138:
experiments on tissue samples, plasma or serum samples and many other sample types. One main focus in antibody array based profiling studies is biomarker discovery, specifically for cancer. For cancer-related research, the development and application of an antibody array comprising 810 different cancer-related antibodies was reported in 2010. Also in 2010, an antibody array comprising 507 cytokines, chemokines, adipokines, growth factors, angiogenic factors, proteases, soluble receptors, soluble adhesion molecules, and other proteins was used to screen the serum of ovarian cancer patients and healthy individuals and found a significant difference in protein expression between normal and cancer samples. More recently, antibody arrays have helped determine specific allergy-related serum proteins whose levels are associated with glioma and can reduce the risk years before diagnosis. Protein profiling with antibody arrays have also proven successful in areas other than cancer research, specifically in neurological diseases such as
Alzheimer's. A number of studies have attempted to identify biomarker panels that can distinguish Alzheimer's patients, and many have used antibody arrays in this process. Jaeger and colleagues measured nearly 600 circulatory proteins to discover biological pathways and networks affected in Alzheimer's and explored the positive and negative relationships of the levels of those individual proteins and networks with the cognitive performance of Alzheimer's patients. Currently the largest commercially available sandwich-based antibody array detects 1000 different proteins. In addition, antibody microarray based protein profiling services are available analyzing protein abundance and protein
127:
antibody, each binding to a unique epitope), confers greater specificity and lower background signal compared with label-based immunodetection (where only 1 capture antibody is used and detection is achieved by chemically labeling all proteins in the starting sample). Sandwich-based antibody arrays usually attain the highest specificity and sensitivity (ng – pg levels) of any array format; their reproducibility also enables quantitative analysis to be performed. Due to the difficulty of developing matched antibody pairs that are compatible with all other antibodies in the panel, small arrays often make use of a sandwich approach. Conversely, high-density arrays are easier to develop at a lower cost using the single antibody label-based approach. In this methodology, one set of specific antibodies is used and all the proteins in a sample are labelled directly by fluorescent dyes or haptens.
123:
number of analytes detected. Other solid supports, such as glass slides and nitrocellulose membranes, were subsequently utilized to develop arrays which could accommodate larger panels of antibodies. Nitrocellulose membrane-based arrays are flexible, easy to handle, and have increased protein binding capacity, but are less amenable to high throughput or automated processing. Chemically derivatized glass slides allow for printing of sub-microliter sized antibody spots, reducing the array surface area without sacrificing spot density. This in turn reduces the volume of sample consumed. Glass slide-based arrays, owing to their smooth and rigid structure, can also be easily fitted to high-throughput liquid handling systems.
79:. Chang coined the term “antibody matrix” and discussed “array” arrangement of minute antibody spots on small glass or plastic surfaces. He demonstrated that a 10×10 (100 in total) and 20×20 (400 in total) grid of antibody spots could be placed on a 1×1 cm surface. He also estimated that if an antibody is coated at a 10 μg/mL concentration, which is optimal for most antibodies, 1 mg of antibody can make 2,000,000 dots of 0.25 mm diameter. Chang's invention focused on the employment of antibody microarrays for the detection and quantification of cells bearing certain surface antigens, such as CD antigens and
134:, studying yeast protein kinases, analyzing autoimmune antibodies, and examining protein-protein interactions. The first approach to simultaneously detect multiple cytokines from physiological samples using antibody array technology was by Ruo-Pan Huang and colleagues in 2001. Their approach used Hybond ECL membranes to detect a small panel of 24 cytokines from cell culture conditioned media and patient's sera and was able to profile cytokine expression at physiological levels. Huang took this technology and started a new business, RayBiotech, Inc., the first to successfully commercialize a planar antibody array.
150:
different blood sample preparations were analyzed using three antibody array platforms: sandwich-based, quantitative, and label-based, and a strong correlation in protein expression was found, suggesting that dried blood spots, which are a more convenient, safe, and inexpensive means of obtaining blood especially in non-hospitalized public health areas, can be used effectively with antibody array analysis for biomarker discovery, protein profiling, and disease screening, diagnosis, and treatment.
31:
92:, Inc. in Houston, Texas in 1986, they purchased the rights on the antibody matrix patents from Centocor as part of the technology base to build their new startup. Their first product in development was an assay, termed “immunosorbent cytometry”, which could be employed to monitor the immune status, i.e., the concentrations and ratios of
158:
Using antibody microarray in different medical diagnostic areas has attracted researchers attention. Digital bioassay is an example of such research domains. In this technology, an array of microwells on a glass/polymer chip are seeded with magnetic beads (coated with fluorescent tagged antibodies),
137:
In the last ten years, the sensitivity of the method was improved by an optimization of the surface chemistry as well as dedicated protocols for their chemical labeling. Currently, the sensitivity of antibody arrays is comparable to that of ELISA and antibody arrays are regularly used for profiling
122:
The first array approaches attempted to miniaturize biochemical and immunobiological assays usually performed in 96-well microtiter plates. While 96-well plate-based antibody arrays have high-throughput capability, the small surface area in each well limits the number of antibody spots and thus, the
114:
The theoretical background for protein microarray-based ligand binding assays was further developed by Roger Ekins and colleagues in the late 1980s. According to the model, antibody microarrays would not only permit simultaneous screening of an analyte panel, but would also be more sensitive and
126:
Most antibody array systems employ 1 of 2 non-competitive methods of immunodetection: single-antibody (label-based) detection and 2-antibody (sandwich-based) detection. The latter method, in which analyte detection requires the binding of 2 distinct antibodies (a capture antibody and a reporter
149:
Antibody arrays are often used for detecting protein expression from many sample types, but also in those with various preparations. Jiang and colleagues illustrated nicely the correlation between array protein expression in two different blood preparations: serum and dried blood spots. These
83:
allotypic antigens, particulate antigens, such as viruses and bacteria, and soluble antigens. The principle of "one sample application, multiple determinations", assay configuration, and mechanics for placing absorbent dots described in the paper and patents should be generally applicable to
163:) for such microwell arrays has been recently demonstrated and the bio-assay model system has been successfully characterised. Furthermore, immunoassays on thiol-ene "synthetic paper" micropillar scaffolds have shown to generate a superior fluorescence signal.
1584:
Lin Y.; Luo S.; Shao N.; Wang S.; Duan C.; Burkholder B.; et al. (2013). "Peeking into the Black Box: How
Cytokine Antibody Arrays Shed Light on Molecular Mechanisms of Breast Cancer Development and its Treatment".
1403:
Wingren
Christer, Ingvarsson Johan, Dexlin Linda, Szul Dominika, Borrebaeck Carl A. K. (2007). "Design of recombinant antibody microarrays for complex proteome analysis: Choice of sample labeling-tag and solid support".
54:
are spotted and fixed on a solid surface such as glass, plastic, membrane, or silicon chip, and the interaction between the antibody and its target antigen is detected. Antibody microarrays are often used for detecting
1093:
Joos T. O., Schrenk M., Hopfl P., Kroger K., Chowdhury U., Stoll D., Schorner D., Durr M., Herick K., Rupp S., Sohn K., Hammerle H. (2000). "A microarray enzyme-linked immunosorbent assay for autoimmune diagnostics".
1359:
Kusnezow W, Banzon V, Schröder C, Schaal R, Hoheisel JD, Rüffer S, Luft P, Duschl A, Syagailo YV (2007). "Antibody microarray-based profiling of complex specimens: systematic evaluation of labeling strategies".
1315:
Kusnezow W, Banzon V, Schröder C, Schaal R, Hoheisel JD, Rüffer S, Luft P, Duschl A, Syagailo YV (2007). "Antibody microarray-based profiling of complex specimens: systematic evaluation of labeling strategies".
1893:
Jiang, W., Mao, Y. Q., Huang, R., Duan, C., Xi, Y., Yang, K., & Huang, R. P. (2014). "Protein expression profiling by antibody array analysis with use of dried blood spot samples on filter paper".
1706:
Huang R., Jiang W., Yang J., Mao Y.Q., Zhang Y., Yang W., Yang D., Burkholder B., Huang R.F., Huang R.P. (2010). "A biotin label-based antibody array for high-content profiling of protein expression".
650:
Jiang, W., Mao, Y. Q., Huang, R., Duan, C., Xi, Y., Yang, K., & Huang, R. P. (2014). "Protein expression profiling by antibody array analysis with use of dried blood spot samples on filter paper".
1305:
R.-P. Huang. (2001). Simultaneous detection of multiple proteins with an array-based enzyme-linked immunosorbant assay (ELISA) and enhanced chemiluminescence (ECL). Clin. Chem. Lab. Med. 39:209-214.
963:
de Wildt, R. M., Mundy, C. R., Gorick, B. D., and
Tomlinson, I. M. (2000) Antibody arrays for high-throughput screening of antibody-antigen interactions. Nature Biotechnol. 18, 989 –994
1036:
Zhu H., Klemic J. F., Chang S., Bertone P., Casamayor A., Klemic K. G., Smith D., Gerstein M., Reed M. A., Snyder M. (2000). "Analysis of yeast protein kinases using protein chips".
59:
from various biofluids including serum, plasma and cell or tissue lysates. Antibody arrays may be used for both basic research and medical and diagnostic applications.
115:
rapid than conventional screening methods. Interest in screening large protein sets only arose as a result of the achievements in genomics by DNA microarrays and the
1655:
Schröder C, Jacob A, Tonack S, Radon TP, Sill M, Zucknick M, Rüffer S, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Bauer A, Fellenberg K, Hoheisel JD (2010).
1092:
1943:
1981:
Guo, W; Vilaplana, L; Hansson, J; Marco, P; van der
Wijngaart, W (2020). "Immunoassays on thiol-ene synthetic paper generate a superior fluorescence signal".
701:"The functional behavior of a macrophage/fibroblast co-culture model derived from normal and diabetic mice with a marine gelatin–oxidized alginate hydrogel"
359:
Lin Y., Huang R.C., Cao X., Wang S.-M., Shi Q., Huang R.-P. (2003). "Detection of multiple cytokines by protein arrays from cell lysate and tissue lysate".
1528:
159:
subjected to targeted antigens and then characterised by a microscope through counting fluorescing wells. A cost-effective fabrication platform (using
1035:
1928:
1733:
1570:
1515:
1447:
1292:
1186:
1137:
1079:
1022:
950:
901:
846:
685:
402:
915:
Lueking A., Horn M., Eickhoff H., Bussow K., Lehrach H., Walter G. (1999). "Protein microarrays for gene expression and antibody screening".
1151:
Walter G., Bussow K., Cahill D., Lueking A., Lehrach H. (2000). "Protein arrays for gene expression and molecular interaction screening".
1705:
218:"A 200-antibody microarray biochip for environmental monitoring: searching for universal microbial biomarkers through immunoprofiling"
1529:
Burkholder B., Burgess R.Y. R., Luo S.H., Jones V.S., Zhang W.J., Lv Z.Q., Gao C.-Y., Wang B.-L., Zhang Y.-M., Huang R.-P. (2014).
1107:
274:
130:
Initial uses of antibody-based array systems included detecting IgGs and specific subclasses, analyzing antigens, screening
2049:
1402:
574:
Ekins RP, Chu FW (November 1991). "Multianalyte microspot immunoassay—microanalytical "compact disk" of the future".
315:
1245:"Stat1 as a Component of Tumor Necrosis Factor Alpha Receptor 1-TRADD Signaling Complex To Inhibit NF-κB Activation"
1944:"Single-Step Imprinting of Femtoliter Microwell Arrays Allows Digital Bioassays with Attomolar Limit of Detection"
17:
1892:
649:
471:"Immunoassay device enclosing matrixes of antibody spots for cell determinations", Priority date March 13, 1987
1460:
Alhamdani, MS; Schröder, C; Hoheisel, JD (Jul 6, 2009). "Oncoproteomic profiling with antibody microarrays".
859:
192:
2044:
1747:
Schwartzbaum J.; Seweryn M.; Holloman C.; Harris R.; Handelman S. K.; Rempala G. A.; et al. (2015).
416:
Chang TW (December 1983). "Binding of cells to matrixes of distinct antibodies coated on solid surface".
1806:
Jaeger P. A.; Lucin K. M.; Britschgi M.; Vardarajan B.; Huang R.-P.; Kirby E. D.; et al. (2016).
1657:"Dual-color proteomic profiling of complex samples with a microarray of 810 cancer-related antibodies"
1473:
610:
345:
1869:
1612:
Huang R.-P. (2007). "An array of possibilities in cancer research using cytokine antibody arrays".
216:
Rivas LA, García-Villadangos M, Moreno-Paz M, Cruz-Gil P, Gómez-Elvira J, Parro V (November 2008).
914:
139:
80:
975:"By-passing selection: Direct screening for antibody–antigen interactions using protein arrays"
456:"Matrix of antibody-coated spots for determination of antigens", Priority date February 2, 1983
131:
486:"Antibody matrix device and method for evaluating immune status", Priority date April 27, 1987
1922:
1727:
1564:
1509:
1441:
1286:
1180:
1131:
1073:
1016:
944:
895:
840:
679:
396:
76:
2039:
1760:
804:
761:
750:"Allogenic iPSC-derived RPE cell transplants induce immune response in pigs: a pilot study"
358:
116:
197:
8:
1764:
765:
2016:
1834:
1807:
1783:
1748:
1683:
1656:
1637:
1429:
1385:
1341:
1225:
1150:
1119:
1061:
782:
749:
725:
716:
700:
521:
384:
333:
298:
Wilson J. J.; Burgess R.; Mao Y. Q.; Luo S.; Tang H.; Jones V. S.; et al. (2015).
177:
47:
1856:
RayBiotech, Inc. Antibody Arrays. (2017). Retrieved from the RayBiotech, Inc. website
1598:
1164:
999:
974:
2020:
2008:
1963:
1910:
1839:
1788:
1715:
1688:
1629:
1552:
1497:
1421:
1377:
1333:
1274:
1269:
1260:
1244:
1217:
1168:
1111:
1053:
1004:
932:
883:
828:
787:
730:
667:
632:
591:
587:
556:
552:
513:
433:
429:
376:
321:
311:
280:
270:
257:
Chaga GS (2008). "Antibody Arrays for
Determination of Relative Protein Abundances".
239:
187:
1808:"Network-driven plasma proteomics expose molecular changes in the Alzheimer's brain"
1641:
1433:
1123:
860:
Mendoza L. G., McQuary P., Mongan A., Gangadharan R., Brignac S., Eggers M. (1999).
525:
388:
71:
in 1983 in a scientific publication and a series of patents, when he was working at
1998:
1990:
1955:
1902:
1829:
1819:
1778:
1768:
1678:
1668:
1621:
1594:
1542:
1487:
1413:
1389:
1369:
1345:
1325:
1264:
1256:
1229:
1209:
1160:
1103:
1065:
1045:
994:
986:
924:
873:
818:
777:
769:
720:
712:
659:
622:
583:
548:
505:
425:
368:
303:
262:
229:
93:
1213:
1773:
972:
823:
806:
627:
56:
1857:
1805:
1547:
1530:
1492:
1475:
266:
1994:
1673:
1242:
1200:
Service R. F. (2000). "Biochemistry: Protein arrays step out of DNA's shadow".
307:
182:
1906:
1824:
1749:"Association between Prediagnostic Allergy-Related Serum Cytokines and Glioma"
663:
482:
467:
452:
2033:
1746:
1625:
1474:
Jones V. S., Huang R. Y., Chen L. P., Chen Z. S., Fu L., Huang R. P. (2016).
160:
68:
67:
The concept and methodology of antibody microarrays were first introduced by
30:
990:
862:"High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA)"
2012:
1967:
1959:
1914:
1843:
1792:
1719:
1692:
1633:
1556:
1501:
1425:
1417:
1381:
1373:
1337:
1329:
1278:
1221:
1172:
1115:
1057:
1008:
936:
928:
887:
791:
734:
671:
380:
325:
284:
243:
1108:
10.1002/1522-2683(20000701)21:13<2641::aid-elps2641>3.0.co;2-5
832:
807:"Mass-sensing, multianalyte microarray immunoassay with imaging detection"
636:
595:
560:
517:
437:
372:
1476:"Cytokines in cancer drug resistance: cues to new therapeutic strategies"
215:
2003:
509:
85:
51:
878:
861:
773:
234:
217:
297:
143:
805:
Silzel J. W., Cercek B., Dodson C., Tsay T., Obremski R. J. (1998).
1531:"Tumor-induced perturbations of cytokines and immune cell networks"
72:
1049:
611:"Ligand assays: from electrophoresis to miniaturized microarrays"
104:
261:. Methods in Molecular Biology. Vol. 441. pp. 129–51.
172:
89:
1358:
1314:
1583:
973:
Holt L. J., Bussow K., Walter G., Tomlinson I. M. (2000).
108:
101:
97:
1980:
1654:
1243:
Wang Y., Wu T. R., Cai S., Welte T., Chin Y. E. (2000).
88:. When Tse Wen Chang and Nancy T. Chang were setting up
34:
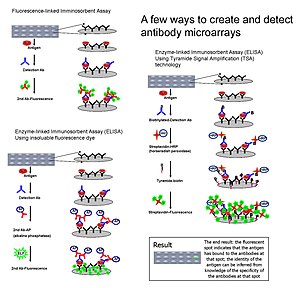
Samples of antibody microarray creations and detections.
1535:
Biochimica et
Biophysica Acta (BBA) - Reviews on Cancer
1480:
Biochimica et
Biophysica Acta (BBA) - Reviews on Cancer
300:
Chapter Seven-Antibody Arrays in
Biomarker Discovery
1870:"Sciomics: Antibody meets microarray - scioPhospho"
198:
Microarray imprinting and surface energy patterning
496:Chang TW (March 1993). "Immunosorbent Cytometry".
2031:
698:
50:. In this technology, a collection of captured
1941:
747:
539:Ekins RP (1989). "Multi-analyte immunoassay".
1858:http://www.raybiotech.com/antibody-array.html
1199:
1927:: CS1 maint: multiple names: authors list (
1732:: CS1 maint: multiple names: authors list (
1569:: CS1 maint: multiple names: authors list (
1514:: CS1 maint: multiple names: authors list (
1446:: CS1 maint: multiple names: authors list (
1291:: CS1 maint: multiple names: authors list (
1185:: CS1 maint: multiple names: authors list (
1136:: CS1 maint: multiple names: authors list (
1078:: CS1 maint: multiple names: authors list (
1021:: CS1 maint: multiple names: authors list (
949:: CS1 maint: multiple names: authors list (
900:: CS1 maint: multiple names: authors list (
845:: CS1 maint: multiple names: authors list (
684:: CS1 maint: multiple names: authors list (
401:: CS1 maint: multiple names: authors list (
1611:
602:
567:
532:
250:
209:
489:
409:
2002:
1833:
1823:
1782:
1772:
1682:
1672:
1546:
1491:
1268:
998:
877:
822:
781:
724:
626:
233:
608:
573:
538:
495:
415:
256:
29:
14:
2032:
1948:ACS Applied Materials & Interfaces
146:status of 1030 proteins in parallel.
748:Sohn Elliott H; et al. (2015).
1661:Molecular & Cellular Proteomics
24:
717:10.1016/j.biomaterials.2010.04.022
302:. Vol. 69. pp. 255–324.
25:
2061:
1599:10.2174/1570164610666131210233343
1895:Journal of Immunological Methods
1261:10.1128/mcb.20.13.4505-4512.2000
652:Journal of Immunological Methods
1974:
1935:
1886:
1862:
1850:
1799:
1740:
1699:
1648:
1605:
1577:
1522:
1467:
1454:
1396:
1352:
1308:
1299:
1236:
1193:
1144:
1086:
1029:
966:
957:
908:
853:
798:
741:
692:
643:
153:
1249:Molecular and Cellular Biology
474:
459:
444:
352:
291:
13:
1:
1983:Biosensors and Bioelectronics
1214:10.1126/science.289.5485.1673
1165:10.1016/s1369-5274(00)00093-x
203:
62:
1774:10.1371/journal.pone.0137503
588:10.1016/0167-7799(94)90111-2
553:10.1016/0731-7085(89)80079-2
430:10.1016/0022-1759(83)90318-6
193:Chemical compound microarray
7:
1812:Molecular Neurodegeneration
1614:Expert Review of Proteomics
1548:10.1016/j.bbcan.2014.01.004
1493:10.1016/j.bbcan.2016.03.005
609:Ekins RP (September 1998).
267:10.1007/978-1-60327-047-2_9
166:
10:
2066:
1995:10.1016/j.bios.2020.112279
1708:Cancer Genomics Proteomics
1674:10.1074/mcp.m900419-mcp200
824:10.1093/clinchem/44.9.2036
628:10.1093/clinchem/44.9.2015
308:10.1016/bs.acc.2015.01.002
27:Form of protein microarray
2050:Reagents for biochemistry
1907:10.1016/j.jim.2013.11.016
1825:10.1186/s13024-016-0105-4
699:Zeng Q., Chen W. (2010).
664:10.1016/j.jim.2013.11.016
1626:10.1586/14789450.4.2.299
46:) is a specific form of
1942:Decrop Deborah (2017).
111:-infected individuals.
1960:10.1021/acsami.6b15415
1418:10.1002/pmic.200700025
1374:10.1002/pmic.200600762
1330:10.1002/pmic.200600762
929:10.1006/abio.1999.4063
132:recombinant antibodies
35:
1153:Curr. Opin. Microbiol
991:10.1093/nar/28.15.e72
483:U.S. patent 5,100,777
468:U.S. patent 4,829,010
453:U.S. patent 4,591,570
373:10.1515/cclm.2003.023
77:Malvern, Pennsylvania
33:
117:Human Genome Project
1954:(12): 10418–10426.
1765:2015PLoSO..1037503S
766:2015NatSR...511791S
541:J Pharm Biomed Anal
510:10.1038/nbt0393-291
418:J. Immunol. Methods
84:different kinds of
40:antibody microarray
2045:1983 introductions
1587:Current Proteomics
754:Scientific Reports
178:Protein microarray
107:, in the blood of
57:protein expression
48:protein microarray
36:
1412:(17): 3055–3065.
1255:(13): 4505–4512.
1102:(13): 2641–2650.
979:Nucleic Acids Res
879:10.2144/99274rr01
774:10.1038/srep11791
711:(22): 5772–5781.
361:Clin Chem Lab Med
276:978-1-58829-679-5
259:Tissue Proteomics
235:10.1021/ac8008093
188:Tissue microarray
16:(Redirected from
2057:
2025:
2024:
2006:
1978:
1972:
1971:
1939:
1933:
1932:
1926:
1918:
1890:
1884:
1883:
1881:
1880:
1866:
1860:
1854:
1848:
1847:
1837:
1827:
1803:
1797:
1796:
1786:
1776:
1744:
1738:
1737:
1731:
1723:
1703:
1697:
1696:
1686:
1676:
1652:
1646:
1645:
1609:
1603:
1602:
1581:
1575:
1574:
1568:
1560:
1550:
1526:
1520:
1519:
1513:
1505:
1495:
1471:
1465:
1458:
1452:
1451:
1445:
1437:
1400:
1394:
1393:
1356:
1350:
1349:
1312:
1306:
1303:
1297:
1296:
1290:
1282:
1272:
1240:
1234:
1233:
1197:
1191:
1190:
1184:
1176:
1148:
1142:
1141:
1135:
1127:
1090:
1084:
1083:
1077:
1069:
1033:
1027:
1026:
1020:
1012:
1002:
970:
964:
961:
955:
954:
948:
940:
912:
906:
905:
899:
891:
881:
857:
851:
850:
844:
836:
826:
817:(9): 2036–2043.
802:
796:
795:
785:
745:
739:
738:
728:
696:
690:
689:
683:
675:
647:
641:
640:
630:
606:
600:
599:
571:
565:
564:
536:
530:
529:
493:
487:
485:
478:
472:
470:
463:
457:
455:
448:
442:
441:
413:
407:
406:
400:
392:
356:
350:
349:
343:
339:
337:
329:
295:
289:
288:
254:
248:
247:
237:
213:
144:ubiquitinylation
21:
2065:
2064:
2060:
2059:
2058:
2056:
2055:
2054:
2030:
2029:
2028:
1979:
1975:
1940:
1936:
1920:
1919:
1891:
1887:
1878:
1876:
1874:www.sciomics.de
1868:
1867:
1863:
1855:
1851:
1804:
1800:
1759:(9): e0137503.
1745:
1741:
1725:
1724:
1704:
1700:
1653:
1649:
1610:
1606:
1582:
1578:
1562:
1561:
1527:
1523:
1507:
1506:
1472:
1468:
1462:Genome medicine
1459:
1455:
1439:
1438:
1401:
1397:
1368:(11): 1786–99.
1357:
1353:
1324:(11): 1786–99.
1313:
1309:
1304:
1300:
1284:
1283:
1241:
1237:
1198:
1194:
1178:
1177:
1149:
1145:
1129:
1128:
1096:Electrophoresis
1091:
1087:
1071:
1070:
1038:Nature Genetics
1034:
1030:
1014:
1013:
971:
967:
962:
958:
942:
941:
913:
909:
893:
892:
858:
854:
838:
837:
803:
799:
746:
742:
697:
693:
677:
676:
648:
644:
607:
603:
582:(11): 1955–67.
572:
568:
537:
533:
494:
490:
481:
479:
475:
466:
464:
460:
451:
449:
445:
424:(1–2): 217–23.
414:
410:
394:
393:
357:
353:
341:
340:
331:
330:
318:
296:
292:
277:
255:
251:
214:
210:
206:
169:
156:
140:phosphorylation
65:
42:(also known as
28:
23:
22:
18:Antibody matrix
15:
12:
11:
5:
2063:
2053:
2052:
2047:
2042:
2027:
2026:
1973:
1934:
1885:
1861:
1849:
1798:
1739:
1698:
1667:(6): 1271–80.
1647:
1620:(2): 299–308.
1604:
1593:(4): 269–277.
1576:
1541:(2): 182–201.
1521:
1486:(2): 255–265.
1466:
1453:
1395:
1351:
1307:
1298:
1235:
1208:(5485): 1673.
1192:
1159:(3): 298–302.
1143:
1085:
1044:(3): 283–289.
1028:
965:
956:
923:(1): 103–111.
907:
872:(4): 778–788.
852:
797:
740:
691:
642:
621:(9): 2015–30.
601:
566:
531:
488:
480:Chang, Tse W.
473:
465:Chang, Tse W.
458:
450:Chang, Tse W.
443:
408:
367:(2): 139–145.
351:
342:|journal=
316:
290:
275:
249:
228:(21): 7970–9.
207:
205:
202:
201:
200:
195:
190:
185:
183:DNA microarray
180:
175:
168:
165:
155:
152:
64:
61:
44:antibody array
26:
9:
6:
4:
3:
2:
2062:
2051:
2048:
2046:
2043:
2041:
2038:
2037:
2035:
2022:
2018:
2014:
2010:
2005:
2000:
1996:
1992:
1988:
1984:
1977:
1969:
1965:
1961:
1957:
1953:
1949:
1945:
1938:
1930:
1924:
1916:
1912:
1908:
1904:
1900:
1896:
1889:
1875:
1871:
1865:
1859:
1853:
1845:
1841:
1836:
1831:
1826:
1821:
1817:
1813:
1809:
1802:
1794:
1790:
1785:
1780:
1775:
1770:
1766:
1762:
1758:
1754:
1750:
1743:
1735:
1729:
1721:
1717:
1714:(3): 129–41.
1713:
1709:
1702:
1694:
1690:
1685:
1680:
1675:
1670:
1666:
1662:
1658:
1651:
1643:
1639:
1635:
1631:
1627:
1623:
1619:
1615:
1608:
1600:
1596:
1592:
1588:
1580:
1572:
1566:
1558:
1554:
1549:
1544:
1540:
1536:
1532:
1525:
1517:
1511:
1503:
1499:
1494:
1489:
1485:
1481:
1477:
1470:
1463:
1457:
1449:
1443:
1435:
1431:
1427:
1423:
1419:
1415:
1411:
1407:
1399:
1391:
1387:
1383:
1379:
1375:
1371:
1367:
1363:
1355:
1347:
1343:
1339:
1335:
1331:
1327:
1323:
1319:
1311:
1302:
1294:
1288:
1280:
1276:
1271:
1266:
1262:
1258:
1254:
1250:
1246:
1239:
1231:
1227:
1223:
1219:
1215:
1211:
1207:
1203:
1196:
1188:
1182:
1174:
1170:
1166:
1162:
1158:
1154:
1147:
1139:
1133:
1125:
1121:
1117:
1113:
1109:
1105:
1101:
1097:
1089:
1081:
1075:
1067:
1063:
1059:
1055:
1051:
1050:10.1038/81576
1047:
1043:
1039:
1032:
1024:
1018:
1010:
1006:
1001:
996:
992:
988:
984:
980:
976:
969:
960:
952:
946:
938:
934:
930:
926:
922:
918:
917:Anal. Biochem
911:
903:
897:
889:
885:
880:
875:
871:
867:
866:BioTechniques
863:
856:
848:
842:
834:
830:
825:
820:
816:
812:
808:
801:
793:
789:
784:
779:
775:
771:
767:
763:
759:
755:
751:
744:
736:
732:
727:
722:
718:
714:
710:
706:
702:
695:
687:
681:
673:
669:
665:
661:
657:
653:
646:
638:
634:
629:
624:
620:
616:
612:
605:
597:
593:
589:
585:
581:
577:
570:
562:
558:
554:
550:
547:(2): 155–68.
546:
542:
535:
527:
523:
519:
515:
511:
507:
503:
499:
498:Biotechnology
492:
484:
477:
469:
462:
454:
447:
439:
435:
431:
427:
423:
419:
412:
404:
398:
390:
386:
382:
378:
374:
370:
366:
362:
355:
347:
335:
327:
323:
319:
317:9780128022658
313:
309:
305:
301:
294:
286:
282:
278:
272:
268:
264:
260:
253:
245:
241:
236:
231:
227:
223:
219:
212:
208:
199:
196:
194:
191:
189:
186:
184:
181:
179:
176:
174:
171:
170:
164:
162:
161:OSTE polymers
151:
147:
145:
141:
135:
133:
128:
124:
120:
118:
112:
110:
106:
103:
99:
95:
91:
87:
82:
78:
74:
70:
69:Tse Wen Chang
60:
58:
53:
49:
45:
41:
32:
19:
2004:10261/211201
1986:
1982:
1976:
1951:
1947:
1937:
1923:cite journal
1901:(1): 79–86.
1898:
1894:
1888:
1877:. Retrieved
1873:
1864:
1852:
1815:
1811:
1801:
1756:
1752:
1742:
1728:cite journal
1711:
1707:
1701:
1664:
1660:
1650:
1617:
1613:
1607:
1590:
1586:
1579:
1565:cite journal
1538:
1534:
1524:
1510:cite journal
1483:
1479:
1469:
1461:
1456:
1442:cite journal
1409:
1405:
1398:
1365:
1361:
1354:
1321:
1317:
1310:
1301:
1287:cite journal
1252:
1248:
1238:
1205:
1201:
1195:
1181:cite journal
1156:
1152:
1146:
1132:cite journal
1099:
1095:
1088:
1074:cite journal
1041:
1037:
1031:
1017:cite journal
982:
978:
968:
959:
945:cite journal
920:
916:
910:
896:cite journal
869:
865:
855:
841:cite journal
814:
810:
800:
757:
753:
743:
708:
705:Biomaterials
704:
694:
680:cite journal
658:(1): 79–86.
655:
651:
645:
618:
614:
604:
579:
575:
569:
544:
540:
534:
504:(3): 291–3.
501:
497:
491:
476:
461:
446:
421:
417:
411:
397:cite journal
364:
360:
354:
299:
293:
258:
252:
225:
221:
211:
157:
154:Applications
148:
136:
129:
125:
121:
113:
66:
43:
39:
37:
2040:Microarrays
985:(15): E72.
86:microarrays
2034:Categories
1989:: 112279.
1879:2018-04-24
1406:Proteomics
1362:Proteomics
1318:Proteomics
811:Clin. Chem
615:Clin. Chem
576:Clin. Chem
222:Anal. Chem
204:References
63:Background
52:antibodies
2021:218688183
1818:(1): 31.
1464:1 (7): 68
760:: 11791.
344:ignored (
334:cite book
2013:32421629
1968:28266828
1915:24287424
1844:27216421
1793:26352148
1753:PLOS ONE
1720:20551245
1693:20164060
1642:30102746
1634:17425464
1557:24440852
1502:26993403
1434:29548647
1426:17787036
1382:17474144
1338:17474144
1279:10848577
1222:11001728
1173:10851162
1124:24008668
1116:10949141
1058:11062466
1009:10908365
937:10328771
888:10524321
792:26138532
735:20452666
672:24287424
526:35328421
389:34616684
381:12666998
326:25934364
285:18370316
244:18837515
167:See also
73:Centocor
1835:4877764
1784:4564184
1761:Bibcode
1684:2877986
1390:9852887
1346:9852887
1230:2753950
1202:Science
1066:9238048
833:9733002
783:4490339
762:Bibcode
726:2876200
637:9733000
596:1934470
561:2488616
518:7765290
438:6606681
105:T cells
2019:
2011:
1966:
1913:
1842:
1832:
1791:
1781:
1718:
1691:
1681:
1640:
1632:
1555:
1500:
1432:
1424:
1388:
1380:
1344:
1336:
1277:
1267:
1228:
1220:
1171:
1122:
1114:
1064:
1056:
1007:
1000:102691
997:
935:
886:
831:
790:
780:
733:
723:
670:
635:
594:
559:
524:
516:
436:
387:
379:
324:
314:
283:
273:
242:
100:, and
2017:S2CID
1638:S2CID
1430:S2CID
1386:S2CID
1342:S2CID
1270:85828
1226:S2CID
1120:S2CID
1062:S2CID
522:S2CID
385:S2CID
173:ELISA
90:Tanox
2009:PMID
1964:PMID
1929:link
1911:PMID
1840:PMID
1789:PMID
1734:link
1716:PMID
1689:PMID
1630:PMID
1571:link
1553:PMID
1539:1845
1516:link
1498:PMID
1484:1865
1448:link
1422:PMID
1378:PMID
1334:PMID
1293:link
1275:PMID
1218:PMID
1187:link
1169:PMID
1138:link
1112:PMID
1080:link
1054:PMID
1023:link
1005:PMID
951:link
933:PMID
902:link
884:PMID
847:link
829:PMID
788:PMID
731:PMID
686:link
668:PMID
633:PMID
592:PMID
557:PMID
514:PMID
434:PMID
403:link
377:PMID
346:help
322:PMID
312:ISBN
281:PMID
271:ISBN
240:PMID
1999:hdl
1991:doi
1987:163
1956:doi
1903:doi
1899:403
1830:PMC
1820:doi
1779:PMC
1769:doi
1679:PMC
1669:doi
1622:doi
1595:doi
1543:doi
1488:doi
1414:doi
1370:doi
1326:doi
1265:PMC
1257:doi
1210:doi
1206:289
1161:doi
1104:doi
1046:doi
995:PMC
987:doi
925:doi
921:270
874:doi
819:doi
778:PMC
770:doi
721:PMC
713:doi
660:doi
656:403
623:doi
584:doi
549:doi
506:doi
426:doi
369:doi
304:doi
263:doi
230:doi
142:or
109:HIV
102:CD8
98:CD4
94:CD3
81:HLA
75:in
38:An
2036::
2015:.
2007:.
1997:.
1985:.
1962:.
1950:.
1946:.
1925:}}
1921:{{
1909:.
1897:.
1872:.
1838:.
1828:.
1816:11
1814:.
1810:.
1787:.
1777:.
1767:.
1757:10
1755:.
1751:.
1730:}}
1726:{{
1710:.
1687:.
1677:.
1663:.
1659:.
1636:.
1628:.
1616:.
1591:10
1589:.
1567:}}
1563:{{
1551:.
1537:.
1533:.
1512:}}
1508:{{
1496:.
1482:.
1478:.
1444:}}
1440:{{
1428:.
1420:.
1408:.
1384:.
1376:.
1364:.
1340:.
1332:.
1320:.
1289:}}
1285:{{
1273:.
1263:.
1253:20
1251:.
1247:.
1224:.
1216:.
1204:.
1183:}}
1179:{{
1167:.
1155:.
1134:}}
1130:{{
1118:.
1110:.
1100:21
1098:.
1076:}}
1072:{{
1060:.
1052:.
1042:26
1040:.
1019:}}
1015:{{
1003:.
993:.
983:28
981:.
977:.
947:}}
943:{{
931:.
919:.
898:}}
894:{{
882:.
870:27
868:.
864:.
843:}}
839:{{
827:.
815:44
813:.
809:.
786:.
776:.
768:.
756:.
752:.
729:.
719:.
709:31
707:.
703:.
682:}}
678:{{
666:.
654:.
631:.
619:44
617:.
613:.
590:.
580:37
578:.
555:.
543:.
520:.
512:.
502:11
500:.
432:.
422:65
420:.
399:}}
395:{{
383:.
375:.
365:41
363:.
338::
336:}}
332:{{
320:.
310:.
279:.
269:.
238:.
226:80
224:.
220:.
119:.
96:,
2023:.
2001::
1993::
1970:.
1958::
1952:9
1931:)
1917:.
1905::
1882:.
1846:.
1822::
1795:.
1771::
1763::
1736:)
1722:.
1712:7
1695:.
1671::
1665:9
1644:.
1624::
1618:4
1601:.
1597::
1573:)
1559:.
1545::
1518:)
1504:.
1490::
1450:)
1436:.
1416::
1410:7
1392:.
1372::
1366:7
1348:.
1328::
1322:7
1295:)
1281:.
1259::
1232:.
1212::
1189:)
1175:.
1163::
1157:3
1140:)
1126:.
1106::
1082:)
1068:.
1048::
1025:)
1011:.
989::
953:)
939:.
927::
904:)
890:.
876::
849:)
835:.
821::
794:.
772::
764::
758:5
737:.
715::
688:)
674:.
662::
639:.
625::
598:.
586::
563:.
551::
545:7
528:.
508::
440:.
428::
405:)
391:.
371::
348:)
328:.
306::
287:.
265::
246:.
232::
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.