78:. There are many shapes of cladograms but they all have lines that branch off from other lines. The lines can be traced back to where they branch off. These branching off points represent a hypothetical ancestor (not an actual entity) which can be inferred to exhibit the traits shared among the terminal taxa above it. This hypothetical ancestor might then provide clues about the order of evolution of various features, adaptation, and other evolutionary narratives about ancestors. Although traditionally such cladograms were generated largely on the basis of morphological characters,
338:
algorithms, such as UPGMA and
Neighbor-Joining, group by overall similarity, and treat both synapomorphies and symplesiomorphies as evidence of grouping, The resulting diagrams are phenograms, not cladograms, Similarly, the results of model-based methods (Maximum Likelihood or Bayesian approaches) that take into account both branching order and "branch length," count both synapomorphies and autapomorphies as evidence for or against grouping, The diagrams resulting from those sorts of analysis are not cladograms, either.
39:
2123:
204:
31:
157:
271:. If a bird, bat, and a winged insect were scored for the character, "presence of wings", a homoplasy would be introduced into the dataset, and this could potentially confound the analysis, possibly resulting in a false hypothesis of relationships. Of course, the only reason a homoplasy is recognizable in the first place is because there are other characters that imply a pattern of relationships that reveal its homoplastic distribution.
285:
104:
239:. States shared between the outgroup and some members of the in-group are symplesiomorphies; states that are present only in a subset of the in-group are synapomorphies. Note that character states unique to a single terminal (autapomorphies) do not provide evidence of grouping. The choice of an outgroup is a crucial step in cladistic analysis because different outgroups can produce trees with profoundly different topologies.
1800:
256:
should not be included as a character in a phylogenetic analysis as they do not contribute anything to our understanding of relationships. However, homoplasy is often not evident from inspection of the character itself (as in DNA sequence, for example), and is then detected by its incongruence (unparsimonious distribution) on a most-parsimonious cladogram. Note that characters that are homoplastic may still contain
2135:
1706:
214:(as shown in this heuristic example), but must be inferred from the pattern of shared states observed in the terminals. Given that each terminal in this example has a unique state, in reality we would not be able to infer anything conclusive about the ancestral states (other than the fact that the existence of unobserved states "A" and "C" would be unparsimonious inferences!)
466:
Besides reflecting the amount of homoplasy, the metric also reflects the number of taxa in the dataset, (to a lesser extent) the number of characters in a dataset, the degree to which each character carries phylogenetic information, and the fashion in which additive characters are coded, rendering it
255:
than common ancestry. The two main types of homoplasy are convergence (evolution of the "same" character in at least two distinct lineages) and reversion (the return to an ancestral character state). Characters that are obviously homoplastic, such as white fur in different lineages of Arctic mammals,
665:
The retention index (RI) was proposed as an improvement of the CI "for certain applications" This metric also purports to measure of the amount of homoplasy, but also measures how well synapomorphies explain the tree. It is calculated taking the (maximum number of changes on a tree minus the number
337:
A cladogram is the diagrammatic result of an analysis, which groups taxa on the basis of synapomorphies alone. There are many other phylogenetic algorithms that treat data somewhat differently, and result in phylogenetic trees that look like cladograms but are not cladograms. For example, phenetic
406:
Because of the astronomical number of possible cladograms, algorithms cannot guarantee that the solution is the overall best solution. A nonoptimal cladogram will be selected if the program settles on a local minimum rather than the desired global minimum. To help solve this problem, many cladogram
431:
The incongruence length difference test (ILD) is a measurement of how the combination of different datasets (e.g. morphological and molecular, plastid and nuclear genes) contributes to a longer tree. It is measured by first calculating the total tree length of each partition and summing them. Then
678:
This measures the amount of homoplasy observed on a tree relative to the maximum amount of homoplasy that could theoretically be present – 1 − (observed homoplasy excess) / (maximum homoplasy excess). A value of 1 indicates no homoplasy; 0 represents as much homoplasy as there would be in a
399:
Algorithms that perform optimization tasks (such as building cladograms) can be sensitive to the order in which the input data (the list of species and their characteristics) is presented. Inputting the data in various orders can cause the same algorithm to produce different "best" cladograms. In
454:
The consistency index (CI) measures the consistency of a tree to a set of data – a measure of the minimum amount of homoplasy implied by the tree. It is calculated by counting the minimum number of changes in a dataset and dividing it by the actual number of changes needed for the cladogram. A
669:
The rescaled consistency index (RC) is obtained by multiplying the CI by the RI; in effect this stretches the range of the CI such that its minimum theoretically attainable value is rescaled to 0, with its maximum remaining at 1. The homoplasy index (HI) is simply 1 − CI.
365:
Some algorithms are useful only when the characteristic data are molecular (DNA, RNA); other algorithms are useful only when the characteristic data are morphological. Other algorithms can be used when the characteristic data includes both molecular and morphological data.
69:
because it does not show how ancestors are related to descendants, nor does it show how much they have changed, so many differing evolutionary trees can be consistent with the same cladogram. A cladogram uses lines that branch off in different directions ending at a
361:
In general, cladogram generation algorithms must be implemented as computer programs, although some algorithms can be performed manually when the data sets are modest (for example, just a few species and a couple of characteristics).
432:
replicates are made by making randomly assembled partitions consisting of the original partitions. The lengths are summed. A p value of 0.01 is obtained for 100 replicates if 99 replicates have longer combined tree lengths.
173:, unicellular, etc.) or molecular (DNA, RNA, or other genetic information). Prior to the advent of DNA sequencing, cladistic analysis primarily used morphological data. Behavioral data (for animals) may also be used.
655:
1658:
Hoyal
Cuthill, Jennifer (2015). "The size of the character state space affects the occurrence and detection of homoplasy: Modelling the probability of incompatibility for unordered phylogenetic characters".
474:
occupies a range from 1 to 1/ in binary characters with an even state distribution; its minimum value is larger when states are not evenly spread. In general, for a binary or non-binary character with
210:
in cladistics. This diagram indicates "A" and "C" as ancestral states, and "B", "D" and "E" as states that are present in terminal taxa. Note that in practice, ancestral conditions are not known
679:
fully random dataset, and negative values indicate more homoplasy still (and tend only to occur in contrived examples). The HER is presented as the best measure of homoplasy currently available.
184:
has become a more and more popular way to infer phylogenetic hypotheses. Using a parsimony criterion is only one of several methods to infer a phylogeny from molecular data. Approaches such as
446:
Some measures attempt to measure the amount of homoplasy in a dataset with reference to a tree, though it is not necessarily clear precisely what property these measures aim to quantify
188:, which incorporate explicit models of sequence evolution, are non-Hennigian ways to evaluate sequence data. Another powerful method of reconstructing phylogenies is the use of genomic
418:
is the direction of the base (or root) of a rooted phylogenetic tree or cladogram. A basal clade is the earliest clade (of a given taxonomic rank) to branch within a larger clade.
403:
Using different algorithms on a single data set can sometimes yield different "best" cladograms, because each algorithm may have a unique definition of what is "best".
513:
235:), because only synapomorphic character states provide evidence of grouping. This determination is usually done by comparison to the character states of one or more
1480:
Archie, J. W. (1989). "Homoplasy Excess Ratios: New
Indices for Measuring Levels of Homoplasy in Phylogenetic Systematics and a Critique of the Consistency Index".
196:
that plagues sequence data. They are also generally assumed to have a low incidence of homoplasies because it was once thought that their integration into the
1522:"A formula for maximum possible steps in multistate characters: Isolating matrix parameter effects on measures of evolutionary convergence"
1448:
Archie, J. W.; Felsenstein, J. (1993). "The Number of
Evolutionary Steps on Random and Minimum Length Trees for Random Evolutionary Data".
666:
of changes on the tree), and dividing by the (maximum number of changes on the tree minus the minimum number of changes in the dataset).
522:
263:
A well-known example of homoplasy due to convergent evolution would be the character, "presence of wings". Although the wings of birds,
1738:
169:
The characteristics used to create a cladogram can be roughly categorized as either morphological (synapsid skull, warm blooded,
918:
354:
to measure how consistent a candidate cladogram is with the data. Most cladogram algorithms use the mathematical techniques of
1269:
1187:
1078:
1024:
969:
944:
880:"Tree thinking, time and topology: Comments on the interpretation of tree diagrams in evolutionary/phylogenetic systematics"
1933:
396:
for a specific kind of cladogram generation algorithm and sometimes as an umbrella term for all phylogenetic algorithms.
1893:
1394:
1361:
393:
378:
324:
143:
1973:
90:
are now very commonly used in the generation of cladograms, either on their own or in combination with morphology.
1978:
1911:
806:
2139:
1988:
1918:
306:
125:
793:
Dayrat, Benoît (Summer 2005). "Ancestor-Descendant
Relationships and the Reconstruction of the Tree of Life".
1094:
Kalersjo, Mari; Albert, Victor A.; Farris, James S. (1999). "Homoplasy
Increases Phylogenetic Structure".
1898:
1805:
1761:
1731:
1710:
87:
355:
302:
121:
17:
2088:
2014:
1336:
Archie, James W. (1996). "Measures of
Homoplasy". In Sanderson, Michael J.; Hufford, Larry (eds.).
1968:
1766:
1058:
295:
114:
2166:
2127:
1851:
1724:
1261:
1255:
1070:
189:
477:
2103:
1781:
698:
415:
181:
1345:
1337:
1285:
Nixon, Kevin C. (1999). "The
Parsimony Ratchet, a New Method for Rapid Parsimony Analysis".
1062:
400:
these situations, the user should input the data in various orders and compare the results.
1940:
1846:
1668:
1569:
Sanderson, M. J.; Donoghue, M. J. (1989). "Patterns of variations in levels of homoplasy".
1413:
Kluge, A. G.; Farris, J. S. (1969). "Quantitative
Phyletics and the Evolution of Anurans".
1377:
Chang, Joseph T.; Kim, Junhyong (1996). "The
Measurement of Homoplasy: A Stochastic View".
1212:
441:
351:
267:, and insects serve the same function, each evolved independently, as can be seen by their
193:
75:
8:
1983:
1865:
408:
257:
1672:
1216:
998:
1906:
1860:
1776:
1640:
1628:
1586:
1551:
1497:
1430:
1386:
1353:
1310:
1298:
1236:
1160:
1111:
1107:
910:
818:
810:
775:
767:
735:
718:
386:
382:
185:
856:
426:
2161:
1834:
1684:
1632:
1594:
1543:
1538:
1521:
1390:
1357:
1302:
1265:
1228:
1183:
1152:
1074:
1063:
1020:
965:
940:
902:
898:
860:
840:
66:
1644:
1555:
1314:
1164:
1115:
914:
879:
822:
779:
411:
approach to increase the likelihood that the selected cladogram is the optimal one.
1923:
1877:
1676:
1624:
1578:
1533:
1489:
1457:
1422:
1382:
1349:
1294:
1240:
1220:
1142:
1103:
994:
894:
852:
802:
759:
750:
Foote, Mike (Spring 1996). "On the Probability of Ancestors in the Fossil Record".
730:
374:
1338:
2024:
1615:
Farris, J. S. (1989). "The retention index and the rescaled consistency index".
200:
was entirely random; this seems at least sometimes not to be the case, however.
1998:
1680:
177:
51:
763:
2155:
1993:
1963:
1870:
1747:
1520:
Hoyal Cuthill, Jennifer F.; Braddy, Simon J.; Donoghue, Philip C. J. (2010).
688:
370:
27:
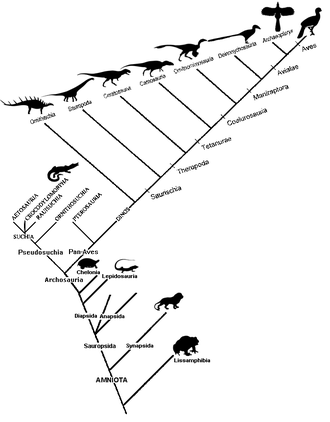
Diagram used to show relations among groups of organisms with common origins
2098:
2044:
2039:
2034:
2019:
1827:
1822:
1688:
1636:
1598:
1547:
1461:
1306:
1156:
1040:
1012:
906:
864:
231:
225:
1232:
251:
is a character state that is shared by two or more taxa due to some cause
2093:
1786:
843:(2001). "Intraspecific gene genealogies: Trees grafting into networks".
1771:
1590:
1501:
1434:
814:
771:
693:
369:
Algorithms for cladograms or other types of phylogenetic trees include
347:
309: in this section. Unsourced material may be challenged and removed.
203:
128: in this section. Unsourced material may be challenged and removed.
62:
38:
1147:
1130:
156:
2108:
2072:
2067:
2062:
1957:
1224:
455:
consistency index can also be calculated for an individual character
248:
207:
170:
30:
1582:
1493:
1426:
284:
103:
1203:
Stewart, Caro-Beth (1993). "The powers and pitfalls of parsimony".
650:{\displaystyle (n.states-1)/(n.taxa-\lceil n.taxa/n.states\rceil )}
427:
Incongruence length difference test (or partition homogeneity test)
65:
to show relations among organisms. A cladogram is not, however, an
350:
available to identify the "best" cladogram. Most algorithms use a
268:
1716:
223:
Researchers must decide which character states are "ancestral" (
1799:
1705:
197:
1928:
1856:
985:
Wenzel, John W. (1992). "Behavioral homology and phylogeny".
71:
1519:
807:
10.1666/0094-8373(2005)031[0347:aratro]2.0.co;2
723:
Journal of Zoological Systematics and Evolutionary Research
1180:
Cladistics: The Theory and Practice of Parsimony Analysis
264:
83:
79:
192:, which are thought to be less prone to the problem of
164:
525:
480:
1795:
1610:
1608:
1475:
1473:
1471:
1093:
218:
1408:
1406:
937:
Biological Systematics: Principles and Applications
649:
507:
1605:
1568:
1515:
1513:
1511:
1468:
1447:
962:Techniques in Molecular Systematics and Evolution
719:"Cladistic analysis or cladistic classification?"
34:A horizontal cladogram, with the root to the left
2153:
1403:
42:Two vertical cladograms, the root at the bottom
1508:
838:
1732:
1657:
1330:
1328:
1326:
1324:
1257:Cladistics: A Practical Course in Systematics
1057:
641:
591:
1412:
1739:
1725:
1321:
274:
1537:
1146:
734:
673:
325:Learn how and when to remove this message
144:Learn how and when to remove this message
93:
1376:
1177:
202:
155:
37:
29:
1202:
959:
14:
2154:
1614:
1479:
1335:
1131:"What is a cladogram and what is not?"
1128:
1065:Developmental Plasticity and Evolution
1039:
1011:
984:
953:
877:
792:
435:
341:
1720:
1284:
1253:
978:
934:
871:
786:
749:
2134:
834:
832:
743:
716:
449:
307:adding citations to reliable sources
278:
126:adding citations to reliable sources
97:
999:10.1146/annurev.es.23.110192.002045
710:
165:Molecular versus morphological data
24:
1629:10.1111/j.1096-0031.1989.tb00573.x
1299:10.1111/j.1096-0031.1999.tb00277.x
1108:10.1111/j.1096-0031.1999.tb00400.x
736:10.1111/j.1439-0469.1974.tb00160.x
660:
392:Biologists sometimes use the term
61:"character") is a diagram used in
25:
2178:
1746:
1698:
928:
845:Trends in Ecology & Evolution
829:
219:Plesiomorphies and synapomorphies
2133:
2122:
2121:
1974:Phylogenetic comparative methods
1798:
1704:
1539:10.1111/j.1096-0031.2009.00270.x
1387:10.1016/b978-012618030-5/50009-5
1354:10.1016/B978-012618030-5/50008-3
924:from the original on 2017-09-21.
899:10.1111/j.1096-0031.2012.00423.x
283:
102:
1979:Phylogenetic niche conservatism
1651:
1562:
1441:
1370:
1278:
1247:
1196:
1171:
1122:
1087:
1069:. Oxford Univ. Press. pp.
1051:
1047:. University of Illinois Press.
1033:
294:needs additional citations for
180:has become cheaper and easier,
113:needs additional citations for
1661:Journal of Theoretical Biology
1450:Theoretical Population Biology
1260:. Oxford Univ. Press. p.
1005:
644:
567:
559:
526:
242:
74:, a group of organisms with a
13:
1:
857:10.1016/S0169-5347(00)02026-7
704:
421:
1129:Brower, Andrew V.Z. (2016).
7:
1899:Phylogenetic reconciliation
1806:Evolutionary biology portal
1762:Computational phylogenetics
1182:. Oxford University Press.
682:
519:occupies a range from 1 to
88:computational phylogenetics
10:
2183:
1681:10.1016/j.jtbi.2014.10.033
935:Schuh, Randall T. (2000).
439:
2117:
2089:Phylogenetic nomenclature
2081:
2055:
2007:
1949:
1886:
1815:
1793:
1754:
764:10.1017/S0094837300016146
229:) and which are derived (
1059:West-Eberhard, Mary Jane
1045:Phylogenetic Systematics
508:{\displaystyle n.states}
1969:Molecular phylogenetics
1919:Distance-matrix methods
1767:Molecular phylogenetics
275:What is not a cladogram
190:retrotransposon markers
1989:Phylogenetics software
1903:Probabilistic methods
1852:Long branch attraction
1462:10.1006/tpbi.1993.1003
1178:Kitching, Ian (1998).
878:Podani, János (2013).
674:Homoplasy Excess Ratio
651:
509:
215:
161:
94:Generating a cladogram
43:
35:
1782:Evolutionary taxonomy
1254:Foley, Peter (1993).
1017:Molecular Systematics
987:Annu. Rev. Ecol. Syst
960:DeSalle, Rob (2002).
699:Basal (phylogenetics)
652:
510:
440:Further information:
206:
182:molecular systematics
159:
41:
33:
1941:Three-taxon analysis
1847:Phylogenetic network
1713:at Wikimedia Commons
1381:. pp. 189–203.
717:Mayr, Ernst (2009).
523:
478:
442:Convergent evolution
303:improve this article
122:improve this article
86:sequencing data and
76:last common ancestor
1984:Phylogenetic signal
1673:2015JThBi.366...24H
1217:1993Natur.361..603S
467:unfit for purpose.
436:Measuring homoplasy
409:simulated annealing
342:Cladogram selection
258:phylogenetic signal
1912:Bayesian inference
1907:Maximum likelihood
1482:Systematic Zoology
1415:Systematic Zoology
841:Crandall, Keith A.
647:
505:
387:Bayesian inference
383:maximum likelihood
358:and minimization.
346:There are several
216:
186:maximum likelihood
162:
160:Cladogram of birds
44:
36:
2149:
2148:
1894:Maximum parsimony
1887:Inference methods
1835:Phylogenetic tree
1709:Media related to
1271:978-0-19-857766-9
1189:978-0-19-850138-1
1148:10.1111/cla.12144
1080:978-0-19-512235-0
1026:978-0-87893-282-5
971:978-3-7643-6257-7
946:978-0-8014-3675-8
450:Consistency index
407:algorithms use a
335:
334:
327:
154:
153:
146:
67:evolutionary tree
16:(Redirected from
2174:
2137:
2136:
2125:
2124:
1924:Neighbor-joining
1878:Ghost population
1808:
1803:
1802:
1741:
1734:
1727:
1718:
1717:
1708:
1693:
1692:
1655:
1649:
1648:
1612:
1603:
1602:
1577:(8): 1781–1795.
1566:
1560:
1559:
1541:
1517:
1506:
1505:
1477:
1466:
1465:
1445:
1439:
1438:
1410:
1401:
1400:
1374:
1368:
1367:
1343:
1332:
1319:
1318:
1282:
1276:
1275:
1251:
1245:
1244:
1225:10.1038/361603a0
1200:
1194:
1193:
1175:
1169:
1168:
1150:
1126:
1120:
1119:
1091:
1085:
1084:
1068:
1055:
1049:
1048:
1037:
1031:
1030:
1009:
1003:
1002:
982:
976:
975:
957:
951:
950:
932:
926:
925:
923:
884:
875:
869:
868:
836:
827:
826:
790:
784:
783:
747:
741:
740:
738:
714:
656:
654:
653:
648:
616:
566:
514:
512:
511:
506:
375:neighbor-joining
330:
323:
319:
316:
310:
287:
279:
149:
142:
138:
135:
129:
106:
98:
21:
2182:
2181:
2177:
2176:
2175:
2173:
2172:
2171:
2152:
2151:
2150:
2145:
2113:
2077:
2051:
2025:Symplesiomorphy
2003:
1945:
1882:
1811:
1804:
1797:
1791:
1755:Relevant fields
1750:
1745:
1701:
1696:
1656:
1652:
1613:
1606:
1583:10.2307/2409392
1567:
1563:
1518:
1509:
1494:10.2307/2992286
1478:
1469:
1446:
1442:
1427:10.2307/2412407
1411:
1404:
1397:
1375:
1371:
1364:
1333:
1322:
1283:
1279:
1272:
1252:
1248:
1211:(6413): 603–7.
1201:
1197:
1190:
1176:
1172:
1127:
1123:
1092:
1088:
1081:
1056:
1052:
1038:
1034:
1027:
1010:
1006:
983:
979:
972:
958:
954:
947:
933:
929:
921:
882:
876:
872:
839:Posada, David;
837:
830:
791:
787:
748:
744:
715:
711:
707:
685:
676:
663:
661:Retention index
612:
562:
524:
521:
520:
518:
479:
476:
475:
473:
462:
452:
444:
438:
429:
424:
344:
331:
320:
314:
311:
300:
288:
277:
245:
221:
167:
150:
139:
133:
130:
119:
107:
96:
28:
23:
22:
15:
12:
11:
5:
2180:
2170:
2169:
2164:
2147:
2146:
2144:
2143:
2131:
2118:
2115:
2114:
2112:
2111:
2106:
2101:
2096:
2091:
2085:
2083:
2079:
2078:
2076:
2075:
2070:
2065:
2059:
2057:
2053:
2052:
2050:
2049:
2048:
2047:
2042:
2037:
2029:
2028:
2027:
2022:
2011:
2009:
2005:
2004:
2002:
2001:
1999:Phylogeography
1996:
1991:
1986:
1981:
1976:
1971:
1966:
1961:
1953:
1951:
1950:Current topics
1947:
1946:
1944:
1943:
1938:
1937:
1936:
1931:
1926:
1916:
1915:
1914:
1909:
1901:
1896:
1890:
1888:
1884:
1883:
1881:
1880:
1875:
1874:
1873:
1863:
1854:
1849:
1844:
1843:
1842:
1832:
1831:
1830:
1819:
1817:
1816:Basic concepts
1813:
1812:
1810:
1809:
1794:
1792:
1790:
1789:
1784:
1779:
1774:
1769:
1764:
1758:
1756:
1752:
1751:
1744:
1743:
1736:
1729:
1721:
1715:
1714:
1700:
1699:External links
1697:
1695:
1694:
1650:
1623:(4): 417–419.
1604:
1561:
1507:
1488:(3): 253–269.
1467:
1440:
1402:
1395:
1369:
1362:
1320:
1293:(4): 407–414.
1277:
1270:
1246:
1195:
1188:
1170:
1141:(5): 573–576.
1121:
1086:
1079:
1050:
1032:
1025:
1004:
977:
970:
964:. Birkhauser.
952:
945:
927:
893:(3): 315–327.
870:
828:
785:
742:
708:
706:
703:
702:
701:
696:
691:
684:
681:
675:
672:
662:
659:
646:
643:
640:
637:
634:
631:
628:
625:
622:
619:
615:
611:
608:
605:
602:
599:
596:
593:
590:
587:
584:
581:
578:
575:
572:
569:
565:
561:
558:
555:
552:
549:
546:
543:
540:
537:
534:
531:
528:
516:
504:
501:
498:
495:
492:
489:
486:
483:
471:
460:
451:
448:
437:
434:
428:
425:
423:
420:
416:basal position
343:
340:
333:
332:
291:
289:
282:
276:
273:
244:
241:
232:synapomorphies
226:plesiomorphies
220:
217:
178:DNA sequencing
166:
163:
152:
151:
110:
108:
101:
95:
92:
26:
9:
6:
4:
3:
2:
2179:
2168:
2167:Phylogenetics
2165:
2163:
2160:
2159:
2157:
2142:
2141:
2132:
2130:
2129:
2120:
2119:
2116:
2110:
2107:
2105:
2102:
2100:
2097:
2095:
2092:
2090:
2087:
2086:
2084:
2080:
2074:
2071:
2069:
2066:
2064:
2061:
2060:
2058:
2054:
2046:
2043:
2041:
2038:
2036:
2033:
2032:
2030:
2026:
2023:
2021:
2018:
2017:
2016:
2013:
2012:
2010:
2006:
2000:
1997:
1995:
1994:Phylogenomics
1992:
1990:
1987:
1985:
1982:
1980:
1977:
1975:
1972:
1970:
1967:
1965:
1964:DNA barcoding
1962:
1960:
1959:
1955:
1954:
1952:
1948:
1942:
1939:
1935:
1934:Least squares
1932:
1930:
1927:
1925:
1922:
1921:
1920:
1917:
1913:
1910:
1908:
1905:
1904:
1902:
1900:
1897:
1895:
1892:
1891:
1889:
1885:
1879:
1876:
1872:
1871:Ghost lineage
1869:
1868:
1867:
1864:
1862:
1858:
1855:
1853:
1850:
1848:
1845:
1841:
1838:
1837:
1836:
1833:
1829:
1826:
1825:
1824:
1821:
1820:
1818:
1814:
1807:
1801:
1796:
1788:
1785:
1783:
1780:
1778:
1775:
1773:
1770:
1768:
1765:
1763:
1760:
1759:
1757:
1753:
1749:
1748:Phylogenetics
1742:
1737:
1735:
1730:
1728:
1723:
1722:
1719:
1712:
1707:
1703:
1702:
1690:
1686:
1682:
1678:
1674:
1670:
1666:
1662:
1654:
1646:
1642:
1638:
1634:
1630:
1626:
1622:
1618:
1611:
1609:
1600:
1596:
1592:
1588:
1584:
1580:
1576:
1572:
1565:
1557:
1553:
1549:
1545:
1540:
1535:
1532:(1): 98–102.
1531:
1527:
1523:
1516:
1514:
1512:
1503:
1499:
1495:
1491:
1487:
1483:
1476:
1474:
1472:
1463:
1459:
1455:
1451:
1444:
1436:
1432:
1428:
1424:
1420:
1416:
1409:
1407:
1398:
1396:9780126180305
1392:
1388:
1384:
1380:
1373:
1365:
1363:9780126180305
1359:
1355:
1351:
1347:
1342:
1341:
1331:
1329:
1327:
1325:
1316:
1312:
1308:
1304:
1300:
1296:
1292:
1288:
1281:
1273:
1267:
1263:
1259:
1258:
1250:
1242:
1238:
1234:
1230:
1226:
1222:
1218:
1214:
1210:
1206:
1199:
1191:
1185:
1181:
1174:
1166:
1162:
1158:
1154:
1149:
1144:
1140:
1136:
1132:
1125:
1117:
1113:
1109:
1105:
1101:
1097:
1090:
1082:
1076:
1072:
1067:
1066:
1060:
1054:
1046:
1042:
1041:Hennig, Willi
1036:
1028:
1022:
1018:
1014:
1013:Hillis, David
1008:
1000:
996:
992:
988:
981:
973:
967:
963:
956:
948:
942:
938:
931:
920:
916:
912:
908:
904:
900:
896:
892:
888:
881:
874:
866:
862:
858:
854:
850:
846:
842:
835:
833:
824:
820:
816:
812:
808:
804:
801:(3): 347–53.
800:
796:
789:
781:
777:
773:
769:
765:
761:
758:(2): 141–51.
757:
753:
746:
737:
732:
728:
724:
720:
713:
709:
700:
697:
695:
692:
690:
689:Phylogenetics
687:
686:
680:
671:
667:
658:
638:
635:
632:
629:
626:
623:
620:
617:
613:
609:
606:
603:
600:
597:
594:
588:
585:
582:
579:
576:
573:
570:
563:
556:
553:
550:
547:
544:
541:
538:
535:
532:
529:
502:
499:
496:
493:
490:
487:
484:
481:
468:
464:
458:
447:
443:
433:
419:
417:
412:
410:
404:
401:
397:
395:
390:
388:
384:
380:
376:
372:
371:least squares
367:
363:
359:
357:
353:
349:
339:
329:
326:
318:
308:
304:
298:
297:
292:This section
290:
286:
281:
280:
272:
270:
266:
261:
259:
254:
250:
240:
238:
234:
233:
228:
227:
213:
209:
205:
201:
199:
195:
191:
187:
183:
179:
174:
172:
158:
148:
145:
137:
127:
123:
117:
116:
111:This section
109:
105:
100:
99:
91:
89:
85:
81:
77:
73:
68:
64:
60:
57:"branch" and
56:
53:
49:
40:
32:
19:
2138:
2126:
2099:Sister group
2082:Nomenclature
2045:Autapomorphy
2040:Synapomorphy
2020:Plesiomorphy
2008:Group traits
1956:
1839:
1828:Cladogenesis
1823:Phylogenesis
1664:
1660:
1653:
1620:
1616:
1574:
1570:
1564:
1529:
1525:
1485:
1481:
1453:
1449:
1443:
1418:
1414:
1378:
1372:
1339:
1334:reviewed in
1290:
1286:
1280:
1256:
1249:
1208:
1204:
1198:
1179:
1173:
1138:
1134:
1124:
1099:
1095:
1089:
1064:
1053:
1044:
1035:
1016:
1007:
990:
986:
980:
961:
955:
936:
930:
890:
886:
873:
851:(1): 37–45.
848:
844:
798:
795:Paleobiology
794:
788:
755:
752:Paleobiology
751:
745:
726:
722:
712:
677:
668:
664:
469:
465:
456:
453:
445:
430:
413:
405:
402:
398:
391:
368:
364:
360:
356:optimization
345:
336:
321:
315:January 2021
312:
301:Please help
296:verification
293:
262:
252:
246:
236:
230:
224:
222:
211:
175:
168:
140:
131:
120:Please help
115:verification
112:
58:
54:
47:
45:
2094:Crown group
2056:Group types
1787:Systematics
1421:(1): 1–32.
1344:. pp.
993:: 361–381.
459:, denoted c
243:Homoplasies
2156:Categories
1772:Cladistics
1711:Cladograms
1617:Cladistics
1526:Cladistics
1287:Cladistics
1135:Cladistics
1096:Cladistics
1019:. Sinaur.
887:Cladistics
729:: 94–128.
705:References
694:Dendrogram
422:Statistics
348:algorithms
134:April 2016
63:cladistics
18:Cladograms
2109:Supertree
2073:Polyphyly
2068:Paraphyly
2063:Monophyly
2035:Apomorphy
2015:Primitive
1958:PhyloCode
1840:Cladogram
1667:: 24–32.
1571:Evolution
1456:: 52–79.
1379:Homoplasy
1340:Homoplasy
1102:: 91–93.
642:⌉
592:⌈
589:−
554:−
394:parsimony
379:parsimony
249:homoplasy
237:outgroups
208:Apomorphy
194:reversion
171:notochord
48:cladogram
2162:Diagrams
2128:Category
2031:Derived
1777:Taxonomy
1689:25451518
1645:84287895
1637:34933481
1599:28564338
1556:53320612
1548:34875753
1315:85720264
1307:34902938
1165:85725091
1157:34740305
1116:85905559
1061:(2003).
1043:(1966).
1015:(1996).
919:Archived
915:53357985
907:34818822
865:11146143
823:54988538
780:89032582
683:See also
212:a priori
2140:Commons
1866:Lineage
1669:Bibcode
1591:2409392
1502:2992286
1435:2412407
1241:4350103
1233:8437621
1213:Bibcode
815:4096939
772:2401114
269:anatomy
1687:
1643:
1635:
1597:
1589:
1554:
1546:
1500:
1433:
1393:
1360:
1348:–188.
1313:
1305:
1268:
1239:
1231:
1205:Nature
1186:
1163:
1155:
1114:
1077:
1073:–376.
1023:
968:
943:
913:
905:
863:
821:
813:
778:
770:
385:, and
352:metric
198:genome
59:gramma
55:clados
50:(from
2104:Basal
1929:UPGMA
1861:Grade
1857:Clade
1641:S2CID
1587:JSTOR
1552:S2CID
1498:JSTOR
1431:JSTOR
1311:S2CID
1237:S2CID
1161:S2CID
1112:S2CID
922:(PDF)
911:S2CID
883:(PDF)
819:S2CID
811:JSTOR
776:S2CID
768:JSTOR
253:other
72:clade
52:Greek
1685:PMID
1633:PMID
1595:PMID
1544:PMID
1391:ISBN
1358:ISBN
1303:PMID
1266:ISBN
1229:PMID
1184:ISBN
1153:PMID
1075:ISBN
1021:ISBN
966:ISBN
941:ISBN
903:PMID
861:PMID
414:The
265:bats
82:and
1859:vs
1677:doi
1665:366
1625:doi
1579:doi
1534:doi
1490:doi
1458:doi
1423:doi
1383:doi
1350:doi
1346:153
1295:doi
1221:doi
1209:361
1143:doi
1104:doi
1071:353
995:doi
895:doi
853:doi
803:doi
760:doi
731:doi
515:, c
305:by
176:As
124:by
84:RNA
80:DNA
2158::
1683:.
1675:.
1663:.
1639:.
1631:.
1619:.
1607:^
1593:.
1585:.
1575:43
1573:.
1550:.
1542:.
1530:26
1528:.
1524:.
1510:^
1496:.
1486:38
1484:.
1470:^
1454:43
1452:.
1429:.
1419:18
1417:.
1405:^
1389:.
1356:.
1323:^
1309:.
1301:.
1291:15
1289:.
1264:.
1262:66
1235:.
1227:.
1219:.
1207:.
1159:.
1151:.
1139:32
1137:.
1133:.
1110:.
1100:15
1098:.
991:23
989:.
939:.
917:.
909:.
901:.
891:29
889:.
885:.
859:.
849:16
847:.
831:^
817:.
809:.
799:31
797:.
774:.
766:.
756:22
754:.
727:12
725:.
721:.
657:.
463:.
389:.
381:,
377:,
373:,
260:.
247:A
46:A
1740:e
1733:t
1726:v
1691:.
1679::
1671::
1647:.
1627::
1621:5
1601:.
1581::
1558:.
1536::
1504:.
1492::
1464:.
1460::
1437:.
1425::
1399:.
1385::
1366:.
1352::
1317:.
1297::
1274:.
1243:.
1223::
1215::
1192:.
1167:.
1145::
1118:.
1106::
1083:.
1029:.
1001:.
997::
974:.
949:.
897::
867:.
855::
825:.
805::
782:.
762::
739:.
733::
645:)
639:s
636:e
633:t
630:a
627:t
624:s
621:.
618:n
614:/
610:a
607:x
604:a
601:t
598:.
595:n
586:a
583:x
580:a
577:t
574:.
571:n
568:(
564:/
560:)
557:1
551:s
548:e
545:t
542:a
539:t
536:s
533:.
530:n
527:(
517:i
503:s
500:e
497:t
494:a
491:t
488:s
485:.
482:n
472:i
470:c
461:i
457:i
328:)
322:(
317:)
313:(
299:.
147:)
141:(
136:)
132:(
118:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.