239:-filtered and have controlled humidity and temperature surroundings, which is typically around 25°C, 50% humidity. Early microarrays were directly printed onto the surface by using printer pins which deposit the samples in a user-defined pattern on the slide. Modern methods are faster, generate less cross-contamination, and produce better spot morphology. The surface to which the probes are printed must be clean, dust free and hydrophobic, for high-density microarrays. Slide coatings include poly-L-lysine, amino silane, epoxy and others, including manufacturers solutions and are chosen based on the type of sample used. Ongoing efforts to advance microarray technology aim to create uniform, dense arrays while reducing the necessary volume of solution and minimizing contamination or damage.
27:
309:-based printing, Electro-printing and droplet dispensing. In contrast to the other methods, non-contact printing does not involve contact between the surface and the stamp, pin, or other used dispenser. The main advantages are reduced contamination, lesser cleaning and higher throughput which increases steadily. Many of the methods are able to load the probes in parallel, allowing multiple arrays to be produced simultaneously.
272:, which in theory covers different, related pattern transfer technologies using patterned polymer monolithic substrates, the most prominent being microstamping. In contrast to pin printing, microstamping is a more parallel deposition method with less individuality. Certain stamps are loaded with reagents and printed with these reagent solutions identically.
254:
as a sample anymore, but also proteins, antibodies, antigens, glycans, cell lysates and other small molecules. All samples used are presynthesized, regularly updated, and more straightforward to maintain. Array fabrication techniques include contact printing, lithography, non-contact and cell free
231:
of a plant were printed on glass slide typically used for light microscopy, modern microarrays on the other hand include now thousands of probes and different carriers with coatings. The fabrication of the microarray requires both biological and physical information, including sample libraries,
317:
In cell free systems, the transcription and translation are carried out in situ, which makes the cloning and expression of proteins in host cells obsolete, because no intact cells are needed. The molecule of interest is directly synthesized onto the surface of a solid area. These assays allow
218:
A large number of technologies underlie the microarray platform, including the material substrates, spotting of biomolecular arrays, and the microfluidic packaging of the arrays. Microarrays can be categorized by how they physically isolate each element of the array, by spotting (making small
232:
printers, and slide substrates. Though all procedures and solutions always dependent on the fabrication technique employed. The basic principle of the microarray is the printing of small stains of solutions containing different species of the probe on a slide several thousand times.
283:
combines various methods like
Photolithography, Interference lithography, laser writing, electron-beam and Dip pen. The most widely used and researched method remains Photolithography, in which photolithographic masks are used to target specific nucleotides to the surface.
267:
microarray contact printing. This technique uses pin types like solid pins, split or quill pins to load and deliver the sample solution directly on solid microarray surfaces. Microstamping offers an alternative to the commonly used pin printing and is also referred as soft
292:
has been blocked, the area will remain protected from the addition of nucleotides, whereas in areas which were exposed to UV light, further nucleotides can be added. With this method high-quality custom arrays can be produced with a very high density of
478:
497:
459:
219:
physical wells), on-chip synthesis (synthesizing the target DNA probes adhered directly on the array), or bead-based (adhering samples to barcoded beads randomly distributed across the array).
242:
For the manufacturing process, a sample library which contains all relevant information is needed. In the early stages of microarray technology, the sole sample used was
646:
Guo, W; Vilaplana, L; Hansson, J; Marco, P; van der
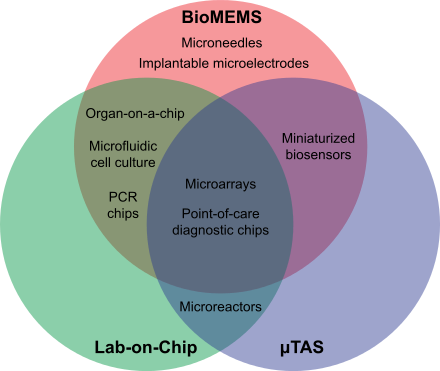
Wijngaart, W (2020). "Immunoassays on thiol-ene synthetic paper generate a superior fluorescence signal".
85:
miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in
511:
Schena, M.; Shalon, D.; Davis, R. W.; Brown, P. O. (1995). "Quantitative
Monitoring of Gene Expression Patterns with a Complementary DNA Microarray".
263:
Contact printing microarray include Pin printing, microstamping or flow printing. Pin printing is the oldest and still widest adopted methodology in
288:
is passed through the mask that acts as a filter to either transmit or block the light from the chemically protected microarray surface. If the
123:
has become the most sophisticated and the most widely used, while the use of protein, peptide and carbohydrate microarrays is expanding.
1073:
Sack, Matej; Hölz, Kathrin; Holik, Ann-Katrin; Kretschy, Nicole; Somoza, Veronika; Stengele, Klaus-Peter; Somoza, Mark M. (2016-03-02).
562:
Wang, D; Carroll, GT; Turro, NJ; Koberstein, JT; Kovác, P; Saksena, R; Adamo, R; Herzenberg, LA; Herzenberg, LA; Steinman, L (2007).
61:. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a
188:
861:
808:
752:
Zhou; et al. (2017). "Thiol–ene–epoxy thermoset for low-temperature bonding to biofunctionalized microarray surfaces".
953:
Romanov, Valentin; Davidoff, S. Nikki; Miles, Adam R.; Grainger, David W.; Gale, Bruce K.; Brooks, Benjamin D. (2014).
895:
Barbulovic-Nad, Irena; Lucente, Michael; Sun, Yu; Zhang, Mingjun; Wheeler, Aaron R.; Bussmann, Markus (January 2006).
1075:"Express photolithographic DNA microarray synthesis with optimized chemistry and high-efficiency photolabile groups"
206:, individual cells can be moved independently and simultaneously on a microarray of magnetic coils. A microarray of
151:
107:
article by the Ron Davis and Pat Brown labs at
Stanford University. With the establishment of companies, such as
848:, Advances in Experimental Medicine and Biology, vol. 593, New York, NY: Springer New York, pp. 1–11,
332:
62:
167:
419:
Tse-Wen Chang, TW (1983). "Binding of cells to matrixes of distinct antibodies coated on solid surface".
207:
492:
473:
454:
318:
high-throughput analysis in a controlled environment without inferences associated with intact cells.
82:
713:
611:
Ham, Donhee; Westervelt, Robert M. (2007). "The silicon that Moves and Feels Small Living Things".
564:"Photogenerated glycan arrays identify immunogenic sugar moieties of Bacillus anthracis exosporium"
788:
708:
203:
70:
43:
20:
202:
People in the field of CMOS biotechnology are developing new kinds of microarrays. Once fed
133:, such as cDNA microarrays, oligonucleotide microarrays, BAC microarrays and SNP microarrays
1193:
966:
520:
327:
183:
26:
8:
172:
161:
86:
970:
524:
1169:
1109:
1074:
1055:
1042:
932:
734:
681:
628:
593:
544:
476:, "Immunoassay device enclosing matrixes of antibody spots for cell determinations"
396:
363:
147:
142:
116:
699:
Barbulovic-Nad; et al. (2008). "Bio-Microarray
Fabrication Techniques—A Review".
362:
Carroll, Gregory T.; Wang, Denong; Turro, Nicholas J.; Koberstein, Jeffrey T. (2008).
1161:
1153:
1114:
1096:
1047:
1029:
990:
982:
924:
916:
867:
857:
814:
804:
769:
726:
685:
673:
585:
536:
436:
432:
401:
383:
228:
227:
The initial publication on microarray production process dates back to 1995, when 48
156:
55:
1173:
936:
738:
632:
597:
1145:
1104:
1086:
1059:
1037:
1021:
1010:"Basic Concepts of Microarrays and Potential Applications in Clinical Microbiology"
974:
908:
849:
796:
761:
718:
663:
655:
620:
575:
548:
528:
428:
391:
375:
195:
103:
843:
532:
90:
78:
853:
842:
Petersen, David W.; Kawasaki, Ernest S. (2007), "Manufacturing of
Microarrays",
800:
896:
659:
624:
337:
306:
130:
120:
1091:
912:
722:
379:
1187:
1157:
1100:
1033:
986:
920:
387:
94:
58:
39:
1133:
1165:
1149:
1118:
1051:
994:
928:
871:
818:
773:
730:
677:
589:
580:
563:
405:
250:
amplification via bacterial vectors. Modern approaches do not include just
177:
31:
1009:
540:
440:
1025:
364:"Photons to illuminate the universe of sugar diversity through bioarrays"
289:
285:
280:
269:
66:
954:
668:
246:, obtained from commonly available clone libraries and acquired through
978:
765:
108:
1134:"Cell-free synthesis-based protein microarrays and their applications"
955:"A critical comparison of protein microarray fabrication technologies"
495:, "Antibody matrix device and method for evaluating immune status"
457:, "Matrix of antibody-coated spots for determination of antigens"
98:
35:
97:
in 1983 in a scientific publication and a series of patents. The "
342:
112:
795:. Methods in Molecular Biology. Vol. 529. pp. 63–79.
136:
16:
Small-scale two-dimensional array of samples on a solid support
894:
74:
952:
236:
213:
561:
361:
297:
features by using a compact device with few moving parts.
294:
264:
251:
247:
243:
645:
101:" industry started to grow significantly after the 1995
34:
outlining and contrasting some aspects of the fields of
510:
1072:
1132:
Chandra, Harini; Srivastava, Sanjeeva (2009-12-01).
19:"Microarrays" redirects here. For the journal, see
1131:
1008:Miller, Melissa B.; Tang, Yi-Wei (October 2009).
1185:
897:"Bio-Microarray Fabrication Techniques—A Review"
841:
845:Microarray Technology and Cancer Gene Profiling
698:
610:
418:
194:Interferometric reflectance imaging sensor (
150:, for detailed analyses or optimization of
1007:
412:
139:, for surveillance of microRNA populations
1108:
1090:
1041:
712:
667:
579:
395:
214:Fabrication and operation of microarrays
189:Reverse phase protein lysate microarrays
25:
793:DNA Microarrays for Biomedical Research
305:Non-contact printing methods vary from
1186:
164:(also called transfection microarrays)
948:
946:
890:
888:
886:
837:
835:
786:
222:
751:
613:IEEE Solid-State Circuits Newsletter
258:
13:
943:
883:
832:
14:
1205:
901:Critical Reviews in Biotechnology
701:Critical Reviews in Biotechnology
210:microcoils is under development.
191:, microarrays of lysates or serum
115:, Applied Microarrays, Arrayjet,
421:Journal of Immunological Methods
119:, and others, the technology of
1125:
1066:
1001:
789:"Fabrication of DNA Microarray"
780:
745:
692:
639:
604:
555:
504:
485:
466:
447:
355:
333:Microarray analysis techniques
300:
275:
126:Types of microarrays include:
1:
1014:Clinical Microbiology Reviews
648:Biosensors and Bioelectronics
168:Chemical compound microarrays
1079:Journal of Nanobiotechnology
533:10.1126/science.270.5235.467
433:10.1016/0022-1759(83)90318-6
312:
152:protein–protein interactions
7:
854:10.1007/978-0-387-39978-2_1
801:10.1007/978-1-59745-538-1_5
321:
10:
1212:
660:10.1016/j.bios.2020.112279
625:10.1109/N-SSC.2007.4785650
208:nuclear magnetic resonance
18:
1092:10.1186/s12951-016-0166-0
913:10.1080/07388550600978358
723:10.1080/07388550600978358
380:10.1007/s10719-007-9052-1
83:high-throughput screening
77:(tests) large amounts of
348:
1150:10.1002/pmic.200900462
581:10.1002/pmic.200600478
368:Glycoconjugate Journal
204:magnetic nanoparticles
71:silicon thin-film cell
47:
493:US patent 5100777
474:US patent 4829010
455:US patent 4591570
184:Phenotype microarrays
180:(carbohydrate arrays)
89:(also referred to as
29:
21:Microarrays (journal)
1026:10.1128/cmr.00019-09
328:Microarray databases
235:Modern printers are
173:Antibody microarrays
162:Cellular microarrays
87:antibody microarrays
971:2014Ana...139.1303R
525:1995Sci...270..467S
148:Peptide microarrays
143:Protein microarrays
79:biological material
979:10.1039/c3an01577g
766:10.1039/C7LC00652G
223:Production process
157:Tissue microarrays
48:
863:978-0-387-39977-5
810:978-1-934115-69-5
787:Dufva, M (2008).
760:(21): 3672–3681.
1201:
1178:
1177:
1129:
1123:
1122:
1112:
1094:
1070:
1064:
1063:
1045:
1005:
999:
998:
965:(6): 1303–1326.
950:
941:
940:
892:
881:
880:
879:
878:
839:
830:
829:
827:
825:
784:
778:
777:
749:
743:
742:
716:
696:
690:
689:
671:
643:
637:
636:
608:
602:
601:
583:
559:
553:
552:
519:(5235): 467–70.
508:
502:
501:
500:
496:
489:
483:
482:
481:
477:
470:
464:
463:
462:
458:
451:
445:
444:
416:
410:
409:
399:
359:
259:Contact printing
104:Science Magazine
1211:
1210:
1204:
1203:
1202:
1200:
1199:
1198:
1184:
1183:
1182:
1181:
1130:
1126:
1071:
1067:
1006:
1002:
951:
944:
893:
884:
876:
874:
864:
840:
833:
823:
821:
811:
785:
781:
750:
746:
714:10.1.1.661.6833
697:
693:
644:
640:
609:
605:
560:
556:
509:
505:
498:
491:
490:
486:
479:
472:
471:
467:
460:
453:
452:
448:
427:(1–2): 217–23.
417:
413:
360:
356:
351:
324:
315:
303:
278:
261:
225:
216:
131:DNA microarrays
121:DNA microarrays
91:antibody matrix
63:solid substrate
24:
17:
12:
11:
5:
1209:
1208:
1197:
1196:
1180:
1179:
1144:(4): 717–730.
1124:
1065:
1020:(4): 611–633.
1000:
942:
907:(4): 237–259.
882:
862:
831:
809:
779:
744:
707:(4): 237–259.
691:
638:
603:
574:(2): 180–184.
554:
503:
484:
465:
446:
411:
353:
352:
350:
347:
346:
345:
340:
338:DNA Microarray
335:
330:
323:
320:
314:
311:
307:Photochemistry
302:
299:
277:
274:
260:
257:
224:
221:
215:
212:
200:
199:
192:
186:
181:
175:
170:
165:
159:
154:
145:
140:
134:
15:
9:
6:
4:
3:
2:
1207:
1206:
1195:
1192:
1191:
1189:
1175:
1171:
1167:
1163:
1159:
1155:
1151:
1147:
1143:
1139:
1135:
1128:
1120:
1116:
1111:
1106:
1102:
1098:
1093:
1088:
1084:
1080:
1076:
1069:
1061:
1057:
1053:
1049:
1044:
1039:
1035:
1031:
1027:
1023:
1019:
1015:
1011:
1004:
996:
992:
988:
984:
980:
976:
972:
968:
964:
960:
956:
949:
947:
938:
934:
930:
926:
922:
918:
914:
910:
906:
902:
898:
891:
889:
887:
873:
869:
865:
859:
855:
851:
847:
846:
838:
836:
820:
816:
812:
806:
802:
798:
794:
790:
783:
775:
771:
767:
763:
759:
755:
748:
740:
736:
732:
728:
724:
720:
715:
710:
706:
702:
695:
687:
683:
679:
675:
670:
665:
661:
657:
653:
649:
642:
634:
630:
626:
622:
618:
614:
607:
599:
595:
591:
587:
582:
577:
573:
569:
565:
558:
550:
546:
542:
538:
534:
530:
526:
522:
518:
514:
507:
494:
488:
475:
469:
456:
450:
442:
438:
434:
430:
426:
422:
415:
407:
403:
398:
393:
389:
385:
381:
377:
373:
369:
365:
358:
354:
344:
341:
339:
336:
334:
331:
329:
326:
325:
319:
310:
308:
298:
296:
291:
287:
282:
273:
271:
266:
256:
253:
249:
245:
240:
238:
233:
230:
220:
211:
209:
205:
197:
193:
190:
187:
185:
182:
179:
178:Glycan arrays
176:
174:
171:
169:
166:
163:
160:
158:
155:
153:
149:
146:
144:
141:
138:
135:
132:
129:
128:
127:
124:
122:
118:
114:
110:
106:
105:
100:
96:
95:Tse Wen Chang
92:
88:
84:
80:
76:
72:
68:
64:
60:
59:lab-on-a-chip
57:
53:
45:
41:
40:lab-on-a-chip
37:
33:
28:
22:
1141:
1137:
1127:
1082:
1078:
1068:
1017:
1013:
1003:
962:
958:
904:
900:
875:, retrieved
844:
824:30 September
822:. Retrieved
792:
782:
757:
753:
747:
704:
700:
694:
669:10261/211201
651:
647:
641:
616:
612:
606:
571:
567:
557:
516:
512:
506:
487:
468:
449:
424:
420:
414:
371:
367:
357:
316:
304:
279:
262:
241:
234:
226:
217:
201:
125:
102:
51:
49:
32:Venn diagram
1194:Microarrays
959:The Analyst
374:(1): 5–10.
301:Non contact
281:Lithography
276:Lithography
270:lithography
255:printing.
67:glass slide
65:—usually a
1138:Proteomics
877:2023-05-18
654:: 112279.
619:(4): 4–9.
568:Proteomics
109:Affymetrix
52:microarray
1158:1615-9853
1101:1477-3155
1085:(1): 14.
1034:0893-8512
987:0003-2654
921:0738-8551
709:CiteSeerX
686:218688183
388:0282-0080
313:Cell free
99:gene chip
56:multiplex
1188:Category
1174:22007600
1166:19953547
1119:26936369
1052:19822891
995:24479125
937:13712888
929:17095434
872:17265711
819:19381969
774:28975170
754:Lab Chip
739:13712888
731:17095434
678:32421629
633:35867338
598:21145793
590:17205603
406:17610157
322:See also
290:UV light
286:UV light
117:Illumina
36:bio-MEMS
1110:4776362
1060:5865637
1043:2772365
967:Bibcode
549:6720459
541:7569999
521:Bibcode
513:Science
441:6606681
397:7088275
343:Biochip
137:MMChips
113:Agilent
1172:
1164:
1156:
1117:
1107:
1099:
1058:
1050:
1040:
1032:
993:
985:
935:
927:
919:
870:
860:
817:
807:
772:
737:
729:
711:
684:
676:
631:
596:
588:
547:
539:
499:
480:
461:
439:
404:
394:
386:
81:using
75:assays
73:—that
1170:S2CID
1056:S2CID
933:S2CID
735:S2CID
682:S2CID
629:S2CID
594:S2CID
545:S2CID
349:Notes
229:cDNAs
93:) by
54:is a
44:μTAS
1162:PMID
1154:ISSN
1115:PMID
1097:ISSN
1048:PMID
1030:ISSN
991:PMID
983:ISSN
925:PMID
917:ISSN
868:PMID
858:ISBN
826:2022
815:PMID
805:ISBN
770:PMID
727:PMID
674:PMID
586:PMID
537:PMID
437:PMID
402:PMID
384:ISSN
237:HEPA
196:IRIS
1146:doi
1105:PMC
1087:doi
1038:PMC
1022:doi
975:doi
963:139
909:doi
850:doi
797:doi
762:doi
719:doi
664:hdl
656:doi
652:163
621:doi
576:doi
529:doi
517:270
429:doi
392:PMC
376:doi
295:DNA
265:DNA
252:DNA
248:DNA
244:DNA
69:or
1190::
1168:.
1160:.
1152:.
1142:10
1140:.
1136:.
1113:.
1103:.
1095:.
1083:14
1081:.
1077:.
1054:.
1046:.
1036:.
1028:.
1018:22
1016:.
1012:.
989:.
981:.
973:.
961:.
957:.
945:^
931:.
923:.
915:.
905:26
903:.
899:.
885:^
866:,
856:,
834:^
813:.
803:.
791:.
768:.
758:17
756:.
733:.
725:.
717:.
705:26
703:.
680:.
672:.
662:.
650:.
627:.
617:12
615:.
592:.
584:.
570:.
566:.
543:.
535:.
527:.
515:.
435:.
425:65
423:.
400:.
390:.
382:.
372:25
370:.
366:.
111:,
50:A
42:,
38:,
30:A
1176:.
1148::
1121:.
1089::
1062:.
1024::
997:.
977::
969::
939:.
911::
852::
828:.
799::
776:.
764::
741:.
721::
688:.
666::
658::
635:.
623::
600:.
578::
572:7
551:.
531::
523::
443:.
431::
408:.
378::
198:)
46:.
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.