862:10.1093/nar/28.21.4051
9850:Nucleic Acids Research
9308:Molecular Microbiology
8986:Molecular Microbiology
8638:10.1098/rstb.2004.1611
8007:10.1038/sj.onc.1206679
7107:Nucleic Acids Research
7067:10.1101/gad.11.21.2801
6775:10.1073/pnas.262672699
6684:10.1006/jmbi.2000.3690
5488:Molecular Microbiology
5211:10.1002/anie.201708228
4902:Nucleic Acids Research
4093:
3990:
3941:transforming principle
3845:
3765:population geneticists
3718:Nanomechanical devices
3698:
3229:chromosome territories
3224:
3058:adenosine triphosphate
2949:
2900:
2835:
2723:
2580:
2500:. A gene is a unit of
2394:
2393:within the chromosomes
2389:Location of eukaryote
2256:
2175:Caenorhabditis elegans
2157:). There is, further,
2056:
2047:Macroscopic appearance
1863:
1734:, and the presence of
1710:
1564:long if straightened.
1537:
1516:homologous chromosomes
1483:, tend to have a high
1303:
1269:5-Dihydroxypentauracil
1145:nucleic acid analogues
1117:heterocyclic compounds
1033:
896:
740:. This information is
738:biological information
729:and the double-ringed
711:phosphodiester linkage
615:. The polymer carries
156:
148:
25:. For other uses, see
15337:Small Cajal Body RNAs
14733:List of genetic codes
14218:
13717:10.1073/pnas.44.7.671
12093:Nature Nanotechnology
9263:10.1007/s000180050259
8812:. Springer. pp.
8535:Stamatoyannopoulos JA
8141:10.1056/NEJMra0804615
7527:Frontiers in Genetics
7027:10.1101/gad.12.8.1073
6939:ndbserver.rutgers.edu
6886:10.1038/news.2010.645
6707:Immunological Reviews
5844:24 April 2008 at the
5144:Frontiers in Genetics
4490:Nature Communications
4238:Nucleic acid sequence
4232:Nucleic acid notation
4091:
3997:started working with
3977:
3824:
3815:Further information:
3744:Further information:
3703:molecular recognition
3695:molecular recognition
3684:
3675:Further information:
3616:nucleic acid sequence
3601:Further information:
3576:Further information:
3562:DNA paternity testing
3487:Further information:
3413:Further information:
3289:Further information:
3241:genetic recombination
3233:chromosomal crossover
3221:
3211:genetic recombination
3177:Genetic recombination
3175:Further information:
3171:Genetic recombination
3125:reverse transcriptase
3024:genetic recombination
2940:
2933:Nucleases and ligases
2928:DNA-modifying enzymes
2905:transcription factors
2895:The lambda repressor
2894:
2884:or being degraded by
2869:transcription factors
2829:
2820:Further information:
2693:
2684:Further information:
2619:Further information:
2571:
2454:Further information:
2388:
2275:and also high-energy
2231:
2214:Further information:
2072:Further information:
2054:
1990:Nucleic acid analogue
1919:Further information:
1857:
1848:Further information:
1844:Quadruplex structures
1811:Further information:
1697:
1680:Further information:
1646:Further information:
1600:Further information:
1532:Further information:
1509:
1371:base pair with three
1331:Further information:
1312:transcription factors
1297:
1027:
882:
502:Deoxyribonucleic acid
442:Personalized medicine
436:Personalized medicine
299:Quantitative genetics
294:Mendelian inheritance
154:
126:
106:A chromosome and its
15390:Subgenomic messenger
15305:Small interfering
15277:Transfer-messenger
15093:alternative splicing
15083:Post-transcriptional
14909:Transcription factor
14632:Behavioural genetics
14312:ENCODE home page at
14250:More spoken articles
14101:. New York: Norton.
14048:Rosenfeld I (2010).
13349:10.1073/pnas.39.2.84
13112:10.1084/jem.79.2.137
12984:Archives de Biologie
12719:Connecticut Medicine
11638:10.1038/nchembio.201
11532:on 12 November 2006.
10616:(5520): 1278a–1278.
9605:Beamer LJ, Pabo CO.
9302:Dame RT (May 2005).
7502:. University of Utah
4674:Biochemical Genetics
4065:. In February 1953,
4003:Cavendish Laboratory
3801:), and insufficient
3761:evolutionary biology
3656:Human Genome Project
3640:homologous sequences
3530:short tandem repeats
3465:genetically modified
3419:Nucleic acid methods
3291:RNA world hypothesis
3235:which occurs during
2965:phosphodiester bonds
2816:DNA-binding proteins
2809:protein interactions
2722:joins them together.
2674:standard amino acids
2629:Protein biosynthesis
2575:(blue) producing an
2537:repetitive sequences
2510:regulatory sequences
2438:in a process called
2407:circular chromosomes
2381:Biological functions
2155:Chromatin remodeling
2078:Chromatin remodeling
1524:mitochondrial genome
1252:α-Putrescinylthymine
979:phosphodiester bonds
861:circular chromosomes
801:Eukaryotic organisms
362:Branches of genetics
27:DNA (disambiguation)
15017:Transfer RNA (tRNA)
14712:Population genomics
14702:Molecular evolution
14662:Genetic engineering
14093:, Watson J (1980).
13972:10.1038/nature01397
13963:2003Natur.421..402O
13708:1958PNAS...44..671M
13626:10.1038/nature01399
13617:2003Natur.421..407M
13539:2023Natur.616..657C
13340:1953PNAS...39...84P
13293:Schwartz J (2008).
13237:10.1085/jgp.36.1.39
13061:Astbury WT (1947).
12803:1934Sci....80..312K
12366:Miescher F (1871).
12148:2008Sci...321.1795A
12105:2009NatNa...4..281I
12052:10.1038/nature07971
12044:2009Natur.459...73A
12000:10.1038/nature04586
11992:2006Natur.440..297R
11772:, Brunak S (2001).
11728:2015PNAS..112.5903T
11506:on 14 December 2006
11460:1985Natur.316...76J
11358:1994PNAS...91.6007C
11149:(4 PT 2): 695–705.
11102:2021Natur.590..537C
10916:2011PNAS..10813995C
10861:2002JMolE..54..134N
10775:2000Natur.407..897V
10724:1993Natur.362..709L
10669:1992PNAS...89.2614S
10518:2002Natur.418..214J
10424:10.1038/nature04885
10416:2006Natur.442..153N
10067:on 26 January 2021.
9907:10.1042/BST20051465
9725:2003PNAS..100.8164L
9365:1997Natur.389..251L
9190:2012PLoSO...741781F
9082:2012PLoSO...751905H
8594:Yin YW, Steitz TA.
8563:10.1038/nature05874
8555:2007Natur.447..799B
8453:2001Natur.409..824W
8363:2001Sci...291.1304V
8176:2011MRRMR.728...12F
7918:1999MRFMM.424....9C
7707:2009Sci...324..929K
7439:2007PhRvE..75b1920B
7372:2019Sci...363..884H
7321:2019Natur.566..436W
7164:2002Natur.417..876P
6766:2002PNAS...9916666O
6599:1978AcCrA..34..751B
6445:1953Natur.171..738W
6377:1953Natur.171..740F
6321:1953AcCry...6..678F
6276:1953AcCry...6..673F
5695:1981Natur.290..457A
5659:10.1038/nature04727
5650:2006Natur.441..315G
5445:1999PNAS...96.7853C
5388:2000BpJ....78.1997C
5376:Biophysical Journal
5246:1980Natur.287..755W
5101:(20): 12655–12687.
5068:10.1021/ja01688a030
4833:2003AcCrD..59..620G
4805:on 5 February 2007.
4577:1953Natur.171..737W
4502:2015NatCo...6.8440I
4048:image, labeled as "
3967:should be equal to
3959:should be equal to
3779:Information storage
3769:ecological genetics
3494:Forensic scientists
3433:restriction digests
3423:Genetic engineering
3409:Genetic engineering
3386:or in interstellar
3374:. Pyrimidine, like
3237:sexual reproduction
2917:signal transduction
2822:DNA-binding protein
2768:Cell-free fetal DNA
2755:may be involved in
2322:naturally occurring
2267:. Mutagens include
2263:, which change the
1747:Patterson functions
1596:Sense and antisense
1450:melting temperature
1408:Hoogsteen base pair
1175:restriction enzymes
1151:Non-canonical bases
1044:interactions among
672:nitrogen-containing
332:Genetic engineering
289:Population genetics
162:Part of a series on
15419:Chloroplast
15262:modified Messenger
15225:Ribonucleic acids
15131:Influential people
15110:Post-translational
14929:Post-transcription
14672:Genetic monitoring
14351:The New York Times
14220:
14007:Micklas D (2003).
13679:on 1 October 2008.
13493:The New York Times
13157:10.1007/BF02173653
12617:"Über Inosinsäure"
12235:(1): REVIEWS0001.
11549:The New York Times
9537:Trends in Genetics
9514:10.1042/BST0290395
9129:Trends in Genetics
8851:10.1002/bies.20604
8765:(1): REVIEWS3002.
8502:10.1093/aob/mci009
8036:The New York Times
7764:10.1002/bies.20342
7626:10.1101/gad.947102
7119:10.1093/nar/gkl655
6974:(2 Pt 1): 405–13.
6587:Acta Crystallogr A
6578:Baianu IC (1978).
6542:10.1007/BF02462372
6524:Baianu IC (1980).
6026:Trends in Genetics
5989:10.1101/gr.2433104
5860:Trends in Genetics
5544:(51): 15996–6010.
4914:10.1093/nar/gkj454
4686:10.1007/BF00485753
4510:10.1038/ncomms9440
4417:Saenger W (1984).
4388:Russell P (2001).
4298:. Merriam-Webster.
4145:Har Gobind Khorana
4094:
3991:
3963:and the amount of
3879:Frederick Griffith
3850:Friedrich Miescher
3846:
3726:gold nanoparticles
3699:
3691:DNA nanotechnology
3677:DNA nanotechnology
3671:DNA nanotechnology
3526:DNA fingerprinting
3404:Uses in technology
3339:. Complex DNA and
3225:
3001:DNA fingerprinting
2950:
2943:restriction enzyme
2901:
2836:
2773:Under the name of
2724:
2581:
2506:open reading frame
2395:
2257:
2057:
1978:uses in technology
1952:Multiple branches
1925:DNA nanotechnology
1864:
1773:molecular modeling
1711:
1538:
1304:
1260:and modifications
1249:α-Glutamythymidine
1226:5-Carboxylcytosine
1173:) is to avoid the
1034:
897:
867:proteins, such as
827:, and some in the
304:Molecular genetics
263:History and topics
157:
155:Simplified diagram
149:
15592:
15591:
15469:Xeno
15431:Complementary
15404:Deoxyribonucleic
15398:
15397:
15375:Small hairpin
15151:
15150:
15035:
15034:
14965:
14964:
14841:Special transfers
14764:
14763:
14687:He Jiankui affair
14677:Genetic genealogy
14667:Genetic diversity
14596:the British Isles
14501:Genetic variation
14216:
14165:Library resources
14127:978-0-09-945184-6
14116:Watson J (2004).
14082:978-0-8090-8947-5
14075:. Hill and Wang.
14059:978-0-231-14271-7
14018:978-0-87969-636-8
13999:978-0-87969-798-3
13663:Crick FH (1955).
13533:(7958): 657–660.
13396:978-0-19-538341-6
13308:978-0-674-02670-4
13001:Burian R (1994).
12677:Levene P (1919).
12545:Kossel A (1886).
12506:Kossel A (1882).
12493:Kossel A (1881).
12476:Kossel A (1881).
12459:Kossel A (1880).
12442:Kossel A (1879).
12142:(5897): 1795–99.
11986:(7082): 297–302.
11879:Mount DM (2004).
11820:978-0-521-58519-4
11783:978-0-262-02506-5
11500:"Colin Pitchfork"
11413:10.1520/JFS14100J
11203:978-1-59745-165-9
11096:(7847): 537–538.
10769:(6806): 897–900.
9901:(Pt 6): 1465–70.
9508:(Pt 4): 395–401.
9478:978-3-540-44208-0
9417:(5532): 1074–80.
8823:978-3-642-12616-1
8687:10.1101/gr.207102
8549:(7146): 799–816.
8413:10.1002/jcb.20519
8357:(5507): 1304–51.
8207:Mutation Research
8164:Mutation Research
8109:978-1-60456-581-2
7906:Mutation Research
7883:10.1021/bi034593c
7417:Physical Review E
7366:(6429): 884–887.
7172:10.1038/nature755
5907:10.1002/jcb.20252
5739:on 13 August 2011
5689:(5806): 457–465.
5550:10.1021/bi048221v
5339:10.1002/bip.22334
5205:(16): 4296–4312.
5199:Angewandte Chemie
4964:978-0-7637-8663-2
4955:Molecular Biology
4953:Tropp BE (2012).
4773:10.1042/bj1200449
4322:978-0-8153-4432-2
4268:Xeno nucleic acid
4214:Genetic genealogy
4157:molecular biology
4046:X-ray diffraction
4042:Rosalind Franklin
3838:Rosalind Franklin
3750:Genetic genealogy
3475:, or be grown in
3415:Molecular biology
3344:organic compounds
3333:organic molecules
3279:Holliday junction
3248:natural selection
3207:Holliday junction
3205:Structure of the
3202:
3201:
3161:RNA polymerase II
2997:molecular cloning
2977:molecular biology
2775:environmental DNA
2736:complementary DNA
2716:Okazaki fragments
2573:T7 RNA polymerase
2450:Genes and genomes
2361:. Others such as
2297:hydrogen peroxide
2273:alkylating agents
2125:
2124:
1956:
1955:
1632:overlapping genes
1577:mitochondrial DNA
1571:, in addition to
1362:
1361:
1349:
1348:
1235:5-Hydroxycytosine
1223:N4-Methylcytosine
833:mitochondrial DNA
795:. Before typical
750:protein sequences
499:
498:
226:Genetic variation
121:
120:
15733:
15696:
15688:
15687:
15686:
15679:
15678:from Wikiversity
15671:
15670:
15669:
15662:
15654:
15653:
15652:
15645:
15637:
15636:
15635:
15625:
15609:
15608:
15600:
15582:
15581:
15559:Yeast
15380:Small temporal
15310:Piwi-interacting
15238:
15237:
15234:
15215:Deoxynucleotides
15178:
15171:
15164:
15155:
15154:
14976:
14975:
14873:
14872:
14791:
14784:
14777:
14768:
14767:
14754:
14753:
14717:Reverse genetics
14692:Medical genetics
14407:
14400:
14393:
14384:
14383:
14240:
14238:
14227:
14226:
14217:
14207:
14205:
14200:
14153:
14131:
14120:. Random House.
14112:
14100:
14086:
14074:
14063:
14044:
14022:
14003:
13988:Olby RC (2009).
13984:
13974:
13957:(6921): 402–05.
13945:(January 2003).
13937:
13915:
13893:
13872:
13853:
13818:
13817:
13807:
13783:
13777:
13776:
13769:Nature Education
13764:
13758:
13757:
13746:
13740:
13739:
13729:
13719:
13687:
13681:
13680:
13678:
13671:
13660:
13654:
13653:
13651:
13628:
13611:(6921): 407–08.
13602:
13593:
13587:
13586:
13575:
13569:
13568:
13550:
13516:
13510:
13509:
13507:
13505:
13483:
13477:
13476:
13474:
13472:
13455:
13449:
13448:
13446:
13444:
13429:
13423:
13422:
13421:on 5 April 2015.
13417:. Archived from
13407:
13401:
13400:
13381:Regis E (2009).
13378:
13372:
13371:
13361:
13351:
13319:
13313:
13312:
13300:
13290:
13284:
13283:
13281:
13279:
13265:
13259:
13258:
13248:
13216:
13210:
13209:
13207:
13183:
13177:
13176:
13140:
13134:
13133:
13123:
13091:
13085:
13082:
13077:. Archived from
13057:
13056:on 14 July 2014.
13055:
13049:. Archived from
13032:
13020:
13014:
13013:
13007:
12998:
12992:
12991:
12979:
12973:
12972:
12962:
12930:
12924:
12923:
12913:
12881:
12875:
12874:
12855:10.1038/35088598
12838:
12832:
12829:
12797:(2075): 312–13.
12784:
12770:
12756:
12741:
12735:
12734:
12716:
12707:
12701:
12700:
12698:
12674:
12668:
12667:
12643:
12637:
12636:
12612:
12606:
12605:
12595:
12571:
12565:
12562:
12519:
12502:
12489:
12472:
12455:
12436:
12430:
12429:
12393:
12387:
12386:
12363:
12357:
12356:
12331:(5): 1479–1490.
12320:
12314:
12313:
12303:
12271:
12265:
12264:
12254:
12244:
12223:Wray GA (2002).
12220:
12214:
12213:
12211:
12209:
12182:
12176:
12175:
12131:
12125:
12124:
12088:
12082:
12081:
12063:
12026:
12020:
12019:
11977:
11968:
11962:
11961:
11943:
11933:
11909:
11903:
11902:
11876:
11870:
11869:
11851:
11831:
11825:
11824:
11802:
11796:
11795:
11766:
11760:
11759:
11749:
11739:
11707:
11701:
11700:
11690:
11666:
11660:
11659:
11649:
11617:
11611:
11610:
11582:
11573:
11572:
11570:
11568:
11540:
11534:
11533:
11522:
11516:
11515:
11513:
11511:
11496:
11490:
11489:
11471:
11469:10.1038/316076a0
11439:
11433:
11432:
11396:
11390:
11389:
11379:
11369:
11337:
11331:
11330:
11328:
11326:
11315:The Conversation
11306:
11300:
11299:
11271:
11265:
11264:
11254:
11222:
11216:
11215:
11181:
11175:
11174:
11138:
11132:
11131:
11113:
11079:
11073:
11072:
11070:
11068:
11051:
11045:
11044:
11042:
11040:
11018:
11012:
11011:
11009:
11007:
10985:
10979:
10978:
10976:
10974:
10954:
10948:
10947:
10937:
10927:
10910:(34): 13995–98.
10895:
10889:
10888:
10844:
10838:
10837:
10809:
10803:
10802:
10783:10.1038/35038060
10758:
10752:
10751:
10732:10.1038/362709a0
10718:(6422): 709–15.
10707:
10701:
10700:
10690:
10680:
10648:
10642:
10641:
10605:
10599:
10598:
10572:
10552:
10546:
10545:
10512:(6894): 214–21.
10501:
10495:
10494:
10476:
10467:(22): 5492–501.
10452:
10446:
10445:
10435:
10410:(7099): 153–58.
10395:
10389:
10388:
10371:(9–10): 587–92.
10360:
10354:
10353:
10317:
10311:
10310:
10274:
10268:
10267:
10248:10.1038/35066075
10231:
10225:
10224:
10222:
10220:
10205:
10199:
10198:
10162:
10156:
10155:
10137:
10113:
10104:
10103:
10075:
10069:
10068:
10066:
10060:. Archived from
10027:
10018:
10012:
10011:
10001:
9969:
9963:
9962:
9952:
9934:
9925:
9919:
9918:
9890:
9884:
9883:
9873:
9841:
9832:
9831:
9821:
9789:
9783:
9782:
9780:
9778:
9763:
9757:
9756:
9746:
9736:
9704:
9698:
9697:
9687:
9663:
9657:
9656:
9628:
9622:
9621:
9619:
9617:
9602:
9596:
9595:
9567:
9561:
9560:
9532:
9526:
9525:
9497:
9491:
9490:
9456:
9450:
9449:
9447:
9408:
9399:
9393:
9392:
9359:(6648): 251–60.
9348:
9342:
9341:
9323:
9299:
9293:
9292:
9274:
9242:
9236:
9235:
9228:
9222:
9221:
9211:
9201:
9169:
9163:
9162:
9152:
9120:
9114:
9113:
9103:
9093:
9061:
9055:
9054:
9026:
9020:
9019:
9009:
8977:
8971:
8970:
8960:
8950:
8941:(11): e1000213.
8926:
8920:
8919:
8909:
8877:
8871:
8870:
8834:
8828:
8827:
8811:
8801:
8795:
8794:
8784:
8774:
8750:
8744:
8743:
8715:
8709:
8708:
8698:
8666:
8660:
8659:
8649:
8632:(1455): 569–79.
8617:
8611:
8610:
8608:
8606:
8591:
8585:
8584:
8574:
8530:
8524:
8523:
8513:
8490:Annals of Botany
8481:
8475:
8474:
8464:
8462:10.1038/35057000
8447:(6822): 824–26.
8432:
8426:
8425:
8415:
8391:
8385:
8384:
8374:
8342:
8336:
8335:
8307:
8301:
8300:
8272:
8266:
8265:
8237:
8231:
8230:
8202:
8196:
8195:
8159:
8153:
8152:
8124:
8118:
8117:
8091:
8085:
8084:
8054:
8048:
8047:
8026:
8020:
8019:
8009:
8000:(37): 5792–812.
7985:
7979:
7978:
7968:
7959:(32): 19633–36.
7944:
7938:
7937:
7901:
7895:
7894:
7865:
7859:
7846:
7840:
7839:
7821:
7792:
7786:
7785:
7775:
7743:
7737:
7736:
7726:
7701:(5929): 929–30.
7686:
7680:
7679:
7645:
7639:
7638:
7628:
7604:
7598:
7597:
7569:
7563:
7562:
7552:
7542:
7518:
7512:
7511:
7509:
7507:
7496:
7490:
7489:
7487:
7485:
7473:
7467:
7466:
7432:
7408:
7402:
7401:
7391:
7349:
7343:
7342:
7332:
7300:
7294:
7293:
7283:
7251:
7245:
7244:
7218:
7198:
7192:
7191:
7158:(6891): 876–80.
7147:
7141:
7140:
7130:
7098:
7089:
7088:
7078:
7046:
7040:
7039:
7029:
7005:
6994:
6993:
6983:
6959:
6950:
6949:
6947:
6945:
6931:
6925:
6924:
6900:
6894:
6893:
6869:
6863:
6862:
6860:
6858:
6838:
6829:
6828:
6826:
6824:
6804:
6798:
6797:
6787:
6777:
6760:(26): 16666–71.
6745:
6739:
6738:
6702:
6696:
6695:
6667:
6661:
6660:
6632:
6626:
6625:
6623:
6621:
6616:on 14 March 2020
6615:
6609:. Archived from
6584:
6575:
6569:
6568:
6560:
6554:
6553:
6530:Bull. Math. Biol
6521:
6515:
6514:
6486:
6480:
6479:
6477:
6453:10.1038/171738a0
6439:(4356): 738–40.
6430:
6421:
6412:
6411:
6409:
6385:10.1038/171740a0
6371:(4356): 740–41.
6362:
6353:
6344:
6341:
6339:
6332:
6309:Acta Crystallogr
6306:
6296:
6294:
6287:
6264:Acta Crystallogr
6261:
6250:
6244:
6243:
6215:
6209:
6208:
6172:
6163:
6162:
6126:
6117:
6116:
6115:on 1 March 2019.
6114:
6108:. Archived from
6075:
6066:
6060:
6059:
6049:
6017:
6011:
6010:
6000:
5968:
5962:
5961:
5933:
5927:
5926:
5890:
5884:
5883:
5855:
5849:
5836:
5830:
5829:
5819:
5795:
5789:
5788:
5760:
5749:
5748:
5746:
5744:
5735:. Archived from
5729:
5723:
5722:
5703:10.1038/290457a0
5678:
5672:
5671:
5661:
5644:(7091): 315–21.
5629:
5623:
5622:
5612:
5602:
5578:
5569:
5568:
5566:
5535:
5526:
5520:
5519:
5483:
5477:
5476:
5466:
5456:
5424:
5418:
5417:
5407:
5382:(4): 1997–2007.
5367:
5361:
5360:
5350:
5318:
5312:
5311:
5283:
5274:
5273:
5254:10.1038/287755a0
5240:(5784): 755–58.
5229:
5223:
5222:
5194:
5188:
5187:
5177:
5159:
5135:
5129:
5128:
5110:
5095:Chemical Reviews
5086:
5080:
5079:
5051:
5045:
5044:
5026:
5002:
4996:
4995:
4993:
4991:
4975:
4969:
4968:
4950:
4944:
4943:
4933:
4893:
4887:
4886:
4884:
4882:
4867:
4861:
4860:
4827:(Pt 4): 620–26.
4816:
4807:
4806:
4801:. Archived from
4792:
4752:
4746:
4745:
4727:
4714:
4713:
4669:
4663:
4662:
4626:
4620:
4619:
4617:
4585:10.1038/171737a0
4571:(4356): 737–38.
4562:
4553:
4540:
4539:
4529:
4481:
4475:
4474:
4442:
4433:
4432:
4414:
4408:
4407:
4395:
4385:
4379:
4378:
4376:
4374:
4360:
4354:
4353:
4337:
4331:
4330:
4306:
4300:
4299:
4286:
4264:
4249:
4208:Genetic disorder
4149:Robert W. Holley
4108:
4027:genetic material
4009:. DNA's role in
3953:Chargaff's rules
3885:discovered that
3628:machine learning
3620:computer science
3473:medical research
3390:and gas clouds.
3209:intermediate in
3198:
3189:
3182:
3056:, predominantly
3016:replication fork
2897:helix-turn-helix
2605:gene duplication
2601:genetic material
2478:DNA condensation
2436:protein sequence
2339:ethidium bromide
2269:oxidizing agents
2166:5-methylcytosine
2115:5-methylcytosine
2104:
2097:
2090:
2083:
1984:Artificial bases
1944:
1937:
1930:
1823:shadow biosphere
1817:For many years,
1771:presented their
1761:Bessel functions
1753:in 1953 for the
1654:DNA supercoiling
1575:, there is also
1563:
1486:
1475:
1471:
1467:
1463:
1447:
1443:
1432:
1405:
1379:
1369:
1358:
1351:
1345:
1338:
1280:Deoxyarchaeosine
1238:5-Methylcytosine
1229:5-Formylcytosine
1157:5-methylcytosine
1138:
1130:
1126:
1114:
1110:
1091:
1087:
1079:
1071:
1063:
1055:
1030:animated version
633:ribonucleic acid
607:composed of two
598:
597:
596:
595:
588:
585:
584:
581:
578:
575:
570:
569:
566:
563:
560:
557:
554:
551:
548:
545:
542:
539:
536:
533:
530:
527:
524:
521:
518:
515:
512:
491:
484:
477:
464:
459:
458:
354:Medical genetics
350:
345:
344:
178:
159:
158:
104:
103:
93:
86:
77:
70:
63:
54:
45:
34:
15741:
15740:
15736:
15735:
15734:
15732:
15731:
15730:
15701:
15700:
15699:
15689:
15684:
15682:
15672:
15667:
15665:
15655:
15650:
15648:
15638:
15633:
15631:
15628:
15624:sister projects
15621:at Knowledge's
15615:
15603:
15595:
15593:
15588:
15570:
15511:Cloning vectors
15505:
15491:Locked
15455:
15405:
15394:
15353:
15281:
15228:
15227:
15219:
15188:
15182:
15152:
15147:
15126:
15061:Transcriptional
15031:
15000:
14961:
14952:Polyadenylation
14923:
14897:
14862:
14856:Protein→Protein
14807:
14800:
14798:Gene expression
14795:
14765:
14760:
14742:
14721:
14620:
14611:the Middle East
14577:Archaeogenetics
14571:
14515:
14454:
14416:
14411:
14254:
14253:
14242:
14236:
14234:
14231:This audio file
14228:
14221:
14212:
14209:
14203:
14202:
14198:
14195:
14194:
14193:
14173:
14172:
14168:
14161:
14156:
14150:
14128:
14109:
14083:
14060:
14041:
14019:
14000:
13934:
13912:
13890:
13879:50 years of DNA
13869:
13850:
13827:
13825:Further reading
13822:
13821:
13784:
13780:
13765:
13761:
13748:
13747:
13743:
13688:
13684:
13676:
13669:
13661:
13657:
13649:
13600:
13594:
13590:
13577:
13576:
13572:
13517:
13513:
13503:
13501:
13484:
13480:
13470:
13468:
13456:
13452:
13442:
13440:
13431:
13430:
13426:
13415:Nature Archives
13409:
13408:
13404:
13397:
13379:
13375:
13320:
13316:
13309:
13291:
13287:
13277:
13275:
13267:
13266:
13262:
13217:
13213:
13198:(24): 172–174.
13184:
13180:
13141:
13137:
13092:
13088:
13081:on 5 July 2014.
13053:
13030:
13021:
13017:
13005:
12999:
12995:
12980:
12976:
12931:
12927:
12882:
12878:
12839:
12835:
12742:
12738:
12714:
12708:
12704:
12675:
12671:
12644:
12640:
12613:
12609:
12572:
12568:
12531:Wayback Machine
12437:
12433:
12394:
12390:
12364:
12360:
12321:
12317:
12272:
12268:
12221:
12217:
12207:
12205:
12183:
12179:
12132:
12128:
12089:
12085:
12038:(7243): 73–76.
12027:
12023:
11975:
11969:
11965:
11910:
11906:
11891:
11877:
11873:
11832:
11828:
11821:
11803:
11799:
11784:
11767:
11763:
11722:(19): 5903–08.
11708:
11704:
11681:(12): 1039–46.
11667:
11663:
11618:
11614:
11583:
11576:
11566:
11564:
11541:
11537:
11524:
11523:
11519:
11509:
11507:
11498:
11497:
11493:
11454:(6023): 76–79.
11440:
11436:
11397:
11393:
11352:(13): 6007–11.
11338:
11334:
11324:
11322:
11307:
11303:
11282:(11): 1105–10.
11272:
11268:
11223:
11219:
11204:
11182:
11178:
11139:
11135:
11080:
11076:
11066:
11064:
11052:
11048:
11038:
11036:
11019:
11015:
11005:
11003:
10986:
10982:
10972:
10970:
10955:
10951:
10896:
10892:
10845:
10841:
10810:
10806:
10759:
10755:
10708:
10704:
10649:
10645:
10606:
10602:
10570:10.1.1.537.7679
10553:
10549:
10526:10.1038/418214a
10502:
10498:
10453:
10449:
10396:
10392:
10361:
10357:
10334:10.1038/nrg1746
10318:
10314:
10291:10.1038/nrg1838
10275:
10271:
10232:
10228:
10218:
10216:
10206:
10202:
10163:
10159:
10114:
10107:
10076:
10072:
10064:
10025:
10019:
10015:
9984:(22): 6321–29.
9970:
9966:
9943:(10): 1849–63.
9932:
9926:
9922:
9891:
9887:
9856:(21): 4051–58.
9842:
9835:
9790:
9786:
9776:
9774:
9764:
9760:
9719:(14): 8164–69.
9705:
9701:
9664:
9660:
9629:
9625:
9615:
9613:
9603:
9599:
9568:
9564:
9533:
9529:
9498:
9494:
9479:
9457:
9453:
9445:
9406:
9400:
9396:
9349:
9345:
9300:
9296:
9257:(12): 1350–64.
9243:
9239:
9230:
9229:
9225:
9170:
9166:
9121:
9117:
9062:
9058:
9027:
9023:
8978:
8974:
8927:
8923:
8892:(21): 6288–93.
8878:
8874:
8835:
8831:
8824:
8802:
8798:
8753:Albà M (2001).
8751:
8747:
8716:
8712:
8675:Genome Research
8667:
8663:
8618:
8614:
8604:
8602:
8592:
8588:
8531:
8527:
8482:
8478:
8433:
8429:
8392:
8388:
8343:
8339:
8318:(17): 1745–80.
8308:
8304:
8273:
8269:
8248:(12): 1489–99.
8238:
8234:
8203:
8199:
8160:
8156:
8135:(15): 1475–85.
8125:
8121:
8110:
8092:
8088:
8073:
8055:
8051:
8027:
8023:
7986:
7982:
7945:
7941:
7902:
7898:
7877:(30): 9221–26.
7866:
7862:
7857:Wayback Machine
7847:
7843:
7793:
7789:
7744:
7740:
7687:
7683:
7668:
7646:
7642:
7605:
7601:
7570:
7566:
7519:
7515:
7505:
7503:
7498:
7497:
7493:
7483:
7481:
7478:"Nucleic Acids"
7474:
7470:
7430:physics/0609135
7409:
7405:
7350:
7346:
7301:
7297:
7252:
7248:
7216:10.1.1.335.2649
7199:
7195:
7148:
7144:
7113:(19): 5402–15.
7099:
7092:
7061:(21): 2801–09.
7047:
7043:
7006:
6997:
6960:
6953:
6943:
6941:
6933:
6932:
6928:
6901:
6897:
6870:
6866:
6856:
6854:
6839:
6832:
6822:
6820:
6805:
6801:
6746:
6742:
6703:
6699:
6668:
6664:
6633:
6629:
6619:
6617:
6613:
6582:
6576:
6572:
6561:
6557:
6522:
6518:
6487:
6483:
6475:
6428:
6422:
6415:
6407:
6360:
6354:
6347:
6337:
6315:(8–9): 678–85.
6304:
6292:
6270:(8–9): 673–77.
6259:
6251:
6247:
6216:
6212:
6173:
6166:
6127:
6120:
6112:
6073:
6071:"DNA mechanics"
6067:
6063:
6018:
6014:
5983:(11): 2268–72.
5977:Genome Research
5969:
5965:
5934:
5930:
5891:
5887:
5856:
5852:
5846:Wayback Machine
5837:
5833:
5796:
5792:
5761:
5752:
5742:
5740:
5731:
5730:
5726:
5679:
5675:
5630:
5626:
5579:
5572:
5564:
5533:
5527:
5523:
5484:
5480:
5439:(14): 7853–58.
5425:
5421:
5368:
5364:
5319:
5315:
5284:
5277:
5230:
5226:
5195:
5191:
5136:
5132:
5087:
5083:
5052:
5048:
5003:
4999:
4989:
4987:
4978:Carr S (1953).
4976:
4972:
4965:
4951:
4947:
4894:
4890:
4880:
4878:
4868:
4864:
4817:
4810:
4753:
4749:
4742:
4728:
4717:
4670:
4666:
4627:
4623:
4615:
4560:
4554:
4543:
4482:
4478:
4459:
4443:
4436:
4429:
4415:
4411:
4404:
4386:
4382:
4372:
4370:
4362:
4361:
4357:
4338:
4334:
4323:
4307:
4303:
4288:
4287:
4283:
4278:
4273:
4262:
4247:
4184:Crystallography
4173:
4106:
4054:Maurice Wilkins
4038:Raymond Gosling
3922:William Astbury
3874:Nikolai Koltsov
3858:Albrecht Kossel
3842:Raymond Gosling
3819:
3811:
3795:storage density
3787:
3781:
3752:
3742:
3679:
3673:
3632:database theory
3605:
3599:
3580:
3574:
3550:double jeopardy
3542:Enderby murders
3496:can use DNA in
3491:
3485:
3449:Recombinant DNA
3425:
3411:
3406:
3311:. This ancient
3301:cell metabolism
3297:history of life
3293:
3287:
3216:
3215:
3214:
3179:
3173:
3165:protein complex
3097:DNA polymerases
3066:
3050:molecular motor
3032:
3006:Enzymes called
2935:
2930:
2861:phosphorylation
2824:
2818:
2805:
2793:
2787:
2752:
2732:DNA replication
2688:
2686:DNA replication
2682:
2631:
2617:
2474:
2452:
2444:DNA replication
2383:
2302:point mutations
2226:
2212:
2135:
2134:
2133:
2080:
2074:DNA methylation
2070:
2065:
2049:
2025:phosphoric acid
2017:
1992:
1986:
1965:
1964:
1963:
1927:
1917:
1905:triple-stranded
1852:
1846:
1815:
1809:
1692:
1678:
1670:DNA replication
1650:
1644:
1617:gene expression
1604:
1598:
1561:
1560:, and would be
1536:
1504:
1495:
1484:
1473:
1469:
1465:
1461:
1457:
1445:
1441:
1430:
1426:
1424:ssDNA vs. dsDNA
1403:
1393:. Purines form
1384:
1383:
1382:
1377:
1367:
1335:
1329:
1292:
1212:7-Methylguanine
1198:N6-methyadenine
1153:
1136:
1128:
1124:
1112:
1108:
1101:
1089:
1085:
1077:
1069:
1061:
1053:
995:glycosidic bond
877:
843:. In contrast,
841:chloroplast DNA
776:(U). Under the
703:phosphate group
649:polysaccharides
591:
590:
572:
509:
505:
495:
454:
447:
446:
437:
429:
428:
427:
426:
375:
367:
366:
358:
336:
317:
309:
308:
264:
256:
255:
242:
241:
240:
186:
117:
116:
115:
105:
97:
96:
95:
94:
89:
87:
80:
78:
73:
71:
66:
64:
57:
55:
50:
46:
30:
17:
12:
11:
5:
15739:
15729:
15728:
15723:
15718:
15713:
15698:
15697:
15680:
15663:
15661:from Wikiquote
15646:
15617:
15614:
15613:
15590:
15589:
15587:
15586:
15575:
15572:
15571:
15569:
15568:
15567:
15566:
15561:
15556:
15551:
15541:
15536:
15531:
15526:
15521:
15515:
15513:
15507:
15506:
15504:
15503:
15498:
15496:Peptide
15493:
15488:
15487:
15486:
15481:
15476:
15474:Glycol
15465:
15463:
15457:
15456:
15454:
15453:
15448:
15443:
15438:
15433:
15428:
15427:
15426:
15421:
15410:
15408:
15400:
15399:
15396:
15395:
15393:
15392:
15387:
15382:
15377:
15372:
15367:
15361:
15359:
15355:
15354:
15352:
15351:
15346:
15345:
15344:
15339:
15334:
15329:
15319:
15314:
15313:
15312:
15307:
15302:
15291:
15289:
15283:
15282:
15280:
15279:
15274:
15269:
15264:
15259:
15258:
15257:
15246:
15244:
15235:
15221:
15220:
15218:
15217:
15212:
15207:
15202:
15196:
15194:
15190:
15189:
15186:nucleic acids
15181:
15180:
15173:
15166:
15158:
15149:
15148:
15146:
15145:
15140:
15138:François Jacob
15134:
15132:
15128:
15127:
15125:
15124:
15123:
15122:
15117:
15107:
15102:
15101:
15100:
15095:
15090:
15080:
15075:
15074:
15073:
15068:
15058:
15057:
15056:
15045:
15043:
15037:
15036:
15033:
15032:
15030:
15029:
15024:
15019:
15014:
15008:
15006:
15002:
15001:
14999:
14998:
14993:
14988:
14982:
14980:
14973:
14967:
14966:
14963:
14962:
14960:
14959:
14954:
14949:
14944:
14939:
14933:
14931:
14925:
14924:
14922:
14921:
14916:
14914:RNA polymerase
14911:
14905:
14903:
14899:
14898:
14896:
14895:
14890:
14885:
14879:
14877:
14870:
14864:
14863:
14861:
14860:
14859:
14858:
14853:
14848:
14838:
14837:
14836:
14818:
14812:
14810:
14802:
14801:
14794:
14793:
14786:
14779:
14771:
14762:
14761:
14759:
14758:
14747:
14744:
14743:
14741:
14740:
14735:
14729:
14727:
14723:
14722:
14720:
14719:
14714:
14709:
14707:Plant genetics
14704:
14699:
14694:
14689:
14684:
14679:
14674:
14669:
14664:
14659:
14654:
14649:
14647:Genome editing
14644:
14639:
14634:
14628:
14626:
14625:Related topics
14622:
14621:
14619:
14618:
14613:
14608:
14603:
14598:
14593:
14588:
14582:
14580:
14573:
14572:
14570:
14569:
14564:
14559:
14554:
14549:
14547:Immunogenetics
14544:
14539:
14534:
14529:
14523:
14521:
14517:
14516:
14514:
14513:
14508:
14503:
14498:
14493:
14488:
14483:
14478:
14473:
14468:
14462:
14460:
14459:Key components
14456:
14455:
14453:
14452:
14447:
14442:
14437:
14432:
14427:
14421:
14418:
14417:
14410:
14409:
14402:
14395:
14387:
14381:
14380:
14370:
14361:
14355:
14343:
14332:
14322:
14316:
14307:
14297:
14287:
14276:
14271:
14266:
14260:
14243:
14229:
14222:
14210:
14197:
14196:
14192:
14191:
14186:
14181:
14175:
14174:
14163:
14162:
14160:
14159:External links
14157:
14155:
14154:
14148:
14132:
14126:
14113:
14107:
14087:
14081:
14064:
14058:
14045:
14039:
14023:
14017:
14004:
13998:
13985:
13939:
13932:
13916:
13910:
13894:
13888:
13873:
13867:
13854:
13848:
13828:
13826:
13823:
13820:
13819:
13778:
13759:
13754:Nobelprize.org
13741:
13682:
13655:
13588:
13583:Nobelprize.org
13570:
13511:
13478:
13450:
13424:
13402:
13395:
13389:. p. 52.
13373:
13314:
13307:
13285:
13260:
13211:
13178:
13151:(6): 201–209.
13135:
13106:(2): 137–158.
13086:
13084:
13083:
13058:
13015:
12993:
12986:(in Italian).
12974:
12945:(3): 563–602.
12925:
12876:
12833:
12831:
12830:
12785:
12771:
12765:(in Russian).
12759:Reprinted in:
12757:
12736:
12702:
12669:
12658:(2): 2474–78.
12638:
12607:
12566:
12564:
12563:
12542:
12520:
12503:
12490:
12473:
12456:
12431:
12398:Human Genetics
12388:
12358:
12315:
12266:
12229:Genome Biology
12215:
12177:
12126:
12083:
12021:
11963:
11904:
11889:
11871:
11849:10.1.1.412.943
11836:Bioinformatics
11826:
11819:
11797:
11782:
11761:
11702:
11661:
11632:(10): 718–20.
11612:
11574:
11535:
11517:
11491:
11434:
11391:
11332:
11301:
11266:
11217:
11202:
11176:
11133:
11074:
11046:
11013:
10980:
10949:
10890:
10839:
10804:
10753:
10702:
10663:(7): 2614–18.
10643:
10600:
10547:
10496:
10447:
10390:
10355:
10312:
10269:
10242:(4): 292–301.
10226:
10200:
10157:
10128:(8): 497–503.
10105:
10070:
10013:
9964:
9920:
9885:
9833:
9784:
9758:
9699:
9658:
9623:
9597:
9562:
9527:
9492:
9477:
9451:
9394:
9343:
9294:
9237:
9223:
9164:
9115:
9076:(12): e51905.
9056:
9037:(5559): 1487.
9021:
8972:
8935:PLOS Pathogens
8921:
8872:
8829:
8822:
8796:
8759:Genome Biology
8745:
8726:(5): 1155–74.
8710:
8661:
8612:
8586:
8525:
8476:
8427:
8386:
8337:
8302:
8267:
8232:
8197:
8170:(1–2): 12–22.
8154:
8119:
8108:
8086:
8071:
8049:
8021:
7980:
7939:
7896:
7860:
7841:
7804:(6): 1129–36.
7787:
7738:
7681:
7666:
7640:
7599:
7564:
7513:
7491:
7468:
7403:
7344:
7295:
7246:
7193:
7142:
7090:
7041:
7020:(8): 1073–85.
6995:
6951:
6926:
6895:
6864:
6830:
6799:
6740:
6697:
6662:
6627:
6570:
6555:
6516:
6481:
6413:
6345:
6343:
6342:
6297:
6245:
6226:(2): 299–309.
6210:
6189:10.1038/nrm831
6164:
6118:
6061:
6012:
5963:
5928:
5885:
5850:
5831:
5790:
5771:(1): 137–140.
5750:
5724:
5673:
5624:
5570:
5521:
5478:
5419:
5362:
5333:(12): 955–68.
5313:
5275:
5224:
5189:
5130:
5081:
5046:
4997:
4970:
4963:
4945:
4888:
4862:
4808:
4747:
4740:
4715:
4680:(3): 367–376.
4664:
4621:
4541:
4476:
4457:
4434:
4427:
4409:
4402:
4380:
4355:
4332:
4321:
4301:
4280:
4279:
4277:
4274:
4272:
4271:
4265:
4256:
4250:
4241:
4235:
4229:
4223:
4217:
4211:
4205:
4202:DNA sequencing
4199:
4196:DNA microarray
4193:
4187:
4181:
4174:
4172:
4169:
4015:Alfred Hershey
3993:Late in 1951,
3949:Erwin Chargaff
3937:Maclyn McCarty
3869:Phoebus Levene
3826:Maclyn McCarty
3810:
3807:
3799:memory latency
3791:storage device
3783:Main article:
3780:
3777:
3741:
3738:
3672:
3669:
3608:Bioinformatics
3603:Bioinformatics
3598:
3597:Bioinformatics
3595:
3583:Deoxyribozymes
3573:
3570:
3566:DNA sequencing
3534:minisatellites
3524:, also called
3484:
3481:
3410:
3407:
3405:
3402:
3398:woolly mammoth
3331:, and related
3286:
3283:
3204:
3203:
3200:
3199:
3191:
3190:
3180:
3172:
3169:
3148:RNA polymerase
3065:
3062:
3035:Topoisomerases
3031:
3028:
3012:lagging strand
2934:
2931:
2929:
2926:
2817:
2814:
2804:
2801:
2789:Main article:
2786:
2783:
2751:
2748:
2744:DNA polymerase
2712:lagging strand
2708:leading strand
2704:DNA polymerase
2681:
2678:
2662:RNA polymerase
2616:
2613:
2559:non-coding RNA
2555:C-value enigma
2451:
2448:
2382:
2379:
2289:thymine dimers
2211:
2208:
2204:kinetoplastids
2170:X-inactivation
2127:
2126:
2123:
2122:
2117:
2112:
2106:
2105:
2098:
2091:
2081:
2069:
2066:
2064:
2061:
2048:
2045:
2023:properties to
2016:
2013:
1988:Main article:
1985:
1982:
1974:nanotechnology
1958:
1957:
1954:
1953:
1950:
1949:Single branch
1946:
1945:
1938:
1928:
1916:
1913:
1888:guanine tetrad
1845:
1842:
1808:
1805:
1677:
1674:
1662:topoisomerases
1643:
1640:
1597:
1594:
1545:nuclear genome
1520:sex chromosome
1503:
1500:
1493:
1455:
1425:
1422:
1395:hydrogen bonds
1373:hydrogen bonds
1364:
1363:
1360:
1359:
1347:
1346:
1336:
1328:
1325:
1291:
1288:
1287:
1286:
1285:
1284:
1281:
1275:
1274:
1273:
1270:
1267:
1264:
1255:
1254:
1253:
1250:
1241:
1240:
1239:
1236:
1233:
1230:
1227:
1224:
1215:
1214:
1213:
1210:
1209:7-Deazaguanine
1201:
1200:
1199:
1196:
1171:bacteriophages
1152:
1149:
1100:
1097:
1038:hydrogen bonds
1003:directionality
952:polynucleotide
909:hydrogen bonds
899:DNA is a long
885:hydrogen bonds
876:
873:
723:hydrogen bonds
709:(known as the
707:covalent bonds
653:macromolecules
609:polynucleotide
497:
496:
494:
493:
486:
479:
471:
468:
467:
466:
465:
449:
448:
445:
444:
438:
435:
434:
431:
430:
425:
424:
419:
414:
409:
404:
402:Immunogenetics
399:
394:
389:
384:
378:
377:
376:
373:
372:
369:
368:
365:
364:
357:
356:
351:
334:
329:
327:DNA sequencing
324:
318:
315:
314:
311:
310:
307:
306:
301:
296:
291:
286:
276:
271:
265:
262:
261:
258:
257:
254:
253:
248:
239:
238:
233:
228:
223:
218:
213:
208:
203:
198:
195:
189:
188:
187:
185:Key components
184:
183:
180:
179:
171:
170:
164:
163:
119:
118:
99:
98:
48:
47:
40:
39:
38:
37:
15:
9:
6:
4:
3:
2:
15738:
15727:
15726:Nucleic acids
15724:
15722:
15719:
15717:
15716:Biotechnology
15714:
15712:
15709:
15708:
15706:
15695:from Wikidata
15694:
15693:
15681:
15677:
15676:
15664:
15660:
15659:
15647:
15643:
15642:
15630:
15629:
15626:
15620:
15612:
15607:
15602:
15601:
15598:
15585:
15577:
15576:
15573:
15565:
15562:
15560:
15557:
15555:
15552:
15550:
15547:
15546:
15545:
15542:
15540:
15537:
15535:
15532:
15530:
15527:
15525:
15522:
15520:
15517:
15516:
15514:
15512:
15508:
15502:
15499:
15497:
15494:
15492:
15489:
15485:
15482:
15480:
15479:Threose
15477:
15475:
15472:
15471:
15470:
15467:
15466:
15464:
15462:
15458:
15452:
15449:
15447:
15444:
15442:
15439:
15437:
15436:Deoxyribozyme
15434:
15432:
15429:
15425:
15424:Mitochondrial
15422:
15420:
15417:
15416:
15415:
15412:
15411:
15409:
15407:
15401:
15391:
15388:
15386:
15383:
15381:
15378:
15376:
15373:
15371:
15368:
15366:
15363:
15362:
15360:
15356:
15350:
15347:
15343:
15340:
15338:
15335:
15333:
15330:
15328:
15325:
15324:
15323:
15320:
15318:
15315:
15311:
15308:
15306:
15303:
15301:
15298:
15297:
15296:
15293:
15292:
15290:
15288:
15284:
15278:
15275:
15273:
15270:
15268:
15265:
15263:
15260:
15256:
15253:
15252:
15251:
15248:
15247:
15245:
15243:
15242:Translational
15239:
15236:
15232:
15226:
15222:
15216:
15213:
15211:
15208:
15206:
15203:
15201:
15198:
15197:
15195:
15191:
15187:
15179:
15174:
15172:
15167:
15165:
15160:
15159:
15156:
15144:
15143:Jacques Monod
15141:
15139:
15136:
15135:
15133:
15129:
15121:
15118:
15116:
15113:
15112:
15111:
15108:
15106:
15105:Translational
15103:
15099:
15096:
15094:
15091:
15089:
15086:
15085:
15084:
15081:
15079:
15076:
15072:
15069:
15067:
15064:
15063:
15062:
15059:
15055:
15052:
15051:
15050:
15047:
15046:
15044:
15042:
15038:
15028:
15025:
15023:
15020:
15018:
15015:
15013:
15010:
15009:
15007:
15003:
14997:
14994:
14992:
14989:
14987:
14984:
14983:
14981:
14977:
14974:
14972:
14968:
14958:
14955:
14953:
14950:
14948:
14945:
14943:
14940:
14938:
14935:
14934:
14932:
14930:
14926:
14920:
14917:
14915:
14912:
14910:
14907:
14906:
14904:
14900:
14894:
14891:
14889:
14886:
14884:
14881:
14880:
14878:
14874:
14871:
14869:
14868:Transcription
14865:
14857:
14854:
14852:
14849:
14847:
14844:
14843:
14842:
14839:
14835:
14831:
14827:
14824:
14823:
14822:
14821:Central dogma
14819:
14817:
14814:
14813:
14811:
14809:
14803:
14799:
14792:
14787:
14785:
14780:
14778:
14773:
14772:
14769:
14757:
14749:
14748:
14745:
14739:
14736:
14734:
14731:
14730:
14728:
14724:
14718:
14715:
14713:
14710:
14708:
14705:
14703:
14700:
14698:
14695:
14693:
14690:
14688:
14685:
14683:
14680:
14678:
14675:
14673:
14670:
14668:
14665:
14663:
14660:
14658:
14655:
14653:
14650:
14648:
14645:
14643:
14640:
14638:
14635:
14633:
14630:
14629:
14627:
14623:
14617:
14614:
14612:
14609:
14607:
14604:
14602:
14599:
14597:
14594:
14592:
14589:
14587:
14584:
14583:
14581:
14578:
14574:
14568:
14565:
14563:
14560:
14558:
14555:
14553:
14550:
14548:
14545:
14543:
14540:
14538:
14535:
14533:
14530:
14528:
14525:
14524:
14522:
14518:
14512:
14509:
14507:
14504:
14502:
14499:
14497:
14494:
14492:
14489:
14487:
14484:
14482:
14479:
14477:
14474:
14472:
14469:
14467:
14464:
14463:
14461:
14457:
14451:
14448:
14446:
14443:
14441:
14438:
14436:
14433:
14431:
14428:
14426:
14423:
14422:
14419:
14415:
14408:
14403:
14401:
14396:
14394:
14389:
14388:
14385:
14378:
14374:
14371:
14369:
14365:
14362:
14359:
14356:
14353:
14352:
14347:
14344:
14342:
14341:
14336:
14333:
14330:
14326:
14323:
14320:
14317:
14315:
14311:
14308:
14306:
14305:
14302:
14298:
14296:
14295:
14292:
14288:
14286:
14285:
14280:
14277:
14275:
14272:
14270:
14267:
14264:
14261:
14259:
14256:
14255:
14251:
14247:
14232:
14190:
14187:
14185:
14182:
14180:
14177:
14176:
14171:
14166:
14151:
14149:0-19-860665-6
14145:
14141:
14137:
14133:
14129:
14123:
14119:
14114:
14110:
14108:0-393-95075-1
14104:
14099:
14098:
14092:
14088:
14084:
14078:
14073:
14072:
14065:
14061:
14055:
14051:
14046:
14042:
14040:0-06-082333-X
14036:
14032:
14028:
14024:
14020:
14014:
14010:
14005:
14001:
13995:
13991:
13986:
13982:
13978:
13973:
13968:
13964:
13960:
13956:
13952:
13948:
13944:
13940:
13935:
13933:0-486-68117-3
13929:
13925:
13921:
13917:
13913:
13911:0-671-22540-5
13907:
13903:
13899:
13895:
13891:
13889:1-4039-1479-6
13885:
13881:
13880:
13874:
13870:
13868:0-12-155089-3
13864:
13860:
13855:
13851:
13849:0-375-41546-7
13845:
13841:
13840:
13835:
13830:
13829:
13815:
13811:
13806:
13801:
13797:
13793:
13789:
13782:
13774:
13770:
13763:
13755:
13751:
13745:
13737:
13733:
13728:
13723:
13718:
13713:
13709:
13705:
13702:(7): 671–82.
13701:
13697:
13693:
13686:
13675:
13668:
13667:
13659:
13648:
13644:
13640:
13636:
13632:
13627:
13622:
13618:
13614:
13610:
13606:
13599:
13592:
13584:
13580:
13574:
13566:
13562:
13558:
13554:
13549:
13544:
13540:
13536:
13532:
13528:
13527:
13522:
13515:
13499:
13495:
13494:
13489:
13482:
13467:
13466:
13461:
13454:
13438:
13434:
13428:
13420:
13416:
13412:
13406:
13398:
13392:
13388:
13384:
13377:
13369:
13365:
13360:
13355:
13350:
13345:
13341:
13337:
13333:
13329:
13325:
13318:
13310:
13304:
13299:
13298:
13289:
13274:
13270:
13264:
13256:
13252:
13247:
13242:
13238:
13234:
13230:
13226:
13222:
13215:
13206:
13201:
13197:
13193:
13189:
13182:
13174:
13170:
13166:
13162:
13158:
13154:
13150:
13146:
13139:
13131:
13127:
13122:
13117:
13113:
13109:
13105:
13101:
13097:
13090:
13080:
13076:
13072:
13068:
13064:
13059:
13052:
13048:
13044:
13040:
13036:
13029:
13024:
13023:
13019:
13011:
13004:
12997:
12989:
12985:
12978:
12970:
12966:
12961:
12956:
12952:
12948:
12944:
12940:
12936:
12929:
12921:
12917:
12912:
12907:
12903:
12899:
12896:(2): 113–59.
12895:
12891:
12887:
12880:
12872:
12868:
12864:
12860:
12856:
12852:
12849:(9): 723–29.
12848:
12844:
12837:
12828:
12826:
12820:
12816:
12812:
12808:
12804:
12800:
12796:
12792:
12786:
12782:
12779:(in German).
12778:
12772:
12768:
12764:
12758:
12754:
12750:
12745:
12744:
12740:
12732:
12728:
12724:
12720:
12713:
12706:
12697:
12692:
12689:(2): 415–24.
12688:
12684:
12680:
12673:
12665:
12661:
12657:
12654:(in German).
12653:
12649:
12642:
12634:
12630:
12626:
12623:(in German).
12622:
12618:
12611:
12603:
12599:
12594:
12589:
12585:
12581:
12577:
12570:
12561:
12556:
12553:(in German).
12552:
12548:
12543:
12540:
12536:
12532:
12528:
12525:
12521:
12517:
12513:
12509:
12504:
12500:
12496:
12491:
12487:
12484:(in German).
12483:
12479:
12474:
12470:
12467:(in German).
12466:
12462:
12457:
12453:
12450:(in German).
12449:
12445:
12440:
12439:
12435:
12427:
12423:
12419:
12415:
12411:
12407:
12404:(6): 565–81.
12403:
12399:
12392:
12385:
12383:
12377:
12374:(in German).
12373:
12369:
12362:
12354:
12350:
12346:
12342:
12338:
12334:
12330:
12326:
12319:
12311:
12307:
12302:
12297:
12293:
12289:
12285:
12281:
12277:
12270:
12262:
12258:
12253:
12248:
12243:
12238:
12234:
12230:
12226:
12219:
12204:
12200:
12196:
12192:
12188:
12181:
12173:
12169:
12165:
12161:
12157:
12153:
12149:
12145:
12141:
12137:
12130:
12122:
12118:
12114:
12110:
12106:
12102:
12099:(5): 281–82.
12098:
12094:
12087:
12079:
12075:
12071:
12067:
12062:
12057:
12053:
12049:
12045:
12041:
12037:
12033:
12025:
12017:
12013:
12009:
12005:
12001:
11997:
11993:
11989:
11985:
11981:
11974:
11967:
11959:
11955:
11951:
11947:
11942:
11937:
11932:
11927:
11923:
11919:
11915:
11908:
11900:
11896:
11892:
11890:0-87969-712-1
11886:
11882:
11875:
11867:
11863:
11859:
11855:
11850:
11845:
11842:(2): 170–79.
11841:
11837:
11830:
11822:
11816:
11812:
11808:
11801:
11793:
11789:
11785:
11779:
11776:. MIT Press.
11775:
11771:
11765:
11757:
11753:
11748:
11743:
11738:
11733:
11729:
11725:
11721:
11717:
11713:
11706:
11698:
11694:
11689:
11684:
11680:
11676:
11672:
11665:
11657:
11653:
11648:
11643:
11639:
11635:
11631:
11627:
11623:
11616:
11608:
11604:
11600:
11596:
11593:(4): 223–29.
11592:
11588:
11581:
11579:
11562:
11558:
11554:
11550:
11546:
11539:
11531:
11527:
11521:
11505:
11501:
11495:
11487:
11483:
11479:
11475:
11470:
11465:
11461:
11457:
11453:
11449:
11445:
11438:
11430:
11426:
11422:
11418:
11414:
11410:
11407:(2): 213–22.
11406:
11402:
11395:
11387:
11383:
11378:
11373:
11368:
11363:
11359:
11355:
11351:
11347:
11343:
11336:
11320:
11316:
11312:
11305:
11297:
11293:
11289:
11285:
11281:
11277:
11270:
11262:
11258:
11253:
11248:
11244:
11240:
11237:(2): 136–41.
11236:
11232:
11228:
11221:
11213:
11209:
11205:
11199:
11195:
11191:
11187:
11180:
11172:
11168:
11164:
11160:
11156:
11152:
11148:
11144:
11137:
11129:
11125:
11121:
11117:
11112:
11107:
11103:
11099:
11095:
11091:
11090:
11085:
11078:
11063:
11062:
11057:
11050:
11034:
11030:
11029:
11024:
11017:
11001:
10997:
10996:
10991:
10984:
10968:
10964:
10960:
10953:
10945:
10941:
10936:
10931:
10926:
10921:
10917:
10913:
10909:
10905:
10901:
10894:
10886:
10882:
10878:
10874:
10870:
10866:
10862:
10858:
10855:(1): 134–37.
10854:
10850:
10843:
10835:
10831:
10827:
10823:
10820:(5): 212–20.
10819:
10815:
10808:
10800:
10796:
10792:
10788:
10784:
10780:
10776:
10772:
10768:
10764:
10757:
10749:
10745:
10741:
10737:
10733:
10729:
10725:
10721:
10717:
10713:
10706:
10698:
10694:
10689:
10684:
10679:
10674:
10670:
10666:
10662:
10658:
10654:
10647:
10639:
10635:
10631:
10627:
10623:
10619:
10615:
10611:
10604:
10596:
10592:
10588:
10584:
10580:
10576:
10571:
10566:
10563:(2): 99–123.
10562:
10558:
10551:
10543:
10539:
10535:
10531:
10527:
10523:
10519:
10515:
10511:
10507:
10500:
10492:
10488:
10484:
10480:
10475:
10470:
10466:
10462:
10458:
10451:
10443:
10439:
10434:
10429:
10425:
10421:
10417:
10413:
10409:
10405:
10401:
10394:
10386:
10382:
10378:
10374:
10370:
10366:
10359:
10351:
10347:
10343:
10339:
10335:
10331:
10327:
10323:
10316:
10308:
10304:
10300:
10296:
10292:
10288:
10285:(5): 337–48.
10284:
10280:
10273:
10265:
10261:
10257:
10253:
10249:
10245:
10241:
10237:
10230:
10215:
10211:
10204:
10196:
10192:
10188:
10184:
10180:
10176:
10173:(6): 925–47.
10172:
10168:
10161:
10153:
10149:
10145:
10141:
10136:
10131:
10127:
10123:
10122:FASEB Journal
10119:
10112:
10110:
10101:
10097:
10093:
10089:
10085:
10081:
10074:
10063:
10059:
10055:
10051:
10047:
10043:
10039:
10035:
10031:
10024:
10017:
10009:
10005:
10000:
9995:
9991:
9987:
9983:
9979:
9975:
9968:
9960:
9956:
9951:
9946:
9942:
9938:
9931:
9924:
9916:
9912:
9908:
9904:
9900:
9896:
9889:
9881:
9877:
9872:
9867:
9863:
9859:
9855:
9851:
9847:
9840:
9838:
9829:
9825:
9820:
9815:
9811:
9807:
9804:(2): 434–50.
9803:
9799:
9795:
9788:
9773:
9769:
9762:
9754:
9750:
9745:
9740:
9735:
9730:
9726:
9722:
9718:
9714:
9710:
9703:
9695:
9691:
9686:
9681:
9678:(2): 157–67.
9677:
9673:
9669:
9662:
9654:
9650:
9646:
9642:
9638:
9634:
9627:
9612:
9608:
9601:
9593:
9589:
9585:
9581:
9578:(3): 141–80.
9577:
9573:
9566:
9558:
9554:
9550:
9546:
9543:(3): 94–100.
9542:
9538:
9531:
9523:
9519:
9515:
9511:
9507:
9503:
9496:
9488:
9484:
9480:
9474:
9470:
9466:
9462:
9455:
9444:
9440:
9436:
9432:
9428:
9424:
9420:
9416:
9412:
9405:
9398:
9390:
9386:
9382:
9378:
9374:
9373:10.1038/38444
9370:
9366:
9362:
9358:
9354:
9347:
9339:
9335:
9331:
9327:
9322:
9317:
9314:(4): 858–70.
9313:
9309:
9305:
9298:
9290:
9286:
9282:
9278:
9273:
9268:
9264:
9260:
9256:
9252:
9248:
9241:
9233:
9227:
9219:
9215:
9210:
9205:
9200:
9195:
9191:
9187:
9184:(8): e41781.
9183:
9179:
9175:
9168:
9160:
9156:
9151:
9146:
9142:
9138:
9134:
9130:
9126:
9119:
9111:
9107:
9102:
9097:
9092:
9087:
9083:
9079:
9075:
9071:
9067:
9060:
9052:
9048:
9044:
9040:
9036:
9032:
9025:
9017:
9013:
9008:
9003:
8999:
8995:
8992:(4): 815–29.
8991:
8987:
8983:
8976:
8968:
8964:
8959:
8954:
8949:
8944:
8940:
8936:
8932:
8925:
8917:
8913:
8908:
8903:
8899:
8895:
8891:
8887:
8883:
8876:
8868:
8864:
8860:
8856:
8852:
8848:
8845:(7): 654–67.
8844:
8840:
8833:
8825:
8819:
8815:
8810:
8809:
8800:
8792:
8788:
8783:
8778:
8773:
8768:
8764:
8760:
8756:
8749:
8741:
8737:
8733:
8729:
8725:
8721:
8714:
8706:
8702:
8697:
8692:
8688:
8684:
8681:(2): 272–80.
8680:
8676:
8672:
8665:
8657:
8653:
8648:
8643:
8639:
8635:
8631:
8627:
8623:
8616:
8601:
8597:
8590:
8582:
8578:
8573:
8568:
8564:
8560:
8556:
8552:
8548:
8544:
8540:
8536:
8529:
8521:
8517:
8512:
8507:
8503:
8499:
8496:(1): 133–46.
8495:
8491:
8487:
8480:
8472:
8468:
8463:
8458:
8454:
8450:
8446:
8442:
8438:
8431:
8423:
8419:
8414:
8409:
8406:(3): 506–21.
8405:
8401:
8397:
8390:
8382:
8378:
8373:
8368:
8364:
8360:
8356:
8352:
8348:
8341:
8333:
8329:
8325:
8321:
8317:
8313:
8306:
8298:
8294:
8290:
8286:
8283:(2): 237–72.
8282:
8278:
8271:
8263:
8259:
8255:
8251:
8247:
8243:
8236:
8228:
8224:
8220:
8216:
8213:(2): 123–60.
8212:
8208:
8201:
8193:
8189:
8185:
8181:
8177:
8173:
8169:
8165:
8158:
8150:
8146:
8142:
8138:
8134:
8130:
8123:
8115:
8111:
8105:
8101:
8097:
8090:
8083:
8078:
8074:
8072:0-8153-4072-9
8068:
8064:
8060:
8053:
8046:
8042:
8038:
8037:
8032:
8025:
8017:
8013:
8008:
8003:
7999:
7995:
7991:
7984:
7976:
7972:
7967:
7962:
7958:
7954:
7950:
7943:
7935:
7931:
7927:
7923:
7919:
7915:
7912:(1–2): 9–21.
7911:
7907:
7900:
7892:
7888:
7884:
7880:
7876:
7872:
7864:
7858:
7854:
7851:
7848:Created from
7845:
7837:
7833:
7829:
7825:
7820:
7815:
7811:
7807:
7803:
7799:
7791:
7783:
7779:
7774:
7769:
7765:
7761:
7758:(3): 309–15.
7757:
7753:
7749:
7742:
7734:
7730:
7725:
7720:
7716:
7712:
7708:
7704:
7700:
7696:
7692:
7685:
7677:
7673:
7669:
7667:3-540-29114-8
7663:
7659:
7655:
7651:
7644:
7636:
7632:
7627:
7622:
7618:
7614:
7610:
7603:
7595:
7591:
7587:
7583:
7579:
7575:
7568:
7560:
7556:
7551:
7546:
7541:
7536:
7532:
7528:
7524:
7517:
7501:
7495:
7479:
7472:
7464:
7460:
7456:
7452:
7448:
7444:
7440:
7436:
7431:
7426:
7423:(2): 021920.
7422:
7418:
7414:
7407:
7399:
7395:
7390:
7385:
7381:
7377:
7373:
7369:
7365:
7361:
7360:
7355:
7348:
7340:
7336:
7331:
7326:
7322:
7318:
7315:(7745): 436.
7314:
7310:
7306:
7299:
7291:
7287:
7282:
7277:
7273:
7269:
7266:(4): 363–71.
7265:
7261:
7257:
7250:
7242:
7238:
7234:
7230:
7226:
7222:
7217:
7212:
7209:(4): 503–14.
7208:
7204:
7197:
7189:
7185:
7181:
7177:
7173:
7169:
7165:
7161:
7157:
7153:
7146:
7138:
7134:
7129:
7124:
7120:
7116:
7112:
7108:
7104:
7097:
7095:
7086:
7082:
7077:
7072:
7068:
7064:
7060:
7056:
7052:
7045:
7037:
7033:
7028:
7023:
7019:
7015:
7011:
7004:
7002:
7000:
6991:
6987:
6982:
6977:
6973:
6969:
6965:
6958:
6956:
6940:
6936:
6930:
6922:
6918:
6914:
6910:
6906:
6899:
6891:
6887:
6883:
6879:
6875:
6868:
6852:
6848:
6844:
6837:
6835:
6818:
6814:
6810:
6803:
6795:
6791:
6786:
6781:
6776:
6771:
6767:
6763:
6759:
6755:
6751:
6744:
6736:
6732:
6728:
6724:
6720:
6716:
6712:
6708:
6701:
6693:
6689:
6685:
6681:
6678:(4): 819–40.
6677:
6673:
6666:
6658:
6654:
6650:
6646:
6642:
6638:
6631:
6612:
6608:
6604:
6600:
6596:
6593:(5): 751–53.
6592:
6588:
6581:
6574:
6566:
6559:
6551:
6547:
6543:
6539:
6536:(4): 137–41.
6535:
6531:
6527:
6520:
6512:
6508:
6504:
6500:
6496:
6492:
6485:
6474:
6470:
6466:
6462:
6458:
6454:
6450:
6446:
6442:
6438:
6434:
6427:
6420:
6418:
6406:
6402:
6398:
6394:
6390:
6386:
6382:
6378:
6374:
6370:
6366:
6359:
6352:
6350:
6336:
6331:
6326:
6322:
6318:
6314:
6310:
6303:
6298:
6291:
6286:
6281:
6277:
6273:
6269:
6265:
6258:
6253:
6252:
6249:
6241:
6237:
6233:
6229:
6225:
6221:
6214:
6206:
6202:
6198:
6194:
6190:
6186:
6183:(6): 430–40.
6182:
6178:
6171:
6169:
6160:
6156:
6152:
6148:
6144:
6140:
6136:
6132:
6125:
6123:
6111:
6107:
6103:
6099:
6095:
6091:
6087:
6083:
6079:
6072:
6065:
6057:
6053:
6048:
6043:
6039:
6035:
6032:(8): 261–66.
6031:
6027:
6023:
6016:
6008:
6004:
5999:
5994:
5990:
5986:
5982:
5978:
5974:
5967:
5959:
5955:
5951:
5947:
5943:
5939:
5932:
5924:
5920:
5916:
5912:
5908:
5904:
5901:(4): 664–71.
5900:
5896:
5889:
5881:
5877:
5873:
5869:
5866:(5): 289–97.
5865:
5861:
5854:
5847:
5843:
5840:
5835:
5827:
5823:
5818:
5813:
5809:
5805:
5801:
5794:
5786:
5782:
5778:
5774:
5770:
5766:
5759:
5757:
5755:
5738:
5734:
5728:
5720:
5716:
5712:
5708:
5704:
5700:
5696:
5692:
5688:
5684:
5677:
5669:
5665:
5660:
5655:
5651:
5647:
5643:
5639:
5635:
5628:
5620:
5616:
5611:
5606:
5601:
5596:
5592:
5588:
5587:BMC Res Notes
5584:
5577:
5575:
5563:
5559:
5555:
5551:
5547:
5543:
5539:
5532:
5525:
5517:
5513:
5509:
5505:
5501:
5497:
5494:(5): 817–24.
5493:
5489:
5482:
5474:
5470:
5465:
5460:
5455:
5450:
5446:
5442:
5438:
5434:
5430:
5423:
5415:
5411:
5406:
5401:
5397:
5393:
5389:
5385:
5381:
5377:
5373:
5366:
5358:
5354:
5349:
5344:
5340:
5336:
5332:
5328:
5324:
5317:
5309:
5305:
5301:
5297:
5293:
5289:
5282:
5280:
5271:
5267:
5263:
5259:
5255:
5251:
5247:
5243:
5239:
5235:
5228:
5220:
5216:
5212:
5208:
5204:
5200:
5193:
5185:
5181:
5176:
5171:
5167:
5163:
5158:
5153:
5149:
5145:
5141:
5134:
5126:
5122:
5118:
5114:
5109:
5104:
5100:
5096:
5092:
5085:
5077:
5073:
5069:
5065:
5061:
5057:
5050:
5042:
5038:
5034:
5030:
5025:
5020:
5016:
5012:
5008:
5001:
4985:
4981:
4974:
4966:
4960:
4956:
4949:
4941:
4937:
4932:
4927:
4923:
4919:
4915:
4911:
4908:(2): 564–74.
4907:
4903:
4899:
4892:
4877:
4873:
4866:
4858:
4854:
4850:
4846:
4842:
4838:
4834:
4830:
4826:
4822:
4815:
4813:
4804:
4800:
4796:
4791:
4786:
4782:
4778:
4774:
4770:
4767:(3): 449–54.
4766:
4762:
4758:
4751:
4743:
4741:0-7167-4955-6
4737:
4733:
4726:
4724:
4722:
4720:
4711:
4707:
4703:
4699:
4695:
4691:
4687:
4683:
4679:
4675:
4668:
4660:
4656:
4652:
4648:
4644:
4640:
4637:(1): 153–61.
4636:
4632:
4625:
4614:
4610:
4606:
4602:
4598:
4594:
4590:
4586:
4582:
4578:
4574:
4570:
4566:
4559:
4552:
4550:
4548:
4546:
4537:
4533:
4528:
4523:
4519:
4515:
4511:
4507:
4503:
4499:
4495:
4491:
4487:
4480:
4472:
4468:
4464:
4460:
4458:0-8153-3218-1
4454:
4450:
4449:
4441:
4439:
4430:
4428:0-387-90762-9
4424:
4420:
4413:
4405:
4403:0-8053-4553-1
4399:
4394:
4393:
4384:
4369:
4365:
4359:
4351:
4347:
4346:Basic Biology
4343:
4336:
4328:
4324:
4318:
4314:
4313:
4305:
4297:
4296:
4291:
4285:
4281:
4269:
4266:
4260:
4257:
4254:
4253:Southern blot
4251:
4245:
4244:Ribosomal DNA
4242:
4239:
4236:
4233:
4230:
4227:
4224:
4221:
4218:
4215:
4212:
4209:
4206:
4203:
4200:
4197:
4194:
4191:
4188:
4185:
4182:
4179:
4176:
4175:
4168:
4165:
4164:Alec Jeffreys
4160:
4158:
4154:
4150:
4146:
4142:
4138:
4134:
4129:
4127:
4122:
4118:
4116:
4112:
4105:
4101:
4100:
4090:
4086:
4084:
4081:
4077:
4072:
4068:
4067:Linus Pauling
4064:
4060:
4055:
4051:
4047:
4043:
4039:
4036:In May 1952,
4034:
4032:
4028:
4024:
4020:
4016:
4012:
4008:
4004:
4000:
3996:
3995:Francis Crick
3988:
3985:
3981:
3976:
3972:
3970:
3966:
3962:
3958:
3954:
3950:
3946:
3942:
3938:
3934:
3933:Colin MacLeod
3930:
3925:
3923:
3918:
3916:
3912:
3908:
3904:
3900:
3895:
3892:
3888:
3884:
3880:
3875:
3870:
3865:
3863:
3859:
3855:
3851:
3843:
3839:
3835:
3831:
3830:Francis Crick
3827:
3823:
3818:
3813:
3806:
3804:
3800:
3796:
3792:
3786:
3776:
3774:
3770:
3766:
3762:
3758:
3751:
3747:
3746:Phylogenetics
3737:
3735:
3731:
3727:
3723:
3719:
3715:
3711:
3710:
3704:
3696:
3692:
3688:
3683:
3678:
3668:
3665:
3664:gene products
3661:
3657:
3653:
3649:
3645:
3641:
3637:
3633:
3629:
3625:
3622:, especially
3621:
3617:
3613:
3609:
3604:
3594:
3592:
3589:selection or
3588:
3584:
3579:
3578:Deoxyribozyme
3569:
3567:
3563:
3558:
3554:
3551:
3545:
3543:
3539:
3538:Alec Jeffreys
3535:
3531:
3527:
3523:
3522:DNA profiling
3519:
3515:
3511:
3507:
3503:
3499:
3495:
3490:
3489:DNA profiling
3483:DNA profiling
3480:
3478:
3474:
3470:
3466:
3462:
3458:
3454:
3450:
3446:
3442:
3438:
3434:
3430:
3424:
3420:
3416:
3401:
3399:
3395:
3391:
3389:
3385:
3381:
3377:
3373:
3369:
3365:
3361:
3357:
3353:
3349:
3345:
3342:
3338:
3334:
3330:
3326:
3321:
3318:
3314:
3310:
3306:
3302:
3298:
3292:
3282:
3280:
3276:
3272:
3268:
3264:
3260:
3256:
3251:
3249:
3244:
3242:
3238:
3234:
3230:
3220:
3212:
3208:
3197:
3193:
3192:
3188:
3184:
3183:
3178:
3168:
3166:
3162:
3158:
3154:
3153:messenger RNA
3149:
3144:
3142:
3138:
3134:
3130:
3127:, which is a
3126:
3121:
3119:
3115:
3111:
3107:
3103:
3098:
3093:
3090:
3086:
3082:
3078:
3074:
3070:
3061:
3059:
3055:
3051:
3047:
3043:
3040:
3036:
3027:
3025:
3021:
3017:
3013:
3009:
3004:
3002:
2998:
2994:
2990:
2986:
2982:
2978:
2974:
2973:endonucleases
2970:
2966:
2962:
2958:
2954:
2947:
2944:
2939:
2925:
2922:
2918:
2912:
2910:
2906:
2898:
2893:
2889:
2887:
2883:
2878:
2873:
2872:chromosomes.
2870:
2866:
2862:
2858:
2854:
2850:
2846:
2842:
2833:
2828:
2823:
2813:
2810:
2800:
2798:
2792:
2782:
2780:
2776:
2771:
2769:
2765:
2762:
2758:
2747:
2745:
2741:
2737:
2733:
2728:
2727:Cell division
2721:
2717:
2713:
2709:
2706:produces the
2705:
2701:
2697:
2692:
2687:
2677:
2675:
2671:
2667:
2663:
2658:
2656:
2652:
2648:
2644:
2640:
2639:messenger RNA
2636:
2630:
2626:
2622:
2612:
2610:
2606:
2602:
2598:
2594:
2590:
2586:
2578:
2574:
2570:
2566:
2564:
2560:
2556:
2552:
2551:
2546:
2542:
2541:noncoding DNA
2538:
2534:
2530:
2526:
2521:
2519:
2515:
2511:
2507:
2503:
2499:
2495:
2491:
2487:
2483:
2479:
2473:
2472:Noncoding DNA
2469:
2465:
2461:
2457:
2447:
2445:
2441:
2437:
2432:
2428:
2424:
2420:
2416:
2412:
2408:
2404:
2400:
2392:
2387:
2378:
2376:
2372:
2368:
2364:
2360:
2356:
2352:
2348:
2344:
2340:
2336:
2332:
2331:
2330:intercalation
2325:
2323:
2319:
2315:
2311:
2307:
2303:
2298:
2294:
2293:free radicals
2290:
2286:
2282:
2278:
2274:
2270:
2266:
2262:
2254:
2253:tobacco smoke
2250:
2246:
2242:
2238:
2235:
2230:
2225:
2221:
2217:
2207:
2205:
2201:
2200:glycosylation
2197:
2193:
2189:
2185:
2181:
2177:
2176:
2171:
2167:
2162:
2160:
2156:
2152:
2148:
2144:
2140:
2131:
2121:
2116:
2111:
2107:
2103:
2099:
2096:
2092:
2089:
2085:
2084:
2079:
2075:
2060:
2053:
2044:
2042:
2039:by repelling
2038:
2034:
2030:
2026:
2022:
2012:
2010:
2006:
2001:
1997:
1996:Hachimoji DNA
1991:
1981:
1979:
1975:
1970:
1961:
1947:
1943:
1939:
1936:
1932:
1931:
1926:
1922:
1912:
1910:
1906:
1900:
1897:
1893:
1889:
1883:
1881:
1877:
1873:
1869:
1861:
1856:
1851:
1841:
1839:
1836:
1832:
1828:
1824:
1820:
1819:exobiologists
1814:
1804:
1802:
1798:
1793:
1788:
1786:
1781:
1778:Although the
1776:
1774:
1770:
1769:Francis Crick
1766:
1762:
1758:
1757:
1752:
1748:
1744:
1739:
1738:in solution.
1737:
1733:
1728:
1724:
1720:
1717:that include
1716:
1715:conformations
1709:
1705:
1701:
1696:
1691:
1690:DNA structure
1687:
1683:
1673:
1671:
1667:
1666:transcription
1663:
1659:
1655:
1649:
1648:DNA supercoil
1639:
1637:
1633:
1629:
1625:
1620:
1618:
1613:
1612:messenger RNA
1609:
1603:
1593:
1591:
1587:
1582:
1578:
1574:
1570:
1565:
1559:
1555:
1551:
1546:
1543:
1535:
1529:
1525:
1521:
1517:
1513:
1508:
1499:
1496:
1488:
1482:
1478:
1458:
1452:(also called
1451:
1438:
1436:
1421:
1419:
1417:
1413:
1409:
1401:
1396:
1392:
1389:
1388:complementary
1380:
1375:. Bottom, an
1374:
1370:
1357:
1353:
1352:
1344:
1340:
1339:
1334:
1324:
1322:
1318:
1313:
1309:
1301:
1300:Hoechst stain
1296:
1282:
1279:
1278:
1276:
1271:
1268:
1265:
1262:
1261:
1259:
1256:
1251:
1248:
1247:
1246:
1242:
1237:
1234:
1231:
1228:
1225:
1222:
1221:
1220:
1216:
1211:
1208:
1207:
1206:
1202:
1197:
1194:
1193:
1192:
1188:
1187:
1186:
1183:
1181:
1176:
1172:
1168:
1167:
1162:
1158:
1148:
1146:
1142:
1134:
1122:
1118:
1106:
1096:
1094:
1083:
1075:
1067:
1059:
1051:
1047:
1043:
1042:base-stacking
1039:
1031:
1026:
1022:
1020:
1016:
1012:
1008:
1004:
998:
996:
992:
988:
984:
980:
976:
972:
969:, which is a
968:
967:2-deoxyribose
964:
960:
955:
953:
949:
945:
941:
937:
933:
929:
924:
922:
918:
914:
910:
906:
902:
894:
890:
886:
881:
872:
870:
866:
862:
858:
854:
850:
846:
842:
838:
834:
830:
826:
822:
818:
814:
810:
806:
802:
798:
797:cell division
794:
789:
787:
783:
779:
775:
771:
770:transcription
767:
763:
760:). It is the
759:
755:
751:
747:
743:
739:
734:
732:
728:
724:
720:
716:
712:
708:
704:
700:
696:
692:
688:
684:
680:
676:
673:
669:
666:units called
665:
660:
658:
654:
650:
646:
642:
638:
637:nucleic acids
634:
630:
626:
623:of all known
622:
618:
614:
610:
606:
602:
594:
587:
503:
492:
487:
485:
480:
478:
473:
472:
470:
469:
463:
453:
452:
451:
450:
443:
440:
439:
433:
432:
423:
420:
418:
415:
413:
410:
408:
405:
403:
400:
398:
395:
393:
390:
388:
385:
383:
380:
379:
371:
370:
363:
360:
359:
355:
352:
348:
339:
335:
333:
330:
328:
325:
323:
320:
319:
313:
312:
305:
302:
300:
297:
295:
292:
290:
287:
284:
280:
277:
275:
272:
270:
267:
266:
260:
259:
252:
249:
247:
244:
243:
237:
234:
232:
229:
227:
224:
222:
219:
217:
214:
212:
209:
207:
204:
202:
199:
196:
194:
191:
190:
182:
181:
177:
173:
172:
169:
166:
165:
161:
160:
153:
146:
142:
138:
134:
130:
125:
113:
109:
92:
84:
76:
69:
61:
53:
44:
36:
35:
32:
28:
24:
19:
15690:
15673:
15656:
15644:from Commons
15639:
15618:
15554:Bacterial
15529:Lambda phage
15403:
15193:Constituents
15120:irreversible
15005:Key elements
14902:Key elements
14825:
14816:Genetic code
14806:Introduction
14657:Genetic code
14591:the Americas
14567:Quantitative
14537:Cytogenetics
14532:Conservation
14470:
14425:Introduction
14349:
14338:
14328:
14304:Forms_of_DNA
14299:
14289:
14282:
14179:Online books
14169:
14139:
14117:
14096:
14070:
14049:
14030:
14008:
13989:
13954:
13950:
13923:
13901:
13878:
13858:
13838:
13798:(2): 20–26.
13795:
13791:
13781:
13772:
13768:
13762:
13753:
13744:
13699:
13695:
13685:
13674:the original
13665:
13658:
13608:
13604:
13591:
13582:
13573:
13530:
13524:
13514:
13502:. Retrieved
13491:
13481:
13469:. Retrieved
13463:
13453:
13441:. Retrieved
13427:
13419:the original
13414:
13405:
13382:
13376:
13334:(2): 84–97.
13331:
13327:
13317:
13296:
13288:
13276:. Retrieved
13272:
13263:
13231:(1): 39–56.
13228:
13224:
13214:
13195:
13191:
13181:
13148:
13144:
13138:
13103:
13099:
13089:
13079:the original
13069:(1): 66–76.
13066:
13051:the original
13038:
13034:
13018:
13009:
12996:
12987:
12983:
12977:
12942:
12938:
12928:
12893:
12889:
12879:
12846:
12842:
12836:
12824:
12822:
12794:
12790:
12783:(6): 345–69.
12780:
12776:
12769:(1): ?.
12766:
12762:
12752:
12748:
12739:
12722:
12718:
12705:
12686:
12682:
12672:
12655:
12651:
12641:
12627:: 1198–203.
12624:
12620:
12610:
12586:(1): 80–97.
12583:
12579:
12569:
12558:
12554:
12550:
12538:
12534:
12515:
12511:
12498:
12494:
12485:
12481:
12468:
12464:
12451:
12447:
12434:
12401:
12397:
12391:
12382:überlassend.
12380:
12379:
12375:
12371:
12361:
12328:
12324:
12318:
12283:
12279:
12269:
12232:
12228:
12218:
12206:. Retrieved
12194:
12190:
12180:
12139:
12135:
12129:
12096:
12092:
12086:
12035:
12031:
12024:
11983:
11979:
11966:
11921:
11918:PLOS Biology
11917:
11907:
11880:
11874:
11839:
11835:
11829:
11806:
11800:
11773:
11764:
11719:
11715:
11705:
11678:
11674:
11664:
11629:
11625:
11615:
11590:
11586:
11565:. Retrieved
11548:
11538:
11530:the original
11520:
11508:. Retrieved
11504:the original
11494:
11451:
11447:
11437:
11404:
11400:
11394:
11349:
11345:
11335:
11323:. Retrieved
11314:
11304:
11279:
11275:
11269:
11234:
11230:
11220:
11185:
11179:
11146:
11142:
11136:
11093:
11087:
11077:
11065:. Retrieved
11059:
11049:
11037:. Retrieved
11026:
11016:
11004:. Retrieved
10995:ScienceDaily
10993:
10983:
10971:. Retrieved
10952:
10907:
10903:
10893:
10852:
10848:
10842:
10817:
10813:
10807:
10766:
10762:
10756:
10715:
10711:
10705:
10660:
10656:
10646:
10613:
10609:
10603:
10560:
10556:
10550:
10509:
10505:
10499:
10464:
10460:
10450:
10407:
10403:
10393:
10368:
10364:
10358:
10328:(1): 45–54.
10325:
10321:
10315:
10282:
10278:
10272:
10239:
10235:
10229:
10217:. Retrieved
10214:www.rcsb.org
10213:
10203:
10170:
10166:
10160:
10125:
10121:
10083:
10079:
10073:
10062:the original
10033:
10029:
10016:
9981:
9977:
9967:
9940:
9936:
9923:
9898:
9894:
9888:
9853:
9849:
9801:
9797:
9787:
9775:. Retrieved
9772:www.rcsb.org
9771:
9761:
9716:
9712:
9702:
9675:
9671:
9661:
9636:
9632:
9626:
9614:. Retrieved
9611:www.rcsb.org
9610:
9600:
9575:
9571:
9565:
9540:
9536:
9530:
9505:
9501:
9495:
9460:
9454:
9414:
9410:
9397:
9356:
9352:
9346:
9311:
9307:
9297:
9254:
9250:
9240:
9226:
9181:
9177:
9167:
9135:(2): 84–91.
9132:
9128:
9118:
9073:
9069:
9059:
9034:
9030:
9024:
8989:
8985:
8975:
8938:
8934:
8924:
8889:
8885:
8875:
8842:
8838:
8832:
8807:
8799:
8762:
8758:
8748:
8723:
8719:
8713:
8678:
8674:
8664:
8629:
8625:
8615:
8603:. Retrieved
8600:www.rcsb.org
8599:
8589:
8546:
8542:
8528:
8493:
8489:
8479:
8444:
8440:
8430:
8403:
8399:
8389:
8354:
8350:
8340:
8315:
8311:
8305:
8280:
8276:
8270:
8245:
8241:
8235:
8210:
8206:
8200:
8167:
8163:
8157:
8132:
8128:
8122:
8099:
8089:
8080:
8062:
8052:
8044:
8034:
8024:
7997:
7993:
7983:
7956:
7952:
7942:
7909:
7905:
7899:
7874:
7871:Biochemistry
7870:
7863:
7844:
7801:
7797:
7790:
7755:
7751:
7741:
7698:
7694:
7684:
7649:
7643:
7616:
7612:
7602:
7580:(2): 89–97.
7577:
7573:
7567:
7530:
7526:
7516:
7504:. Retrieved
7494:
7482:. Retrieved
7471:
7420:
7416:
7406:
7363:
7357:
7347:
7312:
7308:
7298:
7263:
7259:
7249:
7206:
7202:
7196:
7155:
7151:
7145:
7110:
7106:
7058:
7054:
7044:
7017:
7013:
6971:
6967:
6942:. Retrieved
6938:
6929:
6904:
6898:
6877:
6867:
6855:. Retrieved
6846:
6821:. Retrieved
6812:
6802:
6757:
6753:
6743:
6710:
6706:
6700:
6675:
6671:
6665:
6643:(1): 45–63.
6640:
6636:
6630:
6618:. Retrieved
6611:the original
6590:
6586:
6573:
6564:
6558:
6533:
6529:
6519:
6497:(1): 49–72.
6494:
6490:
6484:
6436:
6432:
6368:
6364:
6312:
6308:
6267:
6263:
6248:
6223:
6219:
6213:
6180:
6176:
6134:
6130:
6110:the original
6081:
6077:
6064:
6029:
6025:
6015:
5980:
5976:
5966:
5941:
5937:
5931:
5898:
5894:
5888:
5863:
5859:
5853:
5834:
5810:(1): 29–38.
5807:
5803:
5793:
5768:
5764:
5741:. Retrieved
5737:the original
5727:
5686:
5682:
5676:
5641:
5637:
5627:
5590:
5586:
5541:
5538:Biochemistry
5537:
5524:
5491:
5487:
5481:
5436:
5432:
5422:
5379:
5375:
5365:
5330:
5326:
5316:
5291:
5287:
5237:
5233:
5227:
5202:
5198:
5192:
5147:
5143:
5133:
5098:
5094:
5084:
5059:
5055:
5049:
5014:
5010:
5000:
4988:. Retrieved
4973:
4954:
4948:
4905:
4901:
4891:
4879:. Retrieved
4876:www.rcsb.org
4875:
4865:
4824:
4820:
4803:the original
4764:
4760:
4750:
4732:Biochemistry
4731:
4677:
4673:
4667:
4634:
4630:
4624:
4568:
4564:
4493:
4489:
4479:
4447:
4418:
4412:
4391:
4383:
4371:. Retrieved
4367:
4358:
4345:
4335:
4311:
4304:
4293:
4284:
4161:
4130:
4123:
4119:
4114:
4110:
4103:
4097:
4095:
4071:Robert Corey
4063:antiparallel
4035:
4019:Martha Chase
3999:James Watson
3992:
3929:Oswald Avery
3926:
3919:
3907:cell nucleus
3903:Jean Brachet
3896:
3891:Pneumococcus
3890:
3866:
3847:
3834:James Watson
3812:
3788:
3773:anthropology
3753:
3730:streptavidin
3707:
3700:
3660:gene finding
3652:phylogenetic
3606:
3581:
3559:
3555:
3546:
3525:
3492:
3461:viral vector
3445:biochemistry
3426:
3392:
3350:, including
3322:
3294:
3275:endonuclease
3267:recombinases
3252:
3245:
3226:
3145:
3133:retroviruses
3122:
3102:proofreading
3094:
3080:
3067:
3044:
3039:supercoiling
3033:
3005:
2969:exonucleases
2951:
2924:accessible.
2913:
2902:
2874:
2837:
2806:
2794:
2772:
2766:
2753:
2725:
2702:. Next, one
2670:transfer RNA
2659:
2654:
2651:genetic code
2632:
2621:Genetic code
2582:
2548:
2522:
2490:chloroplasts
2486:mitochondria
2482:cell nucleus
2475:
2456:Cell nucleus
2431:Transmission
2419:human genome
2396:
2371:chemotherapy
2328:
2326:
2265:DNA sequence
2258:
2247:, the major
2173:
2163:
2136:
2058:
2041:nucleophiles
2018:
1993:
1966:
1960:Branched DNA
1921:Branched DNA
1915:Branched DNA
1901:
1892:G-quadruplex
1884:
1865:
1850:G-quadruplex
1816:
1792:right-handed
1789:
1785:paracrystals
1779:
1777:
1765:James Watson
1754:
1750:
1740:
1712:
1651:
1642:Supercoiling
1621:
1608:DNA sequence
1605:
1566:
1550:chromosome 1
1539:
1491:
1489:
1453:
1444:-content (%
1439:
1427:
1420:
1406:-content. A
1391:base pairing
1385:
1376:
1366:
1327:Base pairing
1316:
1308:binding site
1305:
1257:
1244:
1218:
1204:
1190:
1184:
1164:
1154:
1141:methyl group
1102:
1035:
1011:antiparallel
999:
956:
928:double helix
925:
898:
837:chloroplasts
829:mitochondria
821:cell nucleus
790:
778:genetic code
757:
754:antiparallel
735:
719:base pairing
661:
639:. Alongside
621:reproduction
613:double helix
600:
501:
500:
422:Quantitative
392:Cytogenetics
387:Conservation
269:Introduction
129:double helix
67:
31:
18:
15549:P1-derived
15317:Antisense
15210:Nucleotides
15205:Nucleosides
15200:Nucleobases
14971:Translation
14808:to genetics
14637:Epigenetics
14331:Study Guide
14301:Proteopedia
14291:Proteopedia
13145:Experientia
12683:J Biol Chem
11067:17 February
10086:: 283–315.
7619:(1): 6–21.
6905:Nature News
6878:Nature News
6637:Biopolymers
6137:: 369–413.
5944:(1): 1–12.
5327:Biopolymers
5294:: 293–321.
5062:: 2838–44.
4373:21 November
4340:Purcell A.
4143:, allowing
4059:space group
4005:within the
3980:blue plaque
3862:nucleobases
3803:reliability
3709:DNA origami
3518:crime scene
3516:found at a
3477:agriculture
3453:transformed
3394:Ancient DNA
3388:cosmic dust
3370:, found in
3364:outer space
3337:outer space
3307:as part of
3106:exonuclease
3089:active site
3069:Polymerases
3064:Polymerases
3008:DNA ligases
2865:acetylation
2857:methylation
2853:ionic bonds
2797:neutrophils
2680:Replication
2647:translation
2593:pseudogenes
2589:centromeres
2545:genome size
2440:translation
2411:prokaryotes
2399:chromosomes
2391:nuclear DNA
2355:carcinogens
2351:doxorubicin
2281:ultraviolet
2245:benzopyrene
2180:vertebrates
2143:methylation
2130:Deamination
2029:strong acid
1880:human cells
1797:methylation
1573:nuclear DNA
1477:Pribnow box
1416:temperature
1317:(see below)
1121:pyrimidines
905:nucleotides
845:prokaryotes
825:nuclear DNA
793:chromosomes
786:translation
782:amino acids
727:pyrimidines
699:deoxyribose
675:nucleobases
668:nucleotides
83:10 - 10 bp
15705:Categories
15658:Quotations
15501:Morpholino
15414:Organellar
15322:Processual
15287:Regulatory
15231:non-coding
15115:reversible
15078:lac operon
15054:imprinting
15049:Epigenetic
15041:Regulation
14996:Eukaryotic
14942:5' capping
14893:Eukaryotic
14642:Geneticist
14616:South Asia
14562:Population
14542:Ecological
14511:Amino acid
14491:Nucleotide
14466:Chromosome
14246:Audio help
14237:2007-02-12
13443:6 February
13385:. Oxford:
13041:: 109–21.
12557:: 248–64.
12378:: 441–60.
12286:(5): 239.
11924:(3): E73.
11325:22 October
10036:: 133–63.
9639:: 729–49.
8533:Birney E,
7476:Reusch W.
6857:2 December
6823:2 December
6713:: 286–98.
5733:"Untitled"
5593:(1): 106.
5017:: 99–134.
4368:Genome.gov
4276:References
4044:, took an
3899:sea urchin
3883:experiment
3689:at right.
3471:, used in
3384:red giants
3372:meteorites
3368:pyrimidine
3269:, such as
3157:terminator
3137:telomerase
3020:DNA repair
2961:hydrolysis
2882:stem-loops
2849:nucleosome
2720:DNA ligase
2643:amino-acid
2609:divergence
2464:Chromosome
2403:eukaryotes
2347:daunomycin
2306:insertions
2283:light and
2239:between a
2198:, and the
2037:hydrolysis
1876:DNA repair
1872:telomerase
1780:B-DNA form
1736:polyamines
1569:eukaryotes
1562:85 mm
1558:base pairs
1554:chromosome
1528:base pairs
1510:Schematic
1400:GC-content
1302:dye 33258.
1180:epigenetic
1119:, and the
1093:base pairs
948:biopolymer
944:nucleotide
940:nucleoside
936:nucleobase
919:(3.4
875:Properties
746:non-coding
742:replicated
635:(RNA) are
631:. DNA and
417:Population
397:Ecological
322:Geneticist
236:Amino acid
216:Nucleotide
193:Chromosome
145:base pairs
112:base pairs
60:10 - 10 bp
15675:Resources
15461:Analogues
15446:Hachimoji
15229:(coding,
15184:Types of
14986:Bacterial
14883:Bacterial
14557:Molecular
14552:Microbial
14527:Classical
14136:Wilkins M
13898:Judson HF
13832:Berry A,
13775:(1): 100.
13565:258314143
12990:: 519–76.
12825:genonemes
12518:: 422–31.
12488:: 267–71.
12471:: 290–95.
12454:: 284–91.
12280:3 Biotech
11844:CiteSeerX
11557:0362-4331
11276:Biochimie
11120:0028-0836
10973:10 August
10565:CiteSeerX
10365:Biochimie
8839:BioEssays
7819:1874/5219
7752:BioEssays
7211:CiteSeerX
6847:Space.com
6620:29 August
6550:189888972
6205:205496065
6084:: 21–53.
5166:1664-8021
5117:0009-2665
5076:0002-7863
5033:0066-4154
4922:0305-1048
4849:0907-4449
4781:0306-3283
4694:0006-2928
4651:0022-2836
4593:0028-0836
4518:2041-1723
4467:145080076
4392:iGenetics
4220:Haplotype
4080:The Eagle
3984:The Eagle
3927:In 1943,
3920:In 1937,
3915:cytoplasm
3909:and that
3867:In 1909,
3789:DNA as a
3757:phylogeny
3714:polyhedra
3644:mutations
3612:data mine
3439:. Modern
3317:evolution
3313:RNA world
3309:ribozymes
3305:catalysis
3285:Evolution
3141:telomeres
3118:helicases
3114:DNA clamp
3110:replisome
3081:templates
3046:Helicases
2953:Nucleases
2886:nucleases
2877:protein A
2841:chromatin
2718:) before
2635:phenotype
2585:Telomeres
2518:enhancers
2514:promoters
2460:Chromatin
2367:aflatoxin
2359:teratogen
2343:acridines
2310:deletions
2255:, and DNA
2188:mutations
2184:deaminate
2159:crosstalk
2139:chromatin
1896:chelation
1868:telomeres
1835:bacterium
1827:biosphere
1534:Karyotype
1512:karyogram
1481:promoters
1333:Base pair
1245:Thymidine
1243:Modified
1217:Modified
1203:Modified
1189:Modified
959:phosphate
917:ångströms
865:chromatin
857:cytoplasm
664:monomeric
627:and many
625:organisms
412:Molecular
407:Microbial
382:Classical
283:molecular
279:Evolution
15584:Category
15519:Phagemid
15370:Ribozyme
15098:microRNA
15012:Ribosome
14991:Archaeal
14947:Splicing
14919:Promoter
14888:Archaeal
14832: →
14828: →
14756:Category
14682:Heredity
14652:Genomics
14496:Mutation
14486:Heredity
14450:Glossary
14440:Timeline
14414:Genetics
14248: ·
14138:(2003).
14091:Stent GS
14029:(2006).
14027:Ridley M
13981:12540907
13922:(1994).
13900:(1979).
13836:(2003).
13834:Watson J
13814:23386793
13736:16590258
13647:Archived
13635:12540909
13557:37100935
13504:26 April
13498:Archived
13471:25 April
13437:Archived
13368:16578429
13255:12981234
13165:15421335
13130:19871359
13075:20257017
12920:20474956
12871:46277758
12863:11533721
12819:17769043
12602:13103145
12527:Archived
12418:17901982
12353:51905843
12345:30073589
12310:29744271
12261:11806830
12164:18818351
12121:19421208
12070:19424153
12008:16541064
11958:13222080
11950:15024422
11899:55106399
11866:14734307
11792:45951728
11756:25918425
11656:19684594
11567:27 March
11561:Archived
11510:27 March
11429:14511630
11319:Archived
11296:12595138
11261:11950565
11212:17172731
11171:41788896
11128:33597786
11061:CNN News
11033:Archived
11006:9 August
11000:Archived
10967:Archived
10944:21836052
10885:24740859
10877:11734907
10834:15866038
10791:11057666
10638:85976762
10630:11360970
10587:15217990
10534:12110897
10491:39505263
10483:12423347
10442:16838012
10342:16369571
10307:23225873
10299:16619049
10256:11283701
10219:27 March
10195:24946189
10187:12516863
10152:39614573
10100:15952889
10058:26171993
10050:12045093
9959:15128295
9915:16246147
9880:11058099
9777:27 March
9753:12808131
9694:15479634
9653:10966474
9616:27 March
9592:10473346
9522:11497996
9487:12596902
9443:Archived
9431:11498575
9338:26965112
9330:15853876
9289:21101836
9272:11147202
9218:22952587
9178:PLOS ONE
9159:23158400
9110:23300576
9070:PLOS ONE
9051:11859186
9016:20598083
8967:19023416
8916:11591672
8867:32463239
8859:17563084
8791:11178285
8740:12083509
8705:11827946
8656:15905142
8605:27 March
8581:17571346
8520:15596463
8471:11236998
8422:15988757
8381:11181995
8332:11562309
8262:10799645
8192:21600302
8149:19812404
8114:Archived
8077:Archived
8041:Archived
8016:12947387
7994:Oncogene
7934:10064846
7891:12885257
7853:Archived
7850:PDB 1JDG
7836:24801094
7782:16479578
7733:19372393
7676:16570853
7635:11782440
7594:16403636
7559:23133442
7463:17574854
7455:17358380
7398:30792304
7339:30809059
7290:16515737
7233:10338214
7180:12050675
7137:17012276
6921:87341731
6890:Archived
6851:Archived
6817:Archived
6813:BBC News
6794:12486233
6735:20589136
6727:12086319
6692:10891271
6473:Archived
6461:13054693
6405:Archived
6393:13054694
6335:Archived
6290:Archived
6197:12042765
6159:18144189
6151:11395412
6098:16004565
6007:15520290
5958:15680581
5923:23748148
5915:15389973
5880:15851066
5842:Archived
5826:28721182
5668:16710414
5619:30813969
5562:Archived
5558:15609994
5516:24479358
5473:10393911
5414:10733978
5357:23818176
5219:28941008
5184:30619465
5125:27319741
4984:Archived
4940:16449200
4881:27 March
4857:12657780
4710:27950750
4613:Archived
4601:13054692
4536:26455586
4496:: 8440.
4471:Archived
4364:"Uracil"
4350:Archived
4327:Archived
4178:Autosome
4171:See also
4050:Photo 51
4011:heredity
3982:outside
3961:cytosine
3734:aptamers
3587:in vitro
3469:proteins
3457:plasmids
3435:and the
3380:universe
3356:cytosine
3085:hydroxyl
2987:against
2985:bacteria
2979:are the
2971:, while
2845:histones
2832:histones
2761:biofilms
2696:helicase
2666:ribosome
2523:In many
2512:such as
2502:heredity
2498:genotype
2494:nucleoid
2423:sequence
2335:aromatic
2279:such as
2261:mutagens
2243:form of
2234:covalent
2220:Mutation
2147:cytosine
2110:cytosine
1967:In DNA,
1860:telomere
1636:bacteria
1624:plasmids
1590:egg cell
1479:in some
1412:covalent
1219:Cytosine
1058:cytosine
1046:aromatic
1021:in RNA.
932:backbone
869:histones
849:bacteria
817:protists
762:sequence
701:, and a
679:cytosine
641:proteins
462:Category
347:template
338:Genomics
316:Research
221:Mutation
211:Heredity
168:Genetics
108:packaged
15721:Helices
15611:Biology
15524:Plasmid
14851:RNA→DNA
14846:RNA→RNA
14834:Protein
14435:History
14430:Outline
14235: (
14206:minutes
13959:Bibcode
13920:Olby RC
13805:3561883
13704:Bibcode
13643:4428347
13613:Bibcode
13535:Bibcode
13465:AP News
13359:1063734
13336:Bibcode
13246:2147348
13173:2522535
13121:2135445
12969:7968924
12911:2167760
12799:Bibcode
12791:Science
12731:4609088
12593:2599350
12541:: 7–22.
12301:5935598
12208:30 June
12172:2755388
12144:Bibcode
12136:Science
12101:Bibcode
12078:4430815
12040:Bibcode
12016:4316391
11988:Bibcode
11770:Baldi P
11747:4434688
11724:Bibcode
11697:9000012
11647:2746877
11607:9383394
11486:4229883
11478:2989708
11456:Bibcode
11421:9068179
11386:8016106
11354:Bibcode
11252:3481857
11098:Bibcode
11039:5 March
10935:3161613
10912:Bibcode
10857:Bibcode
10799:9879073
10771:Bibcode
10748:4283694
10740:8469282
10720:Bibcode
10697:1372984
10665:Bibcode
10610:Science
10595:4939632
10542:4331004
10514:Bibcode
10433:5607947
10412:Bibcode
10385:9466696
10350:7779574
10264:8547149
10144:7514143
10008:7592405
9828:8336674
9721:Bibcode
9557:8178371
9439:1883924
9411:Science
9389:4328827
9381:9305837
9361:Bibcode
9281:9893710
9209:3430683
9186:Bibcode
9150:4378900
9101:3530553
9078:Bibcode
9031:Science
9007:2962764
8958:2581603
8647:1569473
8572:2212820
8551:Bibcode
8511:4246714
8449:Bibcode
8359:Bibcode
8351:Science
8297:3936066
8227:1881402
8172:Bibcode
7975:9289489
7914:Bibcode
7828:8261512
7773:2754416
7724:3263819
7703:Bibcode
7695:Science
7550:3488762
7533:: 238.
7506:30 June
7484:30 June
7435:Bibcode
7389:6413494
7368:Bibcode
7359:Science
7317:Bibcode
7281:3478329
7188:4422211
7160:Bibcode
7128:1636468
7085:9353250
7036:9553037
6990:3907856
6762:Bibcode
6657:9097733
6595:Bibcode
6511:7441761
6469:4280080
6441:Bibcode
6401:4268222
6373:Bibcode
6317:Bibcode
6272:Bibcode
6240:2482766
6106:1427671
6056:1771674
6047:7173306
5817:5506767
5785:1715276
5743:13 June
5719:4355527
5711:7219534
5691:Bibcode
5646:Bibcode
5610:6391780
5508:7476180
5441:Bibcode
5405:1300792
5384:Bibcode
5348:3844552
5308:6236744
5270:4315465
5262:7432492
5242:Bibcode
5175:6305559
5150:: 640.
5041:9759484
4990:13 July
4931:1360284
4829:Bibcode
4799:5499957
4790:1179624
4702:4991030
4659:7338906
4609:4253007
4573:Bibcode
4527:4608029
4498:Bibcode
4226:Meiosis
4190:DNA Day
4115:in vivo
4111:in vivo
4029:of the
4021:in the
4001:at the
3969:thymine
3965:adenine
3957:guanine
3881:in his
3809:History
3636:aligned
3548:of the
3441:biology
3360:thymine
3329:guanine
3325:adenine
3239:, when
3073:enzymes
2963:of the
2957:enzymes
2909:enzymes
2779:ecology
2742:called
2597:fossils
2550:C-value
2525:species
2377:cells.
2249:mutagen
2194:in the
2151:histone
2120:thymine
2033:protons
2015:Acidity
1980:below.
1969:fraying
1756:in vivo
1660:called
1658:enzymes
1628:viruses
1542:diploid
1365:Top, a
1290:Grooves
1277:Others
1205:Guanine
1191:Adenine
1105:purines
1074:thymine
1066:guanine
1050:adenine
971:pentose
901:polymer
853:archaea
805:animals
731:purines
697:called
691:thymine
687:adenine
683:guanine
629:viruses
617:genetic
605:polymer
603:) is a
274:History
246:Outline
141:element
135:). The
15597:Portal
15539:Fosmid
15534:Cosmid
15484:Hexose
15406:acids
15358:Others
14601:Europe
14586:Africa
14520:Fields
14506:Allele
14481:Genome
14314:Nature
14284:Nature
14167:about
14146:
14124:
14105:
14079:
14056:
14037:
14015:
13996:
13979:
13951:Nature
13943:Olby R
13930:
13908:
13886:
13865:
13846:
13812:
13802:
13734:
13727:528642
13724:
13641:
13633:
13605:Nature
13563:
13555:
13526:Nature
13393:
13366:
13356:
13305:
13278:18 May
13253:
13243:
13171:
13163:
13128:
13118:
13073:
12967:
12960:372978
12957:
12918:
12908:
12869:
12861:
12817:
12729:
12600:
12590:
12426:915930
12424:
12416:
12351:
12343:
12308:
12298:
12259:
12252:150454
12249:
12197:(10).
12170:
12162:
12119:
12076:
12068:
12032:Nature
12014:
12006:
11980:Nature
11956:
11948:
11941:368168
11938:
11897:
11887:
11864:
11846:
11817:
11790:
11780:
11754:
11744:
11695:
11654:
11644:
11605:
11555:
11484:
11476:
11448:Nature
11427:
11419:
11384:
11374:
11294:
11259:
11249:
11210:
11200:
11169:
11163:189942
11161:
11126:
11118:
11089:Nature
10942:
10932:
10883:
10875:
10832:
10797:
10789:
10763:Nature
10746:
10738:
10712:Nature
10695:
10685:
10636:
10628:
10593:
10585:
10567:
10540:
10532:
10506:Nature
10489:
10481:
10440:
10430:
10404:Nature
10383:
10348:
10340:
10305:
10297:
10262:
10254:
10193:
10185:
10150:
10142:
10098:
10056:
10048:
10006:
9999:177480
9996:
9957:
9913:
9878:
9871:113121
9868:
9826:
9819:372918
9816:
9751:
9744:166200
9741:
9692:
9651:
9590:
9555:
9520:
9485:
9475:
9437:
9429:
9387:
9379:
9353:Nature
9336:
9328:
9287:
9279:
9269:
9216:
9206:
9157:
9147:
9108:
9098:
9049:
9014:
9004:
8965:
8955:
8914:
8907:100116
8904:
8865:
8857:
8820:
8789:
8782:150442
8779:
8738:
8703:
8696:155275
8693:
8654:
8644:
8579:
8569:
8543:Nature
8518:
8508:
8469:
8441:Nature
8420:
8379:
8330:
8295:
8260:
8225:
8190:
8147:
8106:
8069:
8014:
7973:
7932:
7889:
7834:
7826:
7780:
7770:
7731:
7721:
7674:
7664:
7633:
7592:
7557:
7547:
7461:
7453:
7396:
7386:
7337:
7309:Nature
7288:
7278:
7241:721901
7239:
7231:
7213:
7186:
7178:
7152:Nature
7135:
7125:
7083:
7076:316649
7073:
7034:
6988:
6944:18 May
6919:
6792:
6785:139201
6782:
6733:
6725:
6690:
6655:
6548:
6509:
6467:
6459:
6433:Nature
6399:
6391:
6365:Nature
6238:
6203:
6195:
6157:
6149:
6104:
6096:
6054:
6044:
6005:
5998:525685
5995:
5956:
5921:
5913:
5878:
5824:
5814:
5783:
5717:
5709:
5683:Nature
5666:
5638:Nature
5617:
5607:
5556:
5514:
5506:
5471:
5461:
5412:
5402:
5355:
5345:
5306:
5268:
5260:
5234:Nature
5217:
5182:
5172:
5164:
5123:
5115:
5074:
5039:
5031:
4961:
4938:
4928:
4920:
4855:
4847:
4797:
4787:
4779:
4738:
4708:
4700:
4692:
4657:
4649:
4607:
4599:
4591:
4565:Nature
4534:
4524:
4516:
4465:
4455:
4425:
4400:
4319:
4151:, and
4141:codons
4099:Nature
3901:eggs,
3887:traits
3630:, and
3544:case.
3510:saliva
3463:. The
3421:, and
3358:, and
3352:uracil
3135:, and
2863:, and
2740:enzyme
2655:codons
2627:, and
2529:genome
2470:, and
2417:; the
2415:genome
2405:, and
2375:cancer
2349:, and
2318:cancer
2285:X-rays
2237:adduct
2222:, and
2210:Damage
2021:acidic
1909:D-loop
1838:GFAJ-1
1801:Z form
1751:et al.
1725:, and
1688:, and
1502:Amount
1474:TATAAT
1321:B form
1266:Uracil
1263:Base J
1258:Uracil
1161:genome
1133:uracil
1072:) and
1019:ribose
991:5′-end
987:3′-end
975:carbon
973:(five-
915:of 34
835:or in
809:plants
774:uracil
645:lipids
460:
374:Fields
231:Allele
206:Genome
131:(type
15641:Media
15564:Human
15342:Y RNA
14979:Types
14876:Types
14726:Lists
14606:Italy
14445:Index
13677:(PDF)
13670:(PDF)
13650:(PDF)
13639:S2CID
13601:(PDF)
13561:S2CID
13169:S2CID
13054:(PDF)
13031:(PDF)
13022:See:
13006:(PDF)
12867:S2CID
12751:[
12715:(PDF)
12497:[
12438:See:
12422:S2CID
12349:S2CID
12168:S2CID
12074:S2CID
12012:S2CID
11976:(PDF)
11954:S2CID
11482:S2CID
11425:S2CID
11377:44126
11167:S2CID
10881:S2CID
10795:S2CID
10744:S2CID
10688:48712
10634:S2CID
10591:S2CID
10538:S2CID
10487:S2CID
10346:S2CID
10303:S2CID
10260:S2CID
10191:S2CID
10148:S2CID
10065:(PDF)
10054:S2CID
10026:(PDF)
9933:(PDF)
9446:(PDF)
9435:S2CID
9407:(PDF)
9385:S2CID
9334:S2CID
9285:S2CID
8863:S2CID
8816:–38.
7832:S2CID
7459:S2CID
7425:arXiv
7237:S2CID
7184:S2CID
6917:S2CID
6731:S2CID
6614:(PDF)
6583:(PDF)
6546:S2CID
6476:(PDF)
6465:S2CID
6429:(PDF)
6408:(PDF)
6397:S2CID
6361:(PDF)
6338:(PDF)
6305:(PDF)
6293:(PDF)
6260:(PDF)
6201:S2CID
6155:S2CID
6113:(PDF)
6102:S2CID
6074:(PDF)
5919:S2CID
5715:S2CID
5565:(PDF)
5534:(PDF)
5512:S2CID
5464:22151
5266:S2CID
4706:S2CID
4616:(PDF)
4605:S2CID
4561:(PDF)
4342:"DNA"
3502:semen
3498:blood
3271:RAD51
3129:viral
2989:phage
2946:EcoRV
2547:, or
2533:exons
2427:genes
2196:brain
1727:Z-DNA
1723:B-DNA
1719:A-DNA
1708:Z-DNA
1398:high
983:atoms
963:sugar
913:pitch
859:, in
813:fungi
758:bases
695:sugar
693:), a
589:
251:Index
137:atoms
133:B-DNA
15692:Data
14375:See
14144:ISBN
14122:ISBN
14103:ISBN
14077:ISBN
14054:ISBN
14035:ISBN
14013:ISBN
13994:ISBN
13977:PMID
13928:ISBN
13906:ISBN
13884:ISBN
13863:ISBN
13844:ISBN
13810:PMID
13732:PMID
13631:PMID
13553:PMID
13506:2023
13473:2023
13445:2011
13391:ISBN
13364:PMID
13303:ISBN
13280:2023
13251:PMID
13161:PMID
13126:PMID
13071:PMID
12965:PMID
12916:PMID
12859:PMID
12815:PMID
12727:PMID
12598:PMID
12414:PMID
12341:PMID
12306:PMID
12257:PMID
12210:2022
12160:PMID
12117:PMID
12066:PMID
12004:PMID
11946:PMID
11895:OCLC
11885:ISBN
11862:PMID
11815:ISBN
11788:OCLC
11778:ISBN
11752:PMID
11693:PMID
11652:PMID
11603:PMID
11569:2023
11553:ISSN
11512:2023
11474:PMID
11417:PMID
11382:PMID
11327:2017
11292:PMID
11257:PMID
11208:PMID
11198:ISBN
11159:PMID
11143:Cell
11124:PMID
11116:ISSN
11069:2021
11041:2015
11028:NASA
11008:2011
10975:2011
10963:NASA
10940:PMID
10873:PMID
10830:PMID
10787:PMID
10736:PMID
10693:PMID
10626:PMID
10583:PMID
10530:PMID
10479:PMID
10438:PMID
10381:PMID
10338:PMID
10295:PMID
10252:PMID
10221:2023
10183:PMID
10140:PMID
10096:PMID
10046:PMID
10004:PMID
9955:PMID
9911:PMID
9876:PMID
9824:PMID
9779:2023
9749:PMID
9690:PMID
9672:Cell
9649:PMID
9618:2023
9588:PMID
9553:PMID
9518:PMID
9483:PMID
9473:ISBN
9427:PMID
9377:PMID
9326:PMID
9277:PMID
9214:PMID
9155:PMID
9106:PMID
9047:PMID
9012:PMID
8963:PMID
8912:PMID
8855:PMID
8818:ISBN
8787:PMID
8736:PMID
8701:PMID
8652:PMID
8607:2023
8577:PMID
8516:PMID
8467:PMID
8418:PMID
8377:PMID
8328:PMID
8293:PMID
8258:PMID
8223:PMID
8188:PMID
8145:PMID
8104:ISBN
8067:ISBN
8012:PMID
7971:PMID
7930:PMID
7887:PMID
7824:PMID
7798:Cell
7778:PMID
7729:PMID
7672:PMID
7662:ISBN
7631:PMID
7590:PMID
7555:PMID
7508:2022
7486:2022
7451:PMID
7394:PMID
7335:PMID
7286:PMID
7229:PMID
7203:Cell
7176:PMID
7133:PMID
7081:PMID
7032:PMID
6986:PMID
6968:Cell
6946:2023
6859:2010
6825:2010
6790:PMID
6723:PMID
6688:PMID
6653:PMID
6622:2019
6507:PMID
6457:PMID
6389:PMID
6236:PMID
6193:PMID
6147:PMID
6094:PMID
6052:PMID
6003:PMID
5954:PMID
5911:PMID
5876:PMID
5822:PMID
5781:PMID
5745:2012
5707:PMID
5664:PMID
5615:PMID
5554:PMID
5504:PMID
5469:PMID
5410:PMID
5353:PMID
5304:PMID
5258:PMID
5215:PMID
5180:PMID
5162:ISSN
5121:PMID
5113:ISSN
5072:ISSN
5037:PMID
5029:ISSN
4992:2016
4959:ISBN
4936:PMID
4918:ISSN
4883:2023
4853:PMID
4845:ISSN
4795:PMID
4777:ISSN
4736:ISBN
4698:PMID
4690:ISSN
4655:PMID
4647:ISSN
4597:PMID
4589:ISSN
4532:PMID
4514:ISSN
4463:OCLC
4453:ISBN
4423:ISBN
4398:ISBN
4375:2019
4317:ISBN
4069:and
4017:and
3935:and
3840:and
3832:and
3748:and
3728:and
3720:and
3706:the
3532:and
3514:hair
3506:skin
3443:and
3348:life
3071:are
3022:and
2999:and
2955:are
2941:The
2698:and
2607:and
2587:and
2577:mRNA
2516:and
2488:and
2468:Gene
2365:and
2076:and
1923:and
1767:and
1732:ions
1706:and
1668:and
1626:and
1586:cell
1127:and
1111:and
1088:and
961:and
946:. A
851:and
815:and
657:life
15711:DNA
15619:DNA
14830:RNA
14826:DNA
14476:RNA
14471:DNA
14340:DNA
14335:PDB
14294:DNA
14170:DNA
13967:doi
13955:421
13800:PMC
13722:PMC
13712:doi
13621:doi
13609:421
13543:doi
13531:616
13354:PMC
13344:doi
13241:PMC
13233:doi
13200:doi
13196:280
13153:doi
13116:PMC
13108:doi
13043:doi
12955:PMC
12947:doi
12906:PMC
12898:doi
12851:doi
12807:doi
12691:doi
12660:doi
12629:doi
12588:PMC
12406:doi
12402:122
12333:doi
12296:PMC
12288:doi
12247:PMC
12237:doi
12199:doi
12152:doi
12140:321
12109:doi
12056:hdl
12048:doi
12036:459
11996:doi
11984:440
11936:PMC
11926:doi
11854:doi
11742:PMC
11732:doi
11720:112
11683:doi
11642:PMC
11634:doi
11595:doi
11464:doi
11452:316
11409:doi
11372:PMC
11362:doi
11284:doi
11247:PMC
11239:doi
11190:doi
11151:doi
11106:doi
11094:590
10930:PMC
10920:doi
10908:108
10865:doi
10822:doi
10779:doi
10767:407
10728:doi
10716:362
10683:PMC
10673:doi
10618:doi
10614:292
10575:doi
10522:doi
10510:418
10469:doi
10465:269
10428:PMC
10420:doi
10408:442
10373:doi
10330:doi
10287:doi
10244:doi
10175:doi
10130:doi
10088:doi
10038:doi
9994:PMC
9986:doi
9982:177
9945:doi
9941:271
9903:doi
9866:PMC
9858:doi
9814:PMC
9806:doi
9739:PMC
9729:doi
9717:100
9680:doi
9676:119
9641:doi
9580:doi
9545:doi
9510:doi
9465:doi
9419:doi
9415:293
9369:doi
9357:389
9316:doi
9267:PMC
9259:doi
9204:PMC
9194:doi
9145:PMC
9137:doi
9096:PMC
9086:doi
9039:doi
9035:295
9002:PMC
8994:doi
8953:PMC
8943:doi
8902:PMC
8894:doi
8890:183
8847:doi
8777:PMC
8767:doi
8728:doi
8724:318
8691:PMC
8683:doi
8642:PMC
8634:doi
8630:360
8567:PMC
8559:doi
8547:447
8506:PMC
8498:doi
8457:doi
8445:409
8408:doi
8367:doi
8355:291
8320:doi
8285:doi
8250:doi
8215:doi
8211:258
8180:doi
8168:728
8137:doi
8133:361
8002:doi
7961:doi
7957:272
7922:doi
7910:424
7879:doi
7814:hdl
7806:doi
7768:PMC
7760:doi
7719:PMC
7711:doi
7699:324
7654:doi
7621:doi
7582:doi
7545:PMC
7535:doi
7443:doi
7384:PMC
7376:doi
7364:363
7325:doi
7313:566
7276:PMC
7268:doi
7221:doi
7168:doi
7156:417
7123:PMC
7115:doi
7071:PMC
7063:doi
7022:doi
6976:doi
6909:doi
6882:doi
6780:PMC
6770:doi
6715:doi
6711:184
6680:doi
6676:300
6645:doi
6603:doi
6538:doi
6499:doi
6495:143
6449:doi
6437:171
6381:doi
6369:171
6325:doi
6280:doi
6228:doi
6185:doi
6139:doi
6086:doi
6042:PMC
6034:doi
5993:PMC
5985:doi
5946:doi
5903:doi
5868:doi
5812:PMC
5773:doi
5769:196
5699:doi
5687:290
5654:doi
5642:441
5605:PMC
5595:doi
5546:doi
5496:doi
5459:PMC
5449:doi
5400:PMC
5392:doi
5343:PMC
5335:doi
5296:doi
5250:doi
5238:287
5207:doi
5170:PMC
5152:doi
5103:doi
5099:116
5064:doi
5019:doi
4926:PMC
4910:doi
4837:doi
4785:PMC
4769:doi
4765:120
4682:doi
4639:doi
4635:152
4581:doi
4569:171
4522:PMC
4506:doi
4083:pub
4076:DNA
3987:pub
3971:.
3947:).
3911:RNA
3854:pus
3771:to
3512:or
3346:of
3341:RNA
3116:or
2409:in
2401:in
2295:or
2251:in
2145:of
2005:RNA
2000:RNA
1567:In
1446:G,C
1431:G,C
1163:of
1090:G-C
1086:A-T
1064:),
1056:),
1015:RNA
839:as
831:as
823:as
766:RNA
689:or
601:DNA
201:RNA
197:DNA
15707::
14579:of
14348:.
14327:–
14281:,
14204:34
13975:.
13965:.
13953:.
13949:.
13808:.
13796:10
13794:.
13790:.
13771:.
13752:.
13730:.
13720:.
13710:.
13700:44
13698:.
13694:.
13645:.
13637:.
13629:.
13619:.
13607:.
13603:.
13581:.
13559:.
13551:.
13541:.
13529:.
13523:.
13496:.
13490:.
13462:.
13413:.
13362:.
13352:.
13342:.
13332:39
13330:.
13326:.
13271:.
13249:.
13239:.
13229:36
13227:.
13223:.
13194:.
13190:.
13167:.
13159:.
13147:.
13124:.
13114:.
13104:79
13102:.
13098:.
13065:.
13037:.
13033:.
12988:44
12963:.
12953:.
12943:58
12941:.
12937:.
12914:.
12904:.
12894:27
12892:.
12888:.
12865:.
12857:.
12845:.
12821:.
12813:.
12805:.
12795:80
12793:.
12781:48
12723:38
12721:.
12717:.
12687:40
12685:.
12681:.
12656:42
12650:.
12625:42
12619:.
12596:.
12584:26
12582:.
12578:.
12555:10
12537:,
12514:.
12420:.
12412:.
12400:.
12347:.
12339:.
12329:45
12327:.
12304:.
12294:.
12282:.
12278:.
12255:.
12245:.
12231:.
12227:.
12193:.
12189:.
12166:.
12158:.
12150:.
12138:.
12115:.
12107:.
12095:.
12072:.
12064:.
12054:.
12046:.
12034:.
12010:.
12002:.
11994:.
11982:.
11978:.
11952:.
11944:.
11934:.
11920:.
11916:.
11893:.
11860:.
11852:.
11840:20
11838:.
11813:.
11809:.
11786:.
11750:.
11740:.
11730:.
11718:.
11714:.
11691:.
11677:.
11673:.
11650:.
11640:.
11628:.
11624:.
11601:.
11589:.
11577:^
11559:.
11551:.
11547:.
11480:.
11472:.
11462:.
11450:.
11446:.
11423:.
11415:.
11405:42
11403:.
11380:.
11370:.
11360:.
11350:91
11348:.
11344:.
11317:.
11313:.
11290:.
11280:84
11278:.
11255:.
11245:.
11235:13
11233:.
11229:.
11206:.
11196:.
11165:.
11157:.
11145:.
11122:.
11114:.
11104:.
11092:.
11086:.
11058:.
11031:.
11025:.
10998:.
10992:.
10965:.
10961:.
10938:.
10928:.
10918:.
10906:.
10902:.
10879:.
10871:.
10863:.
10853:54
10851:.
10828:.
10818:13
10816:.
10793:.
10785:.
10777:.
10765:.
10742:.
10734:.
10726:.
10714:.
10691:.
10681:.
10671:.
10661:89
10659:.
10655:.
10632:.
10624:.
10612:.
10589:.
10581:.
10573:.
10561:39
10559:.
10536:.
10528:.
10520:.
10508:.
10485:.
10477:.
10463:.
10459:.
10436:.
10426:.
10418:.
10406:.
10402:.
10379:.
10369:79
10367:.
10344:.
10336:.
10324:.
10301:.
10293:.
10281:.
10258:.
10250:.
10238:.
10212:.
10189:.
10181:.
10171:50
10169:.
10146:.
10138:.
10124:.
10120:.
10108:^
10094:.
10084:74
10082:.
10052:.
10044:.
10034:71
10032:.
10028:.
10002:.
9992:.
9980:.
9976:.
9953:.
9939:.
9935:.
9909:.
9899:33
9897:.
9874:.
9864:.
9854:28
9852:.
9848:.
9836:^
9822:.
9812:.
9802:57
9800:.
9796:.
9770:.
9747:.
9737:.
9727:.
9715:.
9711:.
9688:.
9674:.
9670:.
9647:.
9637:69
9635:.
9609:.
9586:.
9576:34
9574:.
9551:.
9541:10
9539:.
9516:.
9506:29
9504:.
9481:.
9471:.
9441:.
9433:.
9425:.
9413:.
9409:.
9383:.
9375:.
9367:.
9355:.
9332:.
9324:.
9312:56
9310:.
9306:.
9283:.
9275:.
9265:.
9255:54
9253:.
9249:.
9212:.
9202:.
9192:.
9180:.
9176:.
9153:.
9143:.
9133:29
9131:.
9127:.
9104:.
9094:.
9084:.
9072:.
9068:.
9045:.
9033:.
9010:.
9000:.
8990:77
8988:.
8984:.
8961:.
8951:.
8937:.
8933:.
8910:.
8900:.
8888:.
8884:.
8861:.
8853:.
8843:29
8841:.
8814:25
8785:.
8775:.
8761:.
8757:.
8734:.
8722:.
8699:.
8689:.
8679:12
8677:.
8673:.
8650:.
8640:.
8628:.
8624:.
8598:.
8575:.
8565:.
8557:.
8545:.
8541:.
8514:.
8504:.
8494:95
8492:.
8488:.
8465:.
8455:.
8443:.
8439:.
8416:.
8404:96
8402:.
8398:.
8375:.
8365:.
8353:.
8349:.
8326:.
8314:.
8291:.
8281:28
8279:.
8256:.
8246:59
8244:.
8221:.
8209:.
8186:.
8178:.
8166:.
8143:.
8131:.
8112:.
8075:.
8061:.
8039:.
8033:.
8010:.
7998:22
7996:.
7992:.
7969:.
7955:.
7951:.
7928:.
7920:.
7908:.
7885:.
7875:42
7873:.
7830:.
7822:.
7812:.
7802:75
7800:.
7776:.
7766:.
7756:28
7754:.
7750:.
7727:.
7717:.
7709:.
7697:.
7693:.
7670:.
7660:.
7629:.
7617:16
7615:.
7611:.
7588:.
7578:31
7576:.
7553:.
7543:.
7529:.
7525:.
7457:.
7449:.
7441:.
7433:.
7421:75
7419:.
7415:.
7392:.
7382:.
7374:.
7362:.
7356:.
7333:.
7323:.
7311:.
7307:.
7284:.
7274:.
7264:38
7262:.
7258:.
7235:.
7227:.
7219:.
7207:97
7205:.
7182:.
7174:.
7166:.
7154:.
7131:.
7121:.
7111:34
7109:.
7105:.
7093:^
7079:.
7069:.
7059:11
7057:.
7053:.
7030:.
7018:12
7016:.
7012:.
6998:^
6984:.
6972:43
6970:.
6966:.
6954:^
6937:.
6915:.
6907:.
6888:.
6880:.
6876:.
6849:.
6845:.
6833:^
6815:.
6811:.
6788:.
6778:.
6768:.
6758:99
6756:.
6752:.
6729:.
6721:.
6709:.
6686:.
6674:.
6651:.
6641:44
6639:.
6601:.
6591:34
6589:.
6585:.
6544:.
6534:42
6532:.
6528:.
6505:.
6493:.
6471:.
6463:.
6455:.
6447:.
6435:.
6431:.
6416:^
6403:.
6395:.
6387:.
6379:.
6367:.
6363:.
6348:^
6333:.
6323:.
6311:.
6307:.
6288:.
6278:.
6266:.
6262:.
6234:.
6222:.
6199:.
6191:.
6179:.
6167:^
6153:.
6145:.
6135:70
6133:.
6121:^
6100:.
6092:.
6080:.
6076:.
6050:.
6040:.
6028:.
6024:.
6001:.
5991:.
5981:14
5979:.
5975:.
5952:.
5942:29
5940:.
5917:.
5909:.
5899:93
5897:.
5874:.
5864:21
5862:.
5820:.
5806:.
5802:.
5779:.
5767:.
5753:^
5713:.
5705:.
5697:.
5685:.
5662:.
5652:.
5640:.
5636:.
5613:.
5603:.
5591:12
5589:.
5585:.
5573:^
5560:.
5552:.
5542:43
5540:.
5536:.
5510:.
5502:.
5492:16
5490:.
5467:.
5457:.
5447:.
5437:96
5435:.
5431:.
5408:.
5398:.
5390:.
5380:78
5378:.
5374:.
5351:.
5341:.
5331:99
5329:.
5325:.
5302:.
5292:53
5290:.
5278:^
5264:.
5256:.
5248:.
5236:.
5213:.
5203:57
5201:.
5178:.
5168:.
5160:.
5146:.
5142:.
5119:.
5111:.
5097:.
5093:.
5070:.
5060:47
5058:.
5035:.
5027:.
5015:67
5013:.
5009:.
4934:.
4924:.
4916:.
4906:34
4904:.
4900:.
4874:.
4851:.
4843:.
4835:.
4825:59
4823:.
4811:^
4793:.
4783:.
4775:.
4763:.
4759:.
4718:^
4704:.
4696:.
4688:.
4676:.
4653:.
4645:.
4633:.
4611:.
4603:.
4595:.
4587:.
4579:.
4567:.
4563:.
4544:^
4530:.
4520:.
4512:.
4504:.
4492:.
4488:.
4469:.
4461:.
4437:^
4366:.
4348:.
4344:.
4325:.
4292:.
4159:.
4147:,
4033:.
3978:A
3864:.
3775:.
3716:.
3626:,
3508:,
3504:,
3500:,
3479:.
3417:,
3400:.
3354:,
3327:,
3120:.
3026:.
3003:.
2888:.
2859:,
2623:,
2611:.
2565:.
2466:,
2462:,
2458:,
2429:.
2345:,
2341:,
2308:,
2304:,
2271:,
2232:A
2218:,
2206:.
2011:.
1911:.
1721:,
1702:,
1684:,
1672:.
1606:A
1485:AT
1470:AT
1466:GC
1462:GC
1442:GC
1435:pH
1404:GC
1378:AT
1368:GC
1323:.
1107:,
1095:.
1032:).
997:.
954:.
921:nm
893:3′
889:5'
811:,
807:,
788:.
685:,
681:,
659:.
643:,
599:;
586:-/
583:eɪ
571:,-
562:iː
550:uː
547:nj
544:oʊ
538:aɪ
514:iː
15627::
15599::
15233:)
15177:e
15170:t
15163:v
14790:e
14783:t
14776:v
14406:e
14399:t
14392:v
14252:)
14244:(
14239:)
14208:)
14201:(
14152:.
14130:.
14111:.
14085:.
14062:.
14043:.
14021:.
14002:.
13983:.
13969::
13961::
13936:.
13914:.
13892:.
13871:.
13852:.
13816:.
13773:1
13756:.
13738:.
13714::
13706::
13623::
13615::
13585:.
13567:.
13545::
13537::
13508:.
13475:.
13447:.
13399:.
13370:.
13346::
13338::
13311:.
13282:.
13257:.
13235::
13208:.
13202::
13175:.
13155::
13149:6
13132:.
13110::
13045::
13039:6
12971:.
12949::
12922:.
12900::
12873:.
12853::
12847:2
12809::
12801::
12767:7
12733:.
12699:.
12693::
12666:.
12662::
12635:.
12631::
12604:.
12539:7
12516:6
12486:5
12469:4
12452:3
12428:.
12408::
12376:4
12355:.
12335::
12312:.
12290::
12284:8
12263:.
12239::
12233:3
12212:.
12201::
12195:1
12174:.
12154::
12146::
12123:.
12111::
12103::
12097:4
12080:.
12058::
12050::
12042::
12018:.
11998::
11990::
11960:.
11928::
11922:2
11901:.
11868:.
11856::
11823:.
11794:.
11758:.
11734::
11726::
11699:.
11685::
11679:3
11658:.
11636::
11630:5
11609:.
11597::
11591:1
11571:.
11514:.
11488:.
11466::
11458::
11431:.
11411::
11388:.
11364::
11356::
11329:.
11298:.
11286::
11263:.
11241::
11214:.
11192::
11173:.
11153::
11147:9
11130:.
11108::
11100::
11071:.
11043:.
11010:.
10977:.
10946:.
10922::
10914::
10887:.
10867::
10859::
10836:.
10824::
10801:.
10781::
10773::
10750:.
10730::
10722::
10699:.
10675::
10667::
10640:.
10620::
10597:.
10577::
10544:.
10524::
10516::
10493:.
10471::
10444:.
10422::
10414::
10387:.
10375::
10352:.
10332::
10326:7
10309:.
10289::
10283:7
10266:.
10246::
10240:2
10223:.
10197:.
10177::
10154:.
10132::
10126:8
10102:.
10090::
10040::
10010:.
9988::
9961:.
9947::
9917:.
9905::
9882:.
9860::
9830:.
9808::
9781:.
9755:.
9731::
9723::
9696:.
9682::
9655:.
9643::
9620:.
9594:.
9582::
9559:.
9547::
9524:.
9512::
9489:.
9467::
9421::
9391:.
9371::
9363::
9340:.
9318::
9291:.
9261::
9234:.
9220:.
9196::
9188::
9182:7
9161:.
9139::
9112:.
9088::
9080::
9074:7
9053:.
9041::
9018:.
8996::
8969:.
8945::
8939:4
8918:.
8896::
8869:.
8849::
8826:.
8793:.
8769::
8763:2
8742:.
8730::
8707:.
8685::
8658:.
8636::
8609:.
8583:.
8561::
8553::
8522:.
8500::
8473:.
8459::
8451::
8424:.
8410::
8383:.
8369::
8361::
8334:.
8322::
8316:7
8299:.
8287::
8264:.
8252::
8229:.
8217::
8194:.
8182::
8174::
8151:.
8139::
8018:.
8004::
7977:.
7963::
7936:.
7924::
7916::
7893:.
7881::
7838:.
7816::
7808::
7784:.
7762::
7735:.
7713::
7705::
7678:.
7656::
7637:.
7623::
7596:.
7584::
7561:.
7537::
7531:3
7510:.
7488:.
7465:.
7445::
7437::
7427::
7400:.
7378::
7370::
7341:.
7327::
7319::
7292:.
7270::
7243:.
7223::
7190:.
7170::
7162::
7139:.
7117::
7087:.
7065::
7038:.
7024::
6992:.
6978::
6948:.
6923:.
6911::
6884::
6861:.
6827:.
6796:.
6772::
6764::
6737:.
6717::
6694:.
6682::
6659:.
6647::
6624:.
6605::
6597::
6552:.
6540::
6513:.
6501::
6451::
6443::
6383::
6375::
6327::
6319::
6313:6
6282::
6274::
6268:6
6242:.
6230::
6224:6
6207:.
6187::
6181:3
6161:.
6141::
6088::
6082:7
6058:.
6036::
6030:7
6009:.
5987::
5960:.
5948::
5925:.
5905::
5882:.
5870::
5828:.
5808:9
5787:.
5775::
5747:.
5721:.
5701::
5693::
5670:.
5656::
5648::
5621:.
5597::
5548::
5518:.
5498::
5475:.
5451::
5443::
5416:.
5394::
5386::
5359:.
5337::
5310:.
5298::
5272:.
5252::
5244::
5221:.
5209::
5186:.
5154::
5148:9
5127:.
5105::
5078:.
5066::
5043:.
5021::
4994:.
4967:.
4942:.
4912::
4885:.
4859:.
4839::
4831::
4771::
4744:.
4712:.
4684::
4678:4
4661:.
4641::
4583::
4575::
4538:.
4508::
4500::
4494:6
4431:.
4406:.
4377:.
4107:"
3844:.
1704:B
1700:A
1530:.
1494:m
1492:T
1456:m
1454:T
1137:U
1135:(
1129:T
1125:C
1113:G
1109:A
1078:T
1076:(
1070:G
1068:(
1062:C
1060:(
1054:A
1052:(
847:(
803:(
677:(
580:l
577:k
574:ˌ
568:k
565:ɪ
559:l
556:k
553:ˌ
541:b
535:r
532:ˌ
529:ɪ
526:s
523:k
520:ɒ
517:ˈ
511:d
508:/
504:(
490:e
483:t
476:v
349:)
340:(
285:)
281:(
85:)
81:(
62:)
58:(
29:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.