702:(EMT) allows carcinoma cells to acquire invasive properties. The translational activation of the extracellular matrix component laminin B1 (LAMB1) during EMT has been recently reported, suggesting an IRES-mediated mechanism. The IRES activity of LamB1 was determined by independent bicistronic reporter assays. Strong evidence excludes an impact of cryptic promoter or splice sites on IRES-driven translation of LamB1. Furthermore, no other LamB1 mRNA species arising from alternative transcription start sites or polyadenylation signals were detected that account for its translational control. Mapping of the LamB1 5'-untranslated region (UTR) revealed the minimal LamB1 IRES motif between -293 and -1 upstream of the start codon. RNA affinity purification demonstrated that the La protein interacts with the LamB1 IRES. This interaction and its regulation during EMT were confirmed by ribonucleoprotein immunoprecipitation. La is able to positively modulate LamB1 IRES translation, so LamB1 IRES is activated by binding to La which leads to translational upregulation during hepatocellular EMT.
726:, as well as some primary cell cultures, which can be difficult to propagate on other substrates. Two types of naturally-sourced laminins are commercially available: Laminin-111, extracted from mouse sarcomas, and laminin mixtures from human placenta, which may primarily correspond to laminin-211, 411, or 511, depending on the provider. The various laminin isoforms are practically impossible to isolate from tissues in pure form due to extensive cross-linking and the need for harsh extraction conditions, such as proteolytic enzymes or low pH, that cause degradation. Therefore,
40:
1403:
1249:
1083:
27:
641:
553:. It is also often used as a substrate in cell culture experiments. The presence of laminin-1 can influence how the growth cone responds to other cues. For example, growth cones are repelled by netrin when grown on laminin-111 but are attracted to netrin when grown on fibronectin. This effect of laminin-111 probably occurs through a lowering of intracellular cyclic AMP.
131:. The trimeric proteins intersect, composing a cruciform structure that is able to bind to other molecules of the extracellular matrix and cell membrane. The three short arms have an affinity for binding to other laminin molecules, conducing sheet formation. The long arm is capable of binding to cells and helps anchor organized tissue cells to the basement membrane.
1831:
family proteins have one laminin G domain, the CNTNAP proteins have four laminin G domains, while neurexin 1 and 2 each hold six laminin G domains. On average, approximately one quarter of the proteins that hold laminin G domains is taken up by these laminin G domains themselves. The smallest laminin
493:
glycoprotein complex and
Lutheran blood group glycoprotein. Through these interactions, laminins critically contribute to cell attachment and differentiation, cell shape and movement, maintenance of tissue phenotype, and promotion of tissue survival. Some of these biological functions of laminin have
2626:
Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, Engel J, Engvall E, Hohenester E, Jones JC, Kleinman HK, Marinkovich MP, Martin GR, Mayer U, Meneguzzi G, Miner JH, Miyazaki K, Patarroyo M, Paulsson M, Quaranta V, Sanes JR, Sasaki T, Sekiguchi K, Sorokin LM, Talts JF,
3155:
Wondimu Z, Gorfu G, Kawataki T, Smirnov S, Yurchenco P, Tryggvason K, Patarroyo M (March 2006). "Characterization of commercial laminin preparations from human placenta in comparison to recombinant laminins 2 (alpha2beta1gamma1), 8 (alpha4beta1gamma1), 10 (alpha5beta1gamma1)".
3273:
Miyazaki T, Futaki S, Hasegawa K, Kawasaki M, Sanzen N, Hayashi M, Kawase E, Sekiguchi K, Nakatsuji N, Suemori H (October 2008). "Recombinant human laminin isoforms can support the undifferentiated growth of human embryonic stem cells".
2926:
Ichikawa N, Kasai S, Suzuki N, Nishi N, Oishi S, Fujii N, Kadoya Y, Hatori K, Mizuno Y, Nomizu M, Arikawa-Hirasawa E (April 2005). "Identification of neurite outgrowth active sites on the laminin alpha4 chain G domain".
1788:. It is also known as a 'LE' or 'laminin-type EGF-like' domain. The number of copies of the laminin EGF-like domain in the different forms of laminins is highly variable; from 3 up to 22 copies have been found. In
3455:
Stetefeld J, Mayer U, Timpl R, Huber R (April 1996). "Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin gamma1 chain harboring the nidogen binding site".
1909:
and dystroglycan (and possibly other receptors) recruited to the adherent laminin. This LN domain-dependent self-assembly is considered to be crucial for the integrity of basement membranes, as highlighted by
2881:
Ockleford C, Bright N, Hubbard A, D'Lacey C, Smith J, Gardiner L, Sheikh T, Albentosa M, Turtle K (October 1993). "Micro-trabeculae, macro-plaques or mini-basement membranes in human term fetal membranes?".
124:
genes in humans, respectively. The laminin molecules are named according to their chain composition, e.g. laminin-511 contains α5, β1, and γ1 chains. Fourteen other chain combinations have been identified
2962:
Beckmann G, Hanke J, Bork P, Reich JG (February 1998). "Merging extracellular domains: fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins".
1804:
C1-C3 and C5-C6. Long consecutive arrays of laminin EGF-like domains in laminins form rod-like elements of limited flexibility, which determine the spacing in the formation of laminin networks of
3309:
Rodin S, Domogatskaya A, Ström S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K (June 2010). "Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511".
218:
Laminins were previously numbered as they were discovered, i.e., laminin-1, laminin-2, laminin-3, etc., but the nomenclature was changed to describe which chains are present in each
734:
as they have in the human body. In 2008, two groups independently showed that mouse embryonic stem cells can be grown for months on top of recombinant laminin-511. Later, Rodin
1839:
has remained elusive, and a variety of binding functions has been ascribed to different
Laminin G modules. For example, the laminin alpha1 and alpha2 chains each have five
3494:
Baumgartner R, Czisch M, Mayer U, Pöschl E, Huber R, Timpl R, Holak TA (April 1996). "Structure of the nidogen binding LE module of the laminin gamma1 chain in solution".
134:
Laminins are integral to the structural scaffolding of almost every tissue of an organism—secreted and incorporated into cell-associated extracellular matrices. These
1661:
1507:
1361:
1207:
1041:
932:
823:
738:
showed that recombinant laminin-511 can be used to create a xeno-free and defined cell culture environment to culture human pluripotent ES cells and human iPS cells.
2846:
Smith J, Ockleford CD (January 1994). "Laser scanning confocal examination and comparison of nidogen (entactin) with laminin in term human amniochorion".
3364:"Laminin, a multidomain protein. The A chain has a unique globular domain and homology with the basement membrane proteoglycan and the laminin B chains"
1087:
crystal structure of three consecutive laminin-type epidermal growth factor-like (le) modules of laminin gamma1 chain harboring the nidogen binding site
3696:"Structure of the C-terminal laminin G-like domain pair of the laminin alpha2 chain harbouring binding sites for alpha-dystroglycan and heparin"
541:
Laminin-111 is a major substrate along which nerve axons will grow, both in vivo and in vitro. For example, it lays down a path that developing
3049:"The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy"
1816:
The laminin globular (G) domain, also known as the LNS (Laminin-alpha, Neurexin and Sex hormone-binding globulin) domain, is on average 177
2399:
2181:
1918:
containing the deletion of the LN module from the alpha 2 laminin chain. The laminin N-terminal domain is found in all laminin and netrin
617:
143:
138:
are imperative to the maintenance and vitality of tissues; defective laminins can cause muscles to form improperly, leading to a form of
3743:
Xu H, Wu XR, Wewer UM, Engvall E (November 1994). "Murine muscular dystrophy caused by a mutation in the laminin alpha 2 (Lama2) gene".
2627:
Tryggvason K, Uitto J, Virtanen I, von der Mark K, Wewer UM, Yamada Y, Yurchenco PD (August 2005). "A simplified laminin nomenclature".
4501:
3904:
2767:
2750:
2239:
2247:
2235:
1597:
1463:
158:
In humans, fifteen laminin trimers have been identified. The laminins are combinations of different alpha-, beta-, and gamma-chains.
2387:
2157:
2153:
2081:
2121:
2113:
2481:
2477:
2331:
2327:
2133:
2129:
698:(IRES) are involved in cancer development via corresponding proteins. A crucial event in tumor progression, referred to as the
222:(laminin-111, laminin-211, etc.). In addition, many laminins had common names before either laminin nomenclature was in place.
2678:
2516:
1609:
1309:
1155:
1681:
1527:
1381:
1227:
1061:
952:
843:
699:
659:
651:
4552:
730:
laminins have been produced since the year 2000. This made it possible to test if laminins could have a significant role
1820:
in length and can be found in one to six copies in various laminin family members as well as in a large number of other
1407:
the structure of the ligand-binding domain of neurexin 1beta: regulation of lns domain function by alternative splicing
3673:
2830:
2801:
677:
93:(the protein network foundation for most cells and organs). Laminins are vital to biological activity, influencing
1832:
G domain can be found in one of the collagen proteins (COL24A1; 77 AA) and the largest domain in TSPEAR (219 AA).
620:, characterized by generalized blisters, exuberant granulation tissue of the skin and mucosa, and pitted teeth.
4562:
3897:
2577:
1669:
1515:
1369:
1215:
1049:
940:
831:
4284:
4188:
4163:
582:
4316:
4245:
4183:
4178:
3108:"La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition"
3047:
Hall TE, Bryson-Richardson RJ, Berger S, Jacoby AS, Cole NJ, Hollway GE, Berger J, Currie PD (April 2007).
2506:
695:
3531:"Low nidogen affinity of laminin-5 can be attributed to two serine residues in EGF-like motif gamma 2III4"
4234:
4193:
4080:
4075:
4070:
4064:
3983:
526:
4272:
4208:
4151:
4123:
4113:
4043:
3976:
1875:
1665:
1511:
1365:
1211:
1045:
936:
827:
498:
sequence , which is located on the alpha-chain of laminin, promotes the adhesion of endothelial cells.
3405:"EGF-like domains in extracellular matrix proteins: localized signals for growth and differentiation?"
1602:
4261:
3996:
3958:
3890:
1622:
1468:
1322:
1168:
566:
3797:
94:
200:(note that no known laminin trimer incorporates LAMB4 and its function remains poorly understood).
3234:"Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro"
597:
chains. This laminin's distribution includes the brain and muscle fibers. In muscle, it binds to
616:
Abnormal laminin-332, essential for epithelial cell adhesion to the basement membrane, leads to
1901:
may also associate with this network through heterotypic LN domain interactions. This leads to
1752:
2791:
4330:
2818:
1852:
1709:
542:
494:
been associated with specific amino-acid sequences or fragments of laminin. For example, the
623:
Malfunctional laminin-521 in the kidney filter causes leakage of protein into the urine and
581:
Dysfunctional structure of one particular laminin, laminin-211, is the cause of one form of
3925:
3060:
1648:
1494:
1348:
1194:
1028:
919:
810:
610:
561:
Laminins are enriched at the lesion site after peripheral nerve injury and are secreted by
510:
82:
8:
4157:
2173:
1801:
1720:
727:
3064:
2884:
Philosophical
Transactions of the Royal Society of London. Series B, Biological Sciences
53:
Please help update this article to reflect recent events or newly available information.
4496:
4307:
3768:
3560:
3434:
3344:
3132:
3107:
3083:
3048:
3024:
2999:
2723:
2698:
1915:
1756:
and laminin. Laminin IV domain is not found in short laminin chains (alpha4 or beta3).
1694:
624:
147:
139:
3720:
3695:
3640:
3623:
3380:
3363:
2859:
2793:
Connective tissue and its heritable disorders: molecular, genetic, and medical aspects
2553:
2536:
4633:
4311:
4102:
3855:
3846:
3834:
3825:
3813:
3804:
3760:
3725:
3645:
3601:
3552:
3547:
3530:
3511:
3473:
3426:
3421:
3404:
3385:
3336:
3291:
3255:
3214:
3173:
3137:
3088:
3029:
2980:
2944:
2899:
2863:
2826:
2797:
2772:
2728:
2674:
2644:
2605:
2597:
2558:
1886:
1805:
1793:
1747:
1656:
1614:
1502:
1356:
1314:
1202:
1160:
1036:
927:
818:
570:
506:
86:
3772:
3564:
3438:
3348:
1792:
laminin gamma-1 chain, the seventh LE domain has been shown to be the only one that
1565:
1431:
1277:
1111:
990:
881:
772:
120:) and possess three different chains (α, β, and γ) encoded by five, four, and three
4426:
4421:
4382:
4377:
4372:
4367:
4362:
4107:
3971:
3793:
3752:
3715:
3707:
3635:
3624:"Self-assembly and calcium-binding sites in laminin. A three-arm interaction model"
3591:
3542:
3503:
3465:
3416:
3375:
3326:
3318:
3283:
3245:
3204:
3165:
3127:
3119:
3078:
3068:
3019:
3011:
2998:
Nieuwenhuis, B.; Haenzi, B.; Andrews, M. R.; Verhaagen, J.; Fawcett, J. W. (2018).
2972:
2936:
2891:
2855:
2762:
2718:
2710:
2636:
2589:
2548:
2511:
1740:
1644:
1490:
1344:
1190:
1024:
915:
806:
594:
586:
502:
466:
1578:
1444:
1290:
1124:
4616:
4611:
4606:
4601:
4596:
4591:
4586:
4547:
4404:
4399:
4394:
3990:
3952:
3169:
2640:
1919:
1902:
1867:
1785:
1697:
1590:
1456:
1302:
1136:
1002:
893:
784:
590:
514:
486:
219:
51:. The reason given is: Improve the article with up to date research on the topic.
3663:
3250:
3233:
4034:
3788:
3596:
3579:
3287:
3053:
Proceedings of the
National Academy of Sciences of the United States of America
2714:
1894:
1871:
1836:
1765:
1713:
113:
109:
98:
74:
3867:
3711:
2593:
2535:
Timpl R, Rohde H, Robey PG, Rennard SI, Foidart JM, Martin GR (October 1979).
1716:
I and II from laminin A, B1 and B2 may come together to form a triple helical
4627:
4335:
4299:
3917:
3209:
3192:
2601:
1863:
1821:
1769:
1732:
518:
117:
102:
78:
3073:
4459:
3729:
3529:
Mayer U, Pöschl E, Gerecke DR, Wagman DW, Burgeson RE, Timpl R (May 1995).
3507:
3469:
3340:
3295:
3259:
3218:
3177:
3141:
3092:
3033:
2976:
2948:
2895:
2776:
2732:
2648:
2609:
1855:
1844:
1789:
1743:
719:
598:
562:
490:
135:
90:
3882:
3764:
3649:
3605:
3580:"Structure and function of laminin: anatomy of a multidomain glycoprotein"
3556:
3515:
3477:
3430:
3389:
3000:"Integrins promote axonal regeneration after injury of the nervous system"
2984:
2903:
2867:
2768:
10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R
1618:
1318:
1164:
30:
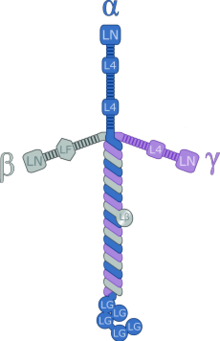
Illustration of the laminin-111 complex depicting the domain organization.
4537:
4466:
4202:
4147:
3123:
2625:
2562:
1827:. For example, all laminin alpha-chains have five laminin G domains, all
1817:
1717:
715:
474:
251:
4168:
3756:
3362:
Sasaki M, Kleinman HK, Huber H, Deutzmann R, Yamada Y (November 1988).
3331:
2699:"Developmental and pathogenic mechanisms of basement membrane assembly"
1890:
1848:
1840:
1781:
1773:
1148:
121:
3015:
2940:
3322:
3191:
Kortesmaa, Jarkko; Yurchenco, Peter; Tryggvason, Karl (19 May 2000).
1906:
1708:
Laminins are trimeric molecules; laminin-1 is an alpha1 beta1 gamma1
723:
530:
1893:
through their N-terminal domain (LN or domain VI) and anchor to the
4579:
4320:
3936:
3668:
1911:
1828:
1780:. The tertiary structure of this domain is remotely similar in its
1777:
1585:
1451:
1297:
1131:
997:
888:
779:
711:
482:
478:
470:
3232:
Domogatskaya A, Rodin S, Boutaud A, Tryggvason K (November 2008).
3231:
4542:
4533:
4481:
4454:
4325:
4289:
4277:
4095:
4090:
4085:
4013:
3913:
3860:
3850:
3839:
3829:
3818:
3808:
3046:
2997:
2880:
2311:
2307:
2303:
2299:
2295:
2291:
2287:
2283:
2279:
2275:
2271:
2267:
2263:
2243:
2231:
2227:
1898:
1859:
1824:
1797:
1736:
1735:
module of unknown function. It is found in a number of different
1143:
522:
495:
127:
4486:
4250:
4223:
4218:
4213:
4138:
4133:
4128:
4118:
4053:
4048:
4006:
4001:
3963:
2411:
2259:
2255:
2251:
2223:
2219:
2215:
2117:
2101:
2097:
2093:
2089:
2085:
1676:
1522:
1402:
1376:
1248:
1222:
1082:
1056:
947:
838:
691:
569:
express integrin receptors that attach to laminins and promote
550:
546:
529:
of the laminin-G domain has been predicted to resemble that of
3272:
1768:
each laminin subunit contains, in its first half, consecutive
47:
Parts of this article (those related to
Pathology) need to be
4511:
4506:
4476:
4471:
4449:
4431:
4409:
4018:
3308:
2668:
2493:
2489:
2485:
2469:
2465:
2461:
2457:
2453:
2449:
2445:
2441:
2437:
2433:
2423:
2415:
2407:
2395:
2391:
2383:
2379:
2375:
2371:
2367:
2363:
2359:
2355:
2351:
2347:
2343:
2339:
2211:
2207:
2203:
2199:
2195:
2185:
2177:
2169:
2165:
2161:
2149:
2145:
2141:
2137:
2109:
2105:
2073:
2069:
2065:
2061:
2057:
2053:
2049:
2045:
2041:
2037:
2033:
2029:
2019:
2015:
2011:
2007:
2003:
1999:
1995:
1991:
1987:
1977:
1973:
1969:
1965:
1961:
1951:
1947:
1943:
1939:
1935:
602:
212:
208:
204:
197:
193:
189:
185:
179:
175:
171:
167:
163:
26:
20:
3845:
Overview of all the structural information available in the
3824:
Overview of all the structural information available in the
3803:
Overview of all the structural information available in the
3493:
3361:
3190:
3106:
Petz M, Them N, Huber H, Beug H, Mikulits W (January 2012).
1925:
694:
pathophysiology. The majority of transcripts that harbor an
4575:
4491:
3154:
2473:
2419:
2403:
2335:
2323:
2319:
2315:
2125:
2077:
1638:
1572:
1560:
1484:
1438:
1426:
1338:
1284:
1272:
1184:
1118:
1106:
1018:
985:
909:
876:
800:
767:
606:
465:
Laminins form independent networks and are associated with
1843:
laminin G domains, where only domains LG4 and LG5 contain
1800:. The binding-sites are located on the surface within the
710:
Together with other major components of the ECM, such as
609:
via the G domain, and via the other end, it binds to the
3693:
3528:
3454:
2925:
2751:"Form and function: the laminin family of heterotrimers"
1253:
laminin alpha 2 chain lg4-5 domain pair, ca1 site mutant
2819:"Extracellular matrix constituents as integrin ligands"
4574:
This article incorporates text from the public domain
3694:
Tisi D, Talts JF, Timpl R, Hohenester E (April 2000).
2961:
2534:
1731:
The laminin B domain (also known as domain IV) is an
690:
Some of the laminin isoforms have been implicated in
1889:
assembly is a cooperative process in which laminins
3276:
2796:(2nd ed.). New York: Wiley-Liss. p. 306.
481:. The proteins also bind to cell membranes through
85:of all animals. They are major constituents of the
2748:
556:
3105:
2537:"Laminin--a glycoprotein from basement membranes"
2214:), cadherin EGF LAG seven-pass G-type receptors (
2084:), cadherin EGF LAG seven-pass G-type receptors (
4625:
3742:
2991:
2096:), cysteine-rich with EGF-like domain proteins (
3577:
2696:
718:, laminins have been used to enhance mammalian
3621:
3617:
3615:
3489:
3487:
3450:
3448:
2845:
1922:except laminin alpha 3A, alpha 4 and gamma 2.
501:Laminin alpha4 is distributed in a variety of
3898:
536:
3687:
3225:
3099:
2919:
2825:. New York: Chapman & Hall. p. 50.
3912:
3736:
3612:
3578:Beck K, Hunter I, Engel J (February 1990).
3571:
3522:
3484:
3445:
3396:
3355:
2955:
2874:
2744:
2742:
2692:
2690:
2621:
2619:
1703:
3905:
3891:
3874:. Imperial College London. April 13, 2011.
2839:
2671:Extracellular matrix: a practical approach
2664:
2662:
2660:
2658:
1862:appear to have a wide variety of roles in
1401:
1247:
1081:
3876:(lecture by Professor Erhard Hoheneseter)
3796:at the U.S. National Library of Medicine
3719:
3639:
3595:
3546:
3420:
3379:
3330:
3266:
3249:
3208:
3131:
3082:
3072:
3023:
2766:
2722:
2552:
1926:Human proteins containing laminin domains
678:Learn how and when to remove this message
2739:
2687:
2669:M. A. Haralson; John R. Hassell (1995).
2616:
722:, especially in the case of pluripotent
146:), and/or defects of the kidney filter (
25:
3935:
2749:Colognato H, Yurchenco PD (June 2000).
2655:
2575:
2394:), chondroitin sulfate proteoglycan 4 (
1776:in length that include eight conserved
4626:
3622:Yurchenco PD, Cheng YS (August 1993).
2104:), multiple EGF-like domain proteins (
705:
3886:
3402:
2789:
2517:List of target antigens in pemphigoid
2402:), growth arrest-specific protein 6 (
1881:
2816:
2528:
2026:Laminin EGF-like (domains III and V)
1835:The exact function of the Laminin G
1076:Laminin EGF-like (Domains III and V)
700:epithelial-to-mesenchymal transition
634:
33:
4553:Cartilage oligomeric matrix protein
3628:The Journal of Biological Chemistry
3368:The Journal of Biological Chemistry
2576:Durbeej, Madeleine (January 2010).
2541:The Journal of Biological Chemistry
1764:Beside different types of globular
1759:
303:Kalinin, epiligrin, nicein, ladsin
16:Protein in the extracellular matrix
13:
2226:), contactin-associated proteins (
741:
650:tone or style may not reflect the
142:, lethal skin blistering disease (
14:
4645:
3859:(Laminin subunit alpha-5) at the
3838:(Laminin subunit alpha-2) at the
3817:(Laminin subunit alpha-1) at the
3782:
3674:European Bioinformatics Institute
2418:), sex hormone-binding globulin (
1897:surface through their G domains.
1712:. It has been suggested that the
630:
3193:"Recombinant Laminin-8 (α4β1γ1)"
2697:Yurchenco PD, Patton BL (2009).
2152:), class F scavenger receptors (
660:guide to writing better articles
639:
618:junctional epidermolysis bullosa
174:(which has three splice forms),
144:junctional epidermolysis bullosa
38:
3868:"How I learned to love laminin"
3656:
3302:
3197:Journal of Biological Chemistry
3184:
3148:
3040:
2910:
557:Role in peripheral nerve repair
2821:. In Elbe, Johannes A. (ed.).
2810:
2783:
2569:
2430:Laminin N-terminal (domain VI)
1750:, a laminin-like protein from
1542:Laminin N-terminal (Domain VI)
1:
3641:10.1016/S0021-9258(19)85334-6
3381:10.1016/S0021-9258(18)37424-6
2860:10.1016/S0143-4004(05)80240-1
2790:Royce, Peter M., ed. (2002).
2703:Current Pharmaceutical Design
2554:10.1016/S0021-9258(19)83607-4
2522:
1633:Available protein structures:
1479:Available protein structures:
1333:Available protein structures:
1179:Available protein structures:
1013:Available protein structures:
904:Available protein structures:
795:Available protein structures:
585:. Laminin-211 is composed of
583:congenital muscular dystrophy
545:follow on their way from the
521:junction, it is required for
4317:Cartilage associated protein
3548:10.1016/0014-5793(95)00438-F
3496:Journal of Molecular Biology
3458:Journal of Molecular Biology
3422:10.1016/0014-5793(89)81417-6
3170:10.1016/j.matbio.2005.10.001
2965:Journal of Molecular Biology
2641:10.1016/j.matbio.2005.05.006
2507:Substrate adhesion molecules
2314:), crumbs homologs 1 and 2 (
2194:: all laminin alpha chains (
2172:), angiopoietin-1 receptor (
1986:: all laminin alpha chains (
1960:: all laminin alpha chains (
1934:: all laminin alpha chains (
1811:
1726:
696:internal ribosome entry site
576:
203:Three gamma-chain isoforms:
7:
3251:10.1634/stemcells.2007-0389
2823:Integrin-ligand interaction
2500:
460:
350:Laminin-321 / Laminin-3A21
336:Laminin-311 / Laminin-3A11
309:Laminin-332 / Laminin-3A32
162:Five alpha-chain isoforms:
10:
4650:
4573:
3597:10.1096/fasebj.4.2.2404817
3288:10.1016/j.bbrc.2008.07.111
2715:10.2174/138161209787846766
2673:. Ithaca, N.Y: IRL Press.
2612:– via Springer Link.
537:Role in neural development
517:, and capillaries; in the
184:Four beta-chain isoforms:
18:
4526:
4442:
4348:
4298:
4027:
3944:
3924:
2594:10.1007/s00441-009-0838-2
1693:Laminins contain several
1675:
1655:
1637:
1632:
1628:
1608:
1596:
1584:
1571:
1559:
1551:
1546:
1541:
1521:
1501:
1483:
1478:
1474:
1462:
1450:
1437:
1425:
1417:
1412:
1400:
1395:
1375:
1355:
1337:
1332:
1328:
1308:
1296:
1283:
1271:
1263:
1258:
1246:
1241:
1221:
1201:
1183:
1178:
1174:
1154:
1142:
1130:
1117:
1105:
1097:
1092:
1080:
1075:
1055:
1035:
1017:
1012:
1008:
996:
984:
976:
971:
966:
946:
926:
908:
903:
899:
887:
875:
867:
862:
857:
837:
817:
799:
794:
790:
778:
766:
758:
753:
748:
567:peripheral nervous system
3798:Medical Subject Headings
3210:10.1074/jbc.275.20.14853
2582:Cell and Tissue Research
1796:with a high affinity to
1704:Laminin I and Laminin II
384:Drosophila-like laminin
153:
19:Not to be confused with
3712:10.1093/emboj/19.7.1432
3074:10.1073/pnas.0700942104
2432:: most laminin chains (
1858:. Laminin G-containing
489:molecules, such as the
341:Laminin-7 / Laminin-7A
327:Laminin-6 / Laminin-6A
300:Laminin-5 / Laminin-5A
3508:10.1006/jmbi.1996.0192
3470:10.1006/jmbi.1996.0191
3112:Nucleic Acids Research
2977:10.1006/jmbi.1997.1510
2896:10.1098/rstb.1993.0142
2755:Developmental Dynamics
2398:), eyes shut homolog (
2338:), NEL-like proteins (
2028:: all laminin chains (
1753:Caenorhabditis elegans
543:retinal ganglion cells
31:
4331:Procollagen peptidase
3403:Engel J (July 1989).
2144:), mucins 3A and 3B (
1984:Laminin B (domain IV)
1853:cell surface receptor
967:Laminin B (Domain IV)
112:proteins with a high
29:
3311:Nature Biotechnology
2817:Kühn, Klaus (1997).
2370:), thrombospondins (
611:extracellular matrix
525:specialisation. The
511:dorsal root ganglion
95:cell differentiation
83:extracellular matrix
3789:The Laminin Protein
3203:(20): 14853–14859.
3065:2007PNAS..104.7092H
2250:), some collagens (
706:Use in cell culture
4497:Matrix gla protein
4308:Prolyl hydroxylase
3757:10.1038/ng1194-297
3664:"Laminin G domain"
3124:10.1093/nar/gkr717
3004:Biological Reviews
2916:Beck et al., 1999.
2358:), slit homologs (
1916:muscular dystrophy
1882:Laminin N-terminal
1806:basement membranes
625:nephrotic syndrome
599:alpha-dystroglycan
234:Chain composition
148:nephrotic syndrome
140:muscular dystrophy
32:
4571:
4570:
4522:
4521:
4344:
4343:
4312:Lysyl hydroxylase
4103:basement membrane
3016:10.1111/brv.12398
2941:10.1021/bi0476228
2680:978-0-19-963220-6
2472:), most netrins (
2322:), fat homologs (
2124:), most netrins (
2018:), and perlecan (
2006:), gamma chains (
1958:Laminin domain II
1887:Basement membrane
1748:basement membrane
1691:
1690:
1687:
1686:
1682:structure summary
1537:
1536:
1533:
1532:
1528:structure summary
1391:
1390:
1387:
1386:
1382:structure summary
1237:
1236:
1233:
1232:
1228:structure summary
1071:
1070:
1067:
1066:
1062:structure summary
962:
961:
958:
957:
953:structure summary
858:Laminin Domain II
853:
852:
849:
848:
844:structure summary
688:
687:
680:
654:used on Knowledge
652:encyclopedic tone
571:neuroregeneration
565:. Neurons of the
507:peripheral nerves
458:
457:
237:New nomenclature
228:Old nomenclature
87:basement membrane
68:
67:
4641:
3942:
3941:
3933:
3932:
3907:
3900:
3893:
3884:
3883:
3875:
3777:
3776:
3740:
3734:
3733:
3723:
3700:The EMBO Journal
3691:
3685:
3684:
3682:
3680:
3660:
3654:
3653:
3643:
3634:(23): 17286–99.
3619:
3610:
3609:
3599:
3575:
3569:
3568:
3550:
3526:
3520:
3519:
3491:
3482:
3481:
3452:
3443:
3442:
3424:
3400:
3394:
3393:
3383:
3374:(32): 16536–44.
3359:
3353:
3352:
3334:
3323:10.1038/nbt.1620
3306:
3300:
3299:
3270:
3264:
3263:
3253:
3229:
3223:
3222:
3212:
3188:
3182:
3181:
3152:
3146:
3145:
3135:
3103:
3097:
3096:
3086:
3076:
3044:
3038:
3037:
3027:
3010:(3): 1339–1362.
2995:
2989:
2988:
2959:
2953:
2952:
2923:
2917:
2914:
2908:
2907:
2890:(1300): 121–36.
2878:
2872:
2871:
2843:
2837:
2836:
2814:
2808:
2807:
2787:
2781:
2780:
2770:
2746:
2737:
2736:
2726:
2694:
2685:
2684:
2666:
2653:
2652:
2623:
2614:
2613:
2573:
2567:
2566:
2556:
2532:
2512:Laminin database
2492:), and usherin (
2192:Laminin G domain
2184:), and usherin (
1932:Laminin domain I
1760:Laminin EGF-like
1741:heparan sulphate
1630:
1629:
1539:
1538:
1476:
1475:
1405:
1396:Laminin G domain
1393:
1392:
1330:
1329:
1251:
1242:Laminin G domain
1239:
1238:
1176:
1175:
1085:
1073:
1072:
1010:
1009:
964:
963:
901:
900:
855:
854:
792:
791:
749:Laminin Domain I
746:
745:
683:
676:
672:
669:
663:
662:for suggestions.
658:See Knowledge's
643:
642:
635:
467:type IV collagen
225:
224:
63:
60:
54:
42:
41:
34:
4649:
4648:
4644:
4643:
4642:
4640:
4639:
4638:
4624:
4623:
4622:
4621:
4572:
4567:
4518:
4438:
4340:
4294:
4177:transmembrane:
4023:
3927:
3920:
3911:
3880:
3866:
3785:
3780:
3745:Nature Genetics
3741:
3737:
3692:
3688:
3678:
3676:
3662:
3661:
3657:
3620:
3613:
3576:
3572:
3541:(2–3): 129–32.
3527:
3523:
3492:
3485:
3453:
3446:
3401:
3397:
3360:
3356:
3307:
3303:
3271:
3267:
3230:
3226:
3189:
3185:
3153:
3149:
3104:
3100:
3045:
3041:
2996:
2992:
2960:
2956:
2935:(15): 5755–62.
2924:
2920:
2915:
2911:
2879:
2875:
2844:
2840:
2833:
2815:
2811:
2804:
2788:
2784:
2747:
2740:
2709:(12): 1277–94.
2695:
2688:
2681:
2667:
2656:
2624:
2617:
2574:
2570:
2533:
2529:
2525:
2503:
2422:) and usherin (
2410:), pikachurin (
2180:), tenascin N (
2076:), attractins (
1928:
1903:cell signalling
1884:
1876:differentiation
1874:, assembly and
1814:
1786:EGF-like module
1784:to that of the
1762:
1729:
1706:
1698:protein domains
1408:
1254:
1088:
744:
742:Laminin domains
708:
684:
673:
667:
664:
657:
648:This section's
644:
640:
633:
603:integrin alpha7
579:
559:
539:
515:skeletal muscle
487:plasma membrane
463:
156:
64:
58:
55:
52:
43:
39:
24:
17:
12:
11:
5:
4647:
4637:
4636:
4620:
4619:
4614:
4609:
4604:
4599:
4594:
4589:
4583:
4569:
4568:
4566:
4565:
4560:
4556:
4555:
4550:
4545:
4540:
4530:
4528:
4524:
4523:
4520:
4519:
4517:
4516:
4515:
4514:
4509:
4499:
4494:
4489:
4484:
4479:
4474:
4469:
4464:
4463:
4462:
4452:
4446:
4444:
4440:
4439:
4437:
4436:
4435:
4434:
4429:
4424:
4414:
4413:
4412:
4407:
4402:
4397:
4387:
4386:
4385:
4380:
4375:
4370:
4365:
4354:
4352:
4346:
4345:
4342:
4341:
4339:
4338:
4333:
4328:
4323:
4314:
4304:
4302:
4296:
4295:
4293:
4292:
4287:
4282:
4281:
4280:
4275:
4265:
4255:
4254:
4253:
4248:
4238:
4228:
4227:
4226:
4221:
4216:
4211:
4197:
4196:
4191:
4186:
4181:
4174:
4173:
4172:
4171:
4166:
4154:
4144:
4143:
4142:
4141:
4136:
4131:
4126:
4121:
4116:
4099:
4098:
4093:
4088:
4083:
4078:
4073:
4068:
4058:
4057:
4056:
4051:
4046:
4031:
4029:
4025:
4024:
4022:
4021:
4016:
4011:
4010:
4009:
4004:
3999:
3987:
3980:
3968:
3967:
3966:
3961:
3948:
3946:
3945:Fibril forming
3939:
3930:
3926:Extracellular
3922:
3921:
3918:scleroproteins
3910:
3909:
3902:
3895:
3887:
3878:
3877:
3864:
3843:
3822:
3801:
3791:
3784:
3783:External links
3781:
3779:
3778:
3751:(3): 297–302.
3735:
3706:(7): 1432–40.
3686:
3655:
3611:
3570:
3521:
3483:
3444:
3395:
3354:
3301:
3265:
3244:(11): 2800–9.
3224:
3183:
3158:Matrix Biology
3147:
3118:(1): 290–302.
3098:
3059:(17): 7092–7.
3039:
2990:
2954:
2918:
2909:
2873:
2838:
2831:
2809:
2802:
2782:
2738:
2686:
2679:
2654:
2629:Matrix Biology
2615:
2588:(1): 259–268.
2568:
2547:(19): 9933–7.
2526:
2524:
2521:
2520:
2519:
2514:
2509:
2502:
2499:
2498:
2497:
2427:
2414:), protein S (
2346:), neurexins (
2189:
2160:), stabilins (
2023:
1981:
1955:
1927:
1924:
1883:
1880:
1813:
1810:
1761:
1758:
1739:that include,
1728:
1725:
1705:
1702:
1689:
1688:
1685:
1684:
1679:
1673:
1672:
1659:
1653:
1652:
1642:
1635:
1634:
1626:
1625:
1612:
1606:
1605:
1600:
1594:
1593:
1588:
1582:
1581:
1576:
1569:
1568:
1563:
1557:
1556:
1553:
1549:
1548:
1544:
1543:
1535:
1534:
1531:
1530:
1525:
1519:
1518:
1505:
1499:
1498:
1488:
1481:
1480:
1472:
1471:
1466:
1460:
1459:
1454:
1448:
1447:
1442:
1435:
1434:
1429:
1423:
1422:
1419:
1415:
1414:
1410:
1409:
1406:
1398:
1397:
1389:
1388:
1385:
1384:
1379:
1373:
1372:
1359:
1353:
1352:
1342:
1335:
1334:
1326:
1325:
1312:
1306:
1305:
1300:
1294:
1293:
1288:
1281:
1280:
1275:
1269:
1268:
1265:
1261:
1260:
1256:
1255:
1252:
1244:
1243:
1235:
1234:
1231:
1230:
1225:
1219:
1218:
1205:
1199:
1198:
1188:
1181:
1180:
1172:
1171:
1158:
1152:
1151:
1146:
1140:
1139:
1134:
1128:
1127:
1122:
1115:
1114:
1109:
1103:
1102:
1099:
1095:
1094:
1090:
1089:
1086:
1078:
1077:
1069:
1068:
1065:
1064:
1059:
1053:
1052:
1039:
1033:
1032:
1022:
1015:
1014:
1006:
1005:
1000:
994:
993:
988:
982:
981:
978:
974:
973:
969:
968:
960:
959:
956:
955:
950:
944:
943:
930:
924:
923:
913:
906:
905:
897:
896:
891:
885:
884:
879:
873:
872:
869:
865:
864:
860:
859:
851:
850:
847:
846:
841:
835:
834:
821:
815:
814:
804:
797:
796:
788:
787:
782:
776:
775:
770:
764:
763:
760:
756:
755:
751:
750:
743:
740:
707:
704:
686:
685:
647:
645:
638:
632:
631:Role in cancer
629:
578:
575:
573:after injury.
558:
555:
538:
535:
462:
459:
456:
455:
452:
449:
447:
443:
442:
439:
436:
434:
431:
430:
427:
424:
422:
418:
417:
414:
411:
409:
405:
404:
401:
398:
396:
392:
391:
388:
385:
382:
378:
377:
374:
371:
369:
365:
364:
361:
358:
356:
352:
351:
348:
345:
342:
338:
337:
334:
331:
328:
324:
323:
320:
317:
315:
311:
310:
307:
304:
301:
297:
296:
293:
290:
287:
283:
282:
279:
276:
273:
269:
268:
265:
262:
259:
255:
254:
249:
246:
243:
239:
238:
235:
232:
229:
216:
215:
201:
182:
155:
152:
116:(~400 to ~900
114:molecular mass
110:heterotrimeric
66:
65:
46:
44:
37:
15:
9:
6:
4:
3:
2:
4646:
4635:
4632:
4631:
4629:
4618:
4615:
4613:
4610:
4608:
4605:
4603:
4600:
4598:
4595:
4593:
4590:
4588:
4585:
4584:
4581:
4577:
4564:
4561:
4558:
4557:
4554:
4551:
4549:
4546:
4544:
4541:
4539:
4535:
4532:
4531:
4529:
4525:
4513:
4510:
4508:
4505:
4504:
4503:
4500:
4498:
4495:
4493:
4490:
4488:
4485:
4483:
4480:
4478:
4475:
4473:
4470:
4468:
4465:
4461:
4458:
4457:
4456:
4453:
4451:
4448:
4447:
4445:
4441:
4433:
4430:
4428:
4425:
4423:
4420:
4419:
4418:
4415:
4411:
4408:
4406:
4403:
4401:
4398:
4396:
4393:
4392:
4391:
4388:
4384:
4381:
4379:
4376:
4374:
4371:
4369:
4366:
4364:
4361:
4360:
4359:
4356:
4355:
4353:
4351:
4347:
4337:
4336:Lysyl oxidase
4334:
4332:
4329:
4327:
4324:
4322:
4318:
4315:
4313:
4309:
4306:
4305:
4303:
4301:
4297:
4291:
4288:
4286:
4283:
4279:
4276:
4274:
4271:
4270:
4269:
4266:
4263:
4259:
4256:
4252:
4249:
4247:
4244:
4243:
4242:
4239:
4236:
4232:
4229:
4225:
4222:
4220:
4217:
4215:
4212:
4210:
4207:
4206:
4205:
4204:
4199:
4198:
4195:
4192:
4190:
4187:
4185:
4182:
4180:
4176:
4175:
4170:
4167:
4165:
4162:
4161:
4160:
4159:
4155:
4153:
4149:
4146:
4145:
4140:
4137:
4135:
4132:
4130:
4127:
4125:
4122:
4120:
4117:
4115:
4112:
4111:
4110:
4109:
4104:
4101:
4100:
4097:
4094:
4092:
4089:
4087:
4084:
4082:
4079:
4077:
4074:
4072:
4069:
4066:
4062:
4059:
4055:
4052:
4050:
4047:
4045:
4042:
4041:
4040:
4036:
4033:
4032:
4030:
4026:
4020:
4017:
4015:
4012:
4008:
4005:
4003:
4000:
3998:
3995:
3994:
3993:
3992:
3988:
3986:
3985:
3981:
3978:
3974:
3973:
3969:
3965:
3962:
3960:
3957:
3956:
3955:
3954:
3950:
3949:
3947:
3943:
3940:
3938:
3934:
3931:
3929:
3923:
3919:
3915:
3908:
3903:
3901:
3896:
3894:
3889:
3888:
3885:
3881:
3873:
3869:
3865:
3862:
3858:
3857:
3852:
3848:
3844:
3841:
3837:
3836:
3831:
3827:
3823:
3820:
3816:
3815:
3810:
3806:
3802:
3799:
3795:
3792:
3790:
3787:
3786:
3774:
3770:
3766:
3762:
3758:
3754:
3750:
3746:
3739:
3731:
3727:
3722:
3717:
3713:
3709:
3705:
3701:
3697:
3690:
3675:
3671:
3670:
3665:
3659:
3651:
3647:
3642:
3637:
3633:
3629:
3625:
3618:
3616:
3607:
3603:
3598:
3593:
3590:(2): 148–60.
3589:
3585:
3584:FASEB Journal
3581:
3574:
3566:
3562:
3558:
3554:
3549:
3544:
3540:
3536:
3532:
3525:
3517:
3513:
3509:
3505:
3502:(3): 658–68.
3501:
3497:
3490:
3488:
3479:
3475:
3471:
3467:
3464:(3): 644–57.
3463:
3459:
3451:
3449:
3440:
3436:
3432:
3428:
3423:
3418:
3414:
3410:
3406:
3399:
3391:
3387:
3382:
3377:
3373:
3369:
3365:
3358:
3350:
3346:
3342:
3338:
3333:
3328:
3324:
3320:
3316:
3312:
3305:
3297:
3293:
3289:
3285:
3281:
3277:
3269:
3261:
3257:
3252:
3247:
3243:
3239:
3235:
3228:
3220:
3216:
3211:
3206:
3202:
3198:
3194:
3187:
3179:
3175:
3171:
3167:
3163:
3159:
3151:
3143:
3139:
3134:
3129:
3125:
3121:
3117:
3113:
3109:
3102:
3094:
3090:
3085:
3080:
3075:
3070:
3066:
3062:
3058:
3054:
3050:
3043:
3035:
3031:
3026:
3021:
3017:
3013:
3009:
3005:
3001:
2994:
2986:
2982:
2978:
2974:
2971:(5): 725–30.
2970:
2966:
2958:
2950:
2946:
2942:
2938:
2934:
2930:
2922:
2913:
2905:
2901:
2897:
2893:
2889:
2885:
2877:
2869:
2865:
2861:
2857:
2854:(1): 95–106.
2853:
2849:
2842:
2834:
2832:9780412138614
2828:
2824:
2820:
2813:
2805:
2803:9780471251859
2799:
2795:
2794:
2786:
2778:
2774:
2769:
2764:
2761:(2): 213–34.
2760:
2756:
2752:
2745:
2743:
2734:
2730:
2725:
2720:
2716:
2712:
2708:
2704:
2700:
2693:
2691:
2682:
2676:
2672:
2665:
2663:
2661:
2659:
2650:
2646:
2642:
2638:
2635:(5): 326–32.
2634:
2630:
2622:
2620:
2611:
2607:
2603:
2599:
2595:
2591:
2587:
2583:
2579:
2572:
2564:
2560:
2555:
2550:
2546:
2542:
2538:
2531:
2527:
2518:
2515:
2513:
2510:
2508:
2505:
2504:
2495:
2491:
2487:
2483:
2479:
2475:
2471:
2467:
2463:
2459:
2455:
2451:
2447:
2443:
2439:
2435:
2431:
2428:
2425:
2421:
2417:
2413:
2409:
2406:), perlecan (
2405:
2401:
2397:
2393:
2389:
2385:
2381:
2377:
2373:
2369:
2365:
2361:
2357:
2353:
2349:
2345:
2341:
2337:
2333:
2329:
2325:
2321:
2317:
2313:
2309:
2305:
2301:
2297:
2293:
2289:
2285:
2281:
2277:
2273:
2269:
2265:
2261:
2257:
2253:
2249:
2245:
2241:
2237:
2233:
2229:
2225:
2221:
2217:
2213:
2209:
2205:
2201:
2197:
2193:
2190:
2187:
2183:
2179:
2176:), perlecan (
2175:
2171:
2167:
2163:
2159:
2155:
2151:
2147:
2143:
2139:
2135:
2131:
2127:
2123:
2119:
2115:
2111:
2107:
2103:
2099:
2095:
2091:
2087:
2083:
2079:
2075:
2071:
2067:
2063:
2059:
2055:
2051:
2047:
2043:
2039:
2035:
2031:
2027:
2024:
2021:
2017:
2013:
2009:
2005:
2001:
1997:
1993:
1989:
1985:
1982:
1979:
1975:
1971:
1967:
1963:
1959:
1956:
1953:
1949:
1945:
1941:
1937:
1933:
1930:
1929:
1923:
1921:
1917:
1913:
1908:
1904:
1900:
1896:
1892:
1888:
1879:
1877:
1873:
1869:
1865:
1864:cell adhesion
1861:
1857:
1854:
1850:
1847:for heparin,
1846:
1845:binding sites
1842:
1838:
1833:
1830:
1826:
1823:
1822:extracellular
1819:
1809:
1807:
1803:
1799:
1795:
1791:
1787:
1783:
1779:
1775:
1771:
1767:
1757:
1755:
1754:
1749:
1745:
1742:
1738:
1734:
1733:extracellular
1724:
1722:
1719:
1715:
1711:
1701:
1699:
1696:
1683:
1680:
1678:
1674:
1671:
1667:
1663:
1660:
1658:
1654:
1650:
1646:
1643:
1640:
1636:
1631:
1627:
1624:
1620:
1616:
1613:
1611:
1607:
1604:
1601:
1599:
1595:
1592:
1589:
1587:
1583:
1580:
1577:
1574:
1570:
1567:
1564:
1562:
1558:
1554:
1550:
1545:
1540:
1529:
1526:
1524:
1520:
1517:
1513:
1509:
1506:
1504:
1500:
1496:
1492:
1489:
1486:
1482:
1477:
1473:
1470:
1467:
1465:
1461:
1458:
1455:
1453:
1449:
1446:
1443:
1440:
1436:
1433:
1430:
1428:
1424:
1420:
1416:
1411:
1404:
1399:
1394:
1383:
1380:
1378:
1374:
1371:
1367:
1363:
1360:
1358:
1354:
1350:
1346:
1343:
1340:
1336:
1331:
1327:
1324:
1320:
1316:
1313:
1311:
1307:
1304:
1301:
1299:
1295:
1292:
1289:
1286:
1282:
1279:
1276:
1274:
1270:
1266:
1262:
1257:
1250:
1245:
1240:
1229:
1226:
1224:
1220:
1217:
1213:
1209:
1206:
1204:
1200:
1196:
1192:
1189:
1186:
1182:
1177:
1173:
1170:
1166:
1162:
1159:
1157:
1153:
1150:
1147:
1145:
1141:
1138:
1135:
1133:
1129:
1126:
1123:
1120:
1116:
1113:
1110:
1108:
1104:
1100:
1096:
1091:
1084:
1079:
1074:
1063:
1060:
1058:
1054:
1051:
1047:
1043:
1040:
1038:
1034:
1030:
1026:
1023:
1020:
1016:
1011:
1007:
1004:
1001:
999:
995:
992:
989:
987:
983:
979:
975:
970:
965:
954:
951:
949:
945:
942:
938:
934:
931:
929:
925:
921:
917:
914:
911:
907:
902:
898:
895:
892:
890:
886:
883:
880:
878:
874:
870:
866:
861:
856:
845:
842:
840:
836:
833:
829:
825:
822:
820:
816:
812:
808:
805:
802:
798:
793:
789:
786:
783:
781:
777:
774:
771:
769:
765:
761:
757:
752:
747:
739:
737:
733:
729:
725:
721:
717:
713:
703:
701:
697:
693:
682:
679:
671:
661:
655:
653:
646:
637:
636:
628:
626:
621:
619:
614:
612:
608:
604:
600:
596:
592:
588:
584:
574:
572:
568:
564:
563:Schwann cells
554:
552:
548:
544:
534:
532:
528:
524:
520:
519:neuromuscular
516:
512:
508:
504:
499:
497:
492:
488:
484:
480:
476:
472:
469:networks via
468:
453:
450:
448:
445:
444:
440:
437:
435:
433:
432:
428:
425:
423:
420:
419:
415:
412:
410:
407:
406:
402:
399:
397:
394:
393:
389:
386:
383:
380:
379:
375:
372:
370:
367:
366:
362:
359:
357:
354:
353:
349:
346:
343:
340:
339:
335:
332:
329:
326:
325:
322:Laminin-3B32
321:
318:
316:
313:
312:
308:
305:
302:
299:
298:
294:
291:
288:
285:
284:
280:
277:
274:
271:
270:
266:
263:
260:
257:
256:
253:
250:
247:
244:
241:
240:
236:
233:
231:Old synonyms
230:
227:
226:
223:
221:
214:
210:
206:
202:
199:
195:
191:
187:
183:
181:
177:
173:
169:
165:
161:
160:
159:
151:
149:
145:
141:
137:
136:glycoproteins
132:
130:
129:
123:
119:
115:
111:
108:Laminins are
106:
104:
100:
96:
92:
89:, namely the
88:
84:
80:
79:glycoproteins
76:
72:
62:
50:
45:
36:
35:
28:
22:
4460:Tropoelastin
4416:
4389:
4357:
4349:
4267:
4257:
4240:
4230:
4201:
4156:
4106:
4060:
4038:
3989:
3982:
3970:
3951:
3879:
3871:
3854:
3833:
3812:
3748:
3744:
3738:
3703:
3699:
3689:
3677:. Retrieved
3667:
3658:
3631:
3627:
3587:
3583:
3573:
3538:
3535:FEBS Letters
3534:
3524:
3499:
3495:
3461:
3457:
3415:(1–2): 1–7.
3412:
3409:FEBS Letters
3408:
3398:
3371:
3367:
3357:
3317:(6): 611–5.
3314:
3310:
3304:
3282:(1): 27–32.
3279:
3275:
3268:
3241:
3237:
3227:
3200:
3196:
3186:
3164:(2): 89–93.
3161:
3157:
3150:
3115:
3111:
3101:
3056:
3052:
3042:
3007:
3003:
2993:
2968:
2964:
2957:
2932:
2929:Biochemistry
2928:
2921:
2912:
2887:
2883:
2876:
2851:
2847:
2841:
2822:
2812:
2792:
2785:
2758:
2754:
2706:
2702:
2670:
2632:
2628:
2585:
2581:
2571:
2544:
2540:
2530:
2429:
2191:
2025:
1983:
1957:
1931:
1885:
1856:dystroglycan
1834:
1815:
1772:of about 60
1763:
1751:
1744:proteoglycan
1730:
1707:
1692:
735:
731:
720:cell culture
709:
689:
674:
665:
649:
622:
615:
580:
560:
540:
505:, including
500:
491:dystroglycan
464:
454:Laminin-523
441:Laminin-522
429:Laminin-423
416:Laminin-213
403:Laminin-521
390:Laminin-511
376:Laminin-421
363:Laminin-411
295:Laminin-221
281:Laminin-121
267:Laminin-211
245:EHS laminin
217:
157:
133:
126:
107:
91:basal lamina
70:
69:
59:October 2023
56:
48:
4538:Cytokeratin
4467:Vitronectin
4148:multiplexin
3679:22 February
3332:10616/40259
1849:sulphatides
1818:amino acids
1774:amino acids
1718:coiled-coil
1547:Identifiers
1421:Laminin_G_2
1413:Identifiers
1267:Laminin_G_1
1259:Identifiers
1101:Laminin_EGF
1093:Identifiers
972:Identifiers
863:Identifiers
754:Identifiers
728:recombinant
716:fibronectin
475:fibronectin
446:Laminin-15
421:Laminin-14
408:Laminin-12
395:Laminin-11
381:Laminin-10
344:KS-laminin
314:Laminin-5B
252:Laminin-111
4169:Endostatin
4158:type XVIII
3238:Stem Cells
2578:"Laminins"
2523:References
2390:), agrin (
2168:), agrin (
1891:polymerise
1868:signalling
1841:C-terminal
1782:N-terminus
1645:structures
1491:structures
1345:structures
1191:structures
1025:structures
916:structures
871:Laminin_II
807:structures
724:stem cells
485:and other
368:Laminin-9
355:Laminin-8
330:K-laminin
289:S-merosin
286:Laminin-4
275:S-laminin
272:Laminin-3
258:Laminin-2
242:Laminin-1
122:paralogous
4617:IPR000034
4612:IPR008211
4607:IPR010307
4602:IPR009254
4597:IPR012680
4592:IPR012679
4587:IPR002049
4548:Reticulin
4241:type VIII
2602:1432-0878
1914:forms of
1907:integrins
1872:migration
1812:Laminin G
1778:cysteines
1727:Laminin B
1721:structure
1695:conserved
1591:IPR008211
1555:Laminin_N
1457:IPR012680
1303:IPR012679
1149:PDOC00021
1137:IPR002049
1003:IPR000034
980:Laminin_B
894:IPR010307
785:IPR009254
762:Laminin_I
712:collagens
668:July 2012
577:Pathology
531:pentraxin
527:structure
483:integrins
99:migration
4634:Laminins
4628:Category
4580:InterPro
4563:diseases
4559:See also
4502:Tectorin
4321:Leprecan
4231:type VII
4061:type XII
3984:type III
3937:Collagen
3773:21549628
3730:10747011
3669:InterPro
3565:21559588
3439:36607427
3349:10801152
3341:20512123
3296:18675790
3260:18757303
3219:10809728
3178:16289578
3142:21896617
3093:17438294
3034:29446228
2949:15823034
2848:Placenta
2777:10842354
2733:19355968
2649:15979864
2610:19693542
2501:See also
2240:CNTNAP3B
1920:subunits
1905:through
1860:proteins
1851:and the
1829:collagen
1825:proteins
1737:proteins
1662:RCSB PDB
1586:InterPro
1508:RCSB PDB
1452:InterPro
1362:RCSB PDB
1298:InterPro
1208:RCSB PDB
1132:InterPro
1042:RCSB PDB
998:InterPro
933:RCSB PDB
889:InterPro
824:RCSB PDB
780:InterPro
732:in vitro
523:synaptic
479:perlecan
471:entactin
461:Function
347:α3Aβ2γ1
333:α3Aβ1γ1
319:α3Bβ3γ2
306:α3Aβ3γ2
261:Merosin
103:adhesion
71:Laminins
4543:Gelatin
4534:Keratin
4482:Decorin
4455:Elastin
4350:Laminin
4326:ADAMTS2
4300:Enzymes
4290:COL28A1
4285:COL27A1
4278:COL11A2
4273:COL11A1
4268:type XI
4262:COL10A1
4203:type VI
4200:other:
4194:COL25A1
4189:COL23A1
4184:COL17A1
4179:COL13A1
4164:COL18A1
4152:COL15A1
4108:type IV
4096:COL22A1
4091:COL21A1
4086:COL20A1
4081:COL19A1
4076:COL16A1
4071:COL14A1
4065:COL12A1
4039:type IX
4019:COL26A1
4014:COL24A1
3972:type II
3914:Protein
3872:YouTube
3861:PDBe-KB
3851:UniProt
3840:PDBe-KB
3830:UniProt
3819:PDBe-KB
3809:UniProt
3794:Laminin
3765:7874173
3650:8349613
3606:2404817
3557:7781764
3516:8648631
3478:8648630
3431:2666164
3390:3182802
3133:3245933
3084:1855385
3061:Bibcode
3025:6055631
2985:9480764
2904:7904354
2868:8208674
2724:2978668
2312:COL27A1
2308:COL24A1
2304:COL22A1
2300:COL21A1
2296:COL20A1
2292:COL19A1
2288:COL18A1
2284:COL16A1
2280:COL15A1
2276:COL14A1
2272:COL12A1
2268:COL11A2
2264:COL11A1
2248:CNTNAP5
2244:CNTNAP4
2236:CNTNAP3
2232:CNTNAP2
2228:CNTNAP1
1912:genetic
1899:Netrins
1837:domains
1798:nidogen
1770:repeats
1766:domains
1714:domains
1566:PF00055
1432:PF02210
1278:PF00054
1144:PROSITE
1112:PF00053
991:PF00052
882:PF06009
773:PF06008
549:to the
503:tissues
496:peptide
451:α5β2γ3
438:α5β2γ2
426:α4β2γ3
413:α2β1γ3
400:α5β2γ1
387:α5β1γ1
373:α4β2γ1
360:α4β1γ1
292:α2β2γ1
278:α1β2γ1
264:α2β1γ1
248:α1β1γ1
220:isoform
128:in vivo
81:of the
49:updated
4487:FAM20C
4258:type X
4251:COL8A2
4246:COL8A1
4235:COL7A1
4224:COL6A5
4219:COL6A3
4214:COL6A2
4209:COL6A1
4139:COL4A6
4134:COL4A5
4129:COL4A4
4124:COL4A3
4119:COL4A2
4114:COL4A1
4054:COL9A3
4049:COL9A2
4044:COL9A1
4007:COL5A3
4002:COL5A2
3997:COL5A1
3991:type V
3977:COL2A1
3964:COL1A2
3959:COL1A1
3953:type I
3928:matrix
3856:O15230
3835:P24043
3814:P19137
3800:(MeSH)
3771:
3763:
3728:
3721:310212
3718:
3648:
3604:
3563:
3555:
3514:
3476:
3437:
3429:
3388:
3347:
3339:
3294:
3258:
3217:
3176:
3140:
3130:
3091:
3081:
3032:
3022:
2983:
2947:
2902:
2866:
2829:
2800:
2775:
2731:
2721:
2677:
2647:
2608:
2600:
2563:114518
2561:
2412:EGFLAM
2388:TSPEAR
2260:COL9A1
2256:COL5A3
2252:COL5A1
2224:CELSR3
2220:CELSR2
2216:CELSR1
2158:SCARF2
2154:SCARF1
2118:MEGF10
2102:CRELD2
2098:CRELD1
2094:CELSR3
2090:CELSR2
2086:CELSR1
2082:ATRNL1
1710:trimer
1677:PDBsum
1651:
1641:
1623:SUPFAM
1579:CL0202
1552:Symbol
1523:PDBsum
1497:
1487:
1445:CL0004
1418:Symbol
1377:PDBsum
1351:
1341:
1323:SUPFAM
1291:CL0004
1264:Symbol
1223:PDBsum
1197:
1187:
1169:SUPFAM
1125:CL0001
1098:Symbol
1057:PDBsum
1031:
1021:
977:Symbol
948:PDBsum
922:
912:
868:Symbol
839:PDBsum
813:
803:
759:Symbol
736:et al.
692:cancer
593:, and
551:tectum
547:retina
477:, and
101:, and
75:family
73:are a
4527:Other
4512:TECTB
4507:TECTA
4477:FREM2
4472:FRAS1
4450:ALCAM
4443:Other
4432:LAMC3
4427:LAMC2
4422:LAMC1
4417:gamma
4410:LAMB4
4405:LAMB3
4400:LAMB2
4395:LAMB1
4383:LAMA5
4378:LAMA4
4373:LAMA3
4368:LAMA2
4363:LAMA1
4358:alpha
4035:FACIT
4028:Other
3769:S2CID
3561:S2CID
3435:S2CID
3345:S2CID
2494:USH2A
2490:NTNG2
2486:NTNG1
2470:LAMC3
2466:LAMC1
2462:LAMB4
2458:LAMB3
2454:LAMB2
2450:LAMB1
2446:LAMA5
2442:LAMA3
2438:LAMA2
2434:LAMA1
2424:USH2A
2416:PROS1
2408:HSPG2
2396:CSPG4
2392:AGRIN
2384:THBS4
2380:THBS3
2376:THBS2
2372:THBS1
2368:SLIT3
2364:SLIT2
2360:SLIT1
2356:NRXN3
2352:NRXN2
2348:NRXN1
2344:NELL2
2340:NELL1
2212:LAMA5
2208:LAMA4
2204:LAMA3
2200:LAMA2
2196:LAMA1
2186:USH2A
2178:HSPG2
2170:AGRIN
2166:STAB2
2162:STAB1
2150:MUC3B
2146:MUC3A
2142:NTNG2
2138:NTNG1
2122:PEAR1
2114:MEGF9
2110:MEGF8
2106:MEGF6
2074:LAMC3
2070:LAMC2
2066:LAMC1
2062:LAMB4
2058:LAMB3
2054:LAMB2
2050:LAMB1
2046:LAMA5
2042:LAMA4
2038:LAMA3
2034:LAMA2
2030:LAMA1
2020:HSPG2
2016:LAMC3
2012:LAMC2
2008:LAMC1
2004:LAMA5
2000:LAMA4
1996:LAMA3
1992:LAMA2
1988:LAMA1
1978:LAMA5
1974:LAMA4
1970:LAMA3
1966:LAMA2
1962:LAMA1
1952:LAMA5
1948:LAMA4
1944:LAMA3
1940:LAMA2
1936:LAMA1
1802:loops
1794:binds
1790:mouse
1746:from
1619:SCOPe
1610:SCOP2
1603:LamNT
1598:SMART
1464:SMART
1319:SCOPe
1310:SCOP2
1165:SCOPe
1156:SCOP2
607:beta1
213:LAMC3
209:LAMC2
205:LAMC1
198:LAMB4
194:LAMB3
190:LAMB2
186:LAMB1
180:LAMA5
176:LAMA4
172:LAMA3
168:LAMA2
164:LAMA1
154:Types
21:Lamin
4578:and
4576:Pfam
4492:ECM1
4390:beta
3849:for
3828:for
3807:for
3761:PMID
3726:PMID
3681:2016
3646:PMID
3602:PMID
3553:PMID
3512:PMID
3474:PMID
3427:PMID
3386:PMID
3337:PMID
3292:PMID
3256:PMID
3215:PMID
3174:PMID
3138:PMID
3089:PMID
3030:PMID
2981:PMID
2945:PMID
2900:PMID
2864:PMID
2827:ISBN
2798:ISBN
2773:PMID
2729:PMID
2675:ISBN
2645:PMID
2606:PMID
2598:ISSN
2559:PMID
2482:NTN4
2478:NTN3
2474:NTN1
2420:SHBG
2404:GAS6
2336:FAT4
2332:FAT3
2328:FAT2
2324:FAT1
2320:CRB2
2316:CRB1
2134:NTN4
2130:NTN3
2126:NTN1
2078:ATRN
1895:cell
1670:PDBj
1666:PDBe
1649:ECOD
1639:Pfam
1615:1klo
1575:clan
1573:Pfam
1561:Pfam
1516:PDBj
1512:PDBe
1495:ECOD
1485:Pfam
1469:TSPN
1441:clan
1439:Pfam
1427:Pfam
1370:PDBj
1366:PDBe
1349:ECOD
1339:Pfam
1315:1qu0
1287:clan
1285:Pfam
1273:Pfam
1216:PDBj
1212:PDBe
1195:ECOD
1185:Pfam
1161:1tle
1121:clan
1119:Pfam
1107:Pfam
1050:PDBj
1046:PDBe
1029:ECOD
1019:Pfam
986:Pfam
941:PDBj
937:PDBe
920:ECOD
910:Pfam
877:Pfam
832:PDBj
828:PDBe
811:ECOD
801:Pfam
768:Pfam
714:and
601:and
3847:PDB
3826:PDB
3805:PDB
3753:doi
3716:PMC
3708:doi
3636:doi
3632:268
3592:doi
3543:doi
3539:365
3504:doi
3500:257
3466:doi
3462:257
3417:doi
3413:251
3376:doi
3372:263
3327:hdl
3319:doi
3284:doi
3280:375
3246:doi
3205:doi
3201:275
3166:doi
3128:PMC
3120:doi
3079:PMC
3069:doi
3057:104
3020:PMC
3012:doi
2973:doi
2969:275
2937:doi
2892:doi
2888:342
2856:doi
2763:doi
2759:218
2719:PMC
2711:doi
2637:doi
2590:doi
2586:339
2549:doi
2545:254
2400:EYS
2182:TNN
2174:TEK
1657:PDB
1503:PDB
1357:PDB
1203:PDB
1037:PDB
928:PDB
819:PDB
150:).
118:kDa
77:of
4630::
4582::
4150::
4105::
4037::
3916::
3870:.
3853::
3832::
3811::
3767:.
3759:.
3747:.
3724:.
3714:.
3704:19
3702:.
3698:.
3672:.
3666:.
3644:.
3630:.
3626:.
3614:^
3600:.
3586:.
3582:.
3559:.
3551:.
3537:.
3533:.
3510:.
3498:.
3486:^
3472:.
3460:.
3447:^
3433:.
3425:.
3411:.
3407:.
3384:.
3370:.
3366:.
3343:.
3335:.
3325:.
3315:28
3313:.
3290:.
3278:.
3254:.
3242:26
3240:.
3236:.
3213:.
3199:.
3195:.
3172:.
3162:25
3160:.
3136:.
3126:.
3116:40
3114:.
3110:.
3087:.
3077:.
3067:.
3055:.
3051:.
3028:.
3018:.
3008:93
3006:.
3002:.
2979:.
2967:.
2943:.
2933:44
2931:.
2898:.
2886:.
2862:.
2852:15
2850:.
2771:.
2757:.
2753:.
2741:^
2727:.
2717:.
2707:15
2705:.
2701:.
2689:^
2657:^
2643:.
2633:24
2631:.
2618:^
2604:.
2596:.
2584:.
2580:.
2557:.
2543:.
2539:.
2488:,
2484:,
2480:,
2476:,
2468:,
2464:,
2460:,
2456:,
2452:,
2448:,
2444:,
2440:,
2436:,
2386:,
2382:,
2378:,
2374:,
2366:,
2362:,
2354:,
2350:,
2342:,
2334:,
2330:,
2326:,
2318:,
2310:,
2306:,
2302:,
2298:,
2294:,
2290:,
2286:,
2282:,
2278:,
2274:,
2270:,
2266:,
2262:,
2258:,
2254:,
2246:,
2242:,
2238:,
2234:,
2230:,
2222:,
2218:,
2210:,
2206:,
2202:,
2198:,
2188:).
2164:,
2156:,
2148:,
2140:,
2136:,
2132:,
2128:,
2120:,
2116:,
2112:,
2108:,
2100:,
2092:,
2088:,
2080:,
2072:,
2068:,
2064:,
2060:,
2056:,
2052:,
2048:,
2044:,
2040:,
2036:,
2032:,
2014:,
2010:,
2002:,
1998:,
1994:,
1990:,
1976:,
1972:,
1968:,
1964:,
1950:,
1946:,
1942:,
1938:,
1878:.
1870:,
1866:,
1808:.
1723:.
1700:.
1668:;
1664:;
1647:/
1621:/
1617:/
1514:;
1510:;
1493:/
1368:;
1364:;
1347:/
1321:/
1317:/
1214:;
1210:;
1193:/
1167:/
1163:/
1048:;
1044:;
1027:/
939:;
935:;
918:/
830:;
826:;
809:/
627:.
613:.
595:γ1
591:β1
589:,
587:α2
533:.
513:,
509:,
473:,
211:,
207:,
196:,
192:,
188:,
178:,
170:,
166:,
105:.
97:,
4536:/
4319:/
4310:/
4264:)
4260:(
4237:)
4233:(
4067:)
4063:(
3979:)
3975:(
3906:e
3899:t
3892:v
3863:.
3842:.
3821:.
3775:.
3755::
3749:8
3732:.
3710::
3683:.
3652:.
3638::
3608:.
3594::
3588:4
3567:.
3545::
3518:.
3506::
3480:.
3468::
3441:.
3419::
3392:.
3378::
3351:.
3329::
3321::
3298:.
3286::
3262:.
3248::
3221:.
3207::
3180:.
3168::
3144:.
3122::
3095:.
3071::
3063::
3036:.
3014::
2987:.
2975::
2951:.
2939::
2906:.
2894::
2870:.
2858::
2835:.
2806:.
2779:.
2765::
2735:.
2713::
2683:.
2651:.
2639::
2592::
2565:.
2551::
2496:)
2426:)
2022:)
1980:)
1954:)
681:)
675:(
670:)
666:(
656:.
605:—
61:)
57:(
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.