326:
225:
384:
311:
435:
359:
417:
216:
hydroxyl radicals, which in turn react with DNA. The location and binding of Iron (II) to DNA may play an important role in determining the substrate and nature of the radical attack on the DNA. The Fenton reaction generates two types of oxidants, Type I and Type II. Type I oxidants are moderately sensitive to peroxides and ethanol. Type I and Type II oxidants preferentially cleave at the specific sequences.
269:
260:. Upon addition of the hydroxyl radical, many stable products can be formed. In general, radical hydroxyl attacks on base moieties do not cause altered sugars or strand breaks except when the modifications labilize the N-glycosyl bond, allowing the formation of baseless sites that are subject to beta-elimination.
316:
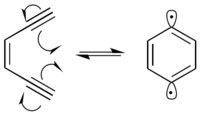
In the presence of DNA, the 1,4-didehydrobenzene diradical abstracts hydrogens from the deoxyribose sugar backbone, predominantly at the C-1’, C-4’ and C-5’ positions. Hydrogen abstraction causes radical formation at the reacted carbon. The carbon radical reacts with molecular oxygen, which leads to
353:
was one of the first such products identified and was originally found in a soil sample taken from
Kerrville, Texas. These compounds are synthesized by bacteria as defense mechanisms due to their ability to cleave DNA through the formation of 1,4-didehydrobenzene from the enediyne component of the
248:
is the repair mechanism used. Hydroxyl radical reactions with the deoxyribose sugar backbone are initiated by hydrogen abstraction from a deoxyribose carbon, and the predominant consequence is eventual strand breakage and base release. The hydroxyl radical reacts with the various hydrogen atoms of
186:
with a free coordination site are capable of reducing peroxides to hydroxyl radicals. Iron is believed to be the metal responsible for the creation of hydroxyl radicals because it exists at the highest concentration of any transition metal in most living organisms. The Fenton reaction is possible
364:
Calicheamicin and other related compounds share several common characteristics. The extended structures attached to the enediyne allow the compound to specifically bind DNA, in most cases to the minor groove of the double helix. Additionally, part of the molecule is known as the “trigger” which,
276:
Hydrogen abstraction from the 1’-deoxyribose carbon by the hydroxyl radical creates a 1 ‘-deoxyribosyl radical. The radical can then react with molecular oxygen, creating a peroxyl radical which can be reduced and dehydrated to yield a 2’-deoxyribonolactone and free base. A deoxyribonolactone is
285:
Radical damage to DNA can also occur through the interaction of DNA with certain natural products known as radiomimetic compounds, molecular compounds which affect DNA in similar ways to radiation exposure. Radiomimetic compounds induce double-strand breaks in DNA via highly specific, concerted
448:
Most enediynes, including the ones listed above, have been used as potent antitumor antibiotics due to their ability to efficiently cleave DNA. Calicheamicin and esperamicin are the two most commonly used types due to their high specificity when binding to DNA, which minimizes unfavorable side
215:
The creation of hydroxyl radicals by iron(II) catalysis is important because iron(II) can be found coordinated with, and therefore in close proximity to, DNA. This reaction allows for hydrogen peroxide created by radiolysis of water to diffuse to the nucleus and react with Iron (II) to produce
467:
generates a free radical under anoxic conditions instead of the trigger mechanism of an enediyne. The free radical then continues on to cleave DNA in a similar manner to 1,4-didehydrobenzene in order to treat cancerous cells. It is currently in Phase III trials.
317:
a strand break in the DNA through a variety of mechanisms. 1,4-Didehydrobenzene is able to position itself in such a way that it can abstract proximal hydrogens from both strands of DNA. This produces a double-strand break in the DNA, which can lead to cellular
87:
damage is caused by hydroxyl radicals, yet hydroxyl radicals are so reactive that they can only diffuse one or two molecular diameters before reacting with cellular components. Thus, hydroxyl radicals must be formed immediately adjacent to
331:
Enediynes generally undergo the
Bergman cyclization at temperatures exceeding 200 °C. However, incorporating the enediyne into a 10-membered cyclic hydrocarbon makes the reaction more thermodynamically favorable by releasing the
336:
of the reactants. This allows for the
Bergman cyclization to occur at 37 °C, the biological temperature of humans. Molecules which incorporate enediynes into these larger ring structures have been found to be extremely
92:
in order to react. Radiolysis of water creates peroxides that can act as diffusable, latent forms of hydroxyl radicals. Some metal ions in the vicinity of DNA generate the hydroxyl radicals from peroxide.
349:
Enediynes are present in many complicated natural products. They were originally discovered in the early 1980s during a search for new anticancer products produced by microorganisms.
187:
because transition metals can exist in more than one oxidation state and their valence electrons may be unpaired, allowing them to participate in one-electron redox reactions.
778:
Steenken S (1989). "Purine bases, nuclesides and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts".
52:
can be caused by indirect DNA damage because it is found in parts of the body not exposed to sunlight. DNA is vulnerable to radical attack because of the very
249:
the deoxyribose in the order 5′ H > 4′ H > 3′ H ≈ 2′ H ≈ 1′ H. This order of reactivity parallels the exposure to solvent of the deoxyribose hydrogens.
1089:
Zein N, Sinha AM, McGahren WJ, Ellestad GA (May 1988). "Calicheamicin gamma 1I: an antitumor antibiotic that cleaves double-stranded DNA site specifically".
684:
Pogozelski WK, Tullius TD (May 1998). "Oxidative Strand
Scission of Nucleic Acids: Routes Initiated by Hydrogen Abstraction from the Sugar Moiety".
440:
The chromophore is unreactive when bound to the apoprotein. Upon its release, it reacts to form 1,4-didehydrobenzene and subsequently cleaves DNA.
1052:"Selective ablation of acute myeloid leukemia using antibody-targeted chemotherapy: a phase I study of an anti-CD33 calicheamicin immunoconjugate"
1051:
252:
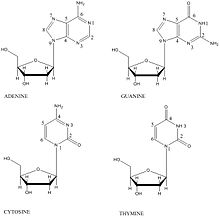
Hydroxyl radicals react with DNA bases via addition to the electron-rich, pi bonds. These pi bonds in the bases are located between C5-C6 of
379:
The calicheamicin types are defined by a methyl trisulfide group that is involved in triggering the molecule by the following mechanism.
840:
Povirk LF (1996). "DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes".
721:"DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone"
953:
Ellestad GA (September 2011). "Structural and conformational features relevant to the anti-tumor activity of calicheamicin γ 1I".
1050:
Sievers EL, Appelbaum FR, Spielberger RT, Forman SJ, Flowers D, Smith FO, Shannon-Dorcy K, Berger MS, Bernstein ID (June 1999).
306:
diradical. The 1,4-didehydrobenzene diradical is highly reactive, and will abstract hydrogens from any possible hydrogen-donor.
178:
results in the creation of hydroxyl radicals from hydrogen peroxide and an Iron (II) catalyst. Iron(III) is regenerated via the
365:
under specific physiological conditions, activates the enediyne, known as the “warhead” and 1,4-didehydrobenzene is generated.
383:
244:. Cells have developed complex and efficient repair mechanisms to fix the lesions. In the case of free radical attack on DNA,
555:
456:
Additionally, calicheamicin is able to cleave DNA at low concentrations, proving to be up to 1000 times more effective than
434:
310:
400:
and enediyne core. The anthraquinone component allows for specific binding of DNA at the 3’ side of purine bases through
818:
606:
993:"DNA intercalation and cleavage of an antitumor antibiotic dynemicin that contains anthracycline and enediyne cores"
591:
DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability
488:
20:
404:, a site that is different from calicheamicin. Its ability to cleave DNA is greatly increased in the presence of
491:
caused by oxidative free radicals has been hypothesized to be a major driving force in the evolution of meiosis
401:
1246:
232:
Hydroxyl radicals can attack the deoxyribose DNA backbone and bases, potentially causing a plethora of
53:
325:
224:
179:
805:
Lhomme J, Constant JF, Demeunynck M (1999). "Abasic DNA structure, reactivity, and recognition".
450:
61:
1098:
1004:
732:
245:
593:. Progress in Nucleic Acid Research and Molecular Biology. Vol. 35. pp. 95–125.
8:
1241:
882:
Kraka E, Cremer D (2000). "Computer design of anticancer drugs. A new enediyne warhead".
480:
299:
45:
1102:
1008:
736:
166:
Free radical damage to DNA is thought to cause mutations that may lead to some cancers.
1207:
1182:
1158:
1133:
661:
566:
531:
463:
The free radical mechanism to treat certain types of cancers extends beyond enediynes.
49:
33:
598:
1212:
1163:
1114:
1071:
1032:
1027:
992:
970:
932:
857:
853:
822:
760:
755:
720:
701:
653:
612:
602:
571:
551:
183:
48:
because the radicals formed can diffuse throughout the body and affect other organs.
16:
Damage to DNA as a result of exposure to ionizing radiation or radiomimetic compounds
665:
1202:
1194:
1153:
1145:
1106:
1063:
1022:
1012:
962:
922:
891:
849:
814:
787:
750:
740:
693:
643:
594:
561:
543:
80:
175:
927:
910:
547:
511:
Barbusinski K (2009). "Fenton
Reaction – Controversy Concerning the Chemistry".
997:
Proceedings of the
National Academy of Sciences of the United States of America
725:
Proceedings of the
National Academy of Sciences of the United States of America
427:
393:
have been used as anticancer drugs due to their high toxicity and specificity.
1149:
1067:
1230:
648:
631:
397:
373:
350:
1110:
1017:
911:"A new macromolecular antitumor antibiotic, C-1027. III. Antitumor activity"
745:
277:
mutagenic and resistant to repair enzymes. Thus, an abasic site is created.
1236:
1216:
1198:
1167:
1075:
974:
826:
705:
632:"Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide"
575:
464:
89:
41:
1118:
1036:
936:
861:
764:
657:
616:
412:
compounds. This compound has also found prominence as an antitumor agent.
423:
390:
369:
333:
286:
free-radical attacks on the deoxyribose moieties in both strands of DNA.
791:
966:
484:
457:
253:
72:
24:
895:
697:
368:
Three classes of enediynes have since been identified: calicheamicin,
842:
Mutation
Research/Fundamental and Molecular Mechanisms of Mutagenesis
449:
reactions. They have been shown to be especially useful for treating
338:
318:
241:
237:
57:
396:
Dynemicin and its relatives are characterized by the presence of an
295:
76:
909:
Zhen YS, Ming XY, Yu B, Otani T, Saito H, Yamada Y (August 1989).
1049:
476:
56:
that can be abstracted and the prevalence of double bonds in the
718:
358:
280:
819:
10.1002/1097-0282(1999)52:2<65::aid-bip1>3.3.co;2-l
257:
233:
416:
409:
405:
303:
719:
Balasubramanian B, Pogozelski WK, Tullius TD (August 1998).
268:
1088:
422:
Chromoprotein enediynes are characterized by an unstable
84:
37:
1134:"The oxidative damage initiation hypothesis for meiosis"
990:
804:
75:
of intracellular water by ionizing radiation creates
991:Sugiura Y, Shiraki T, Konishi M, Oki T (May 1990).
683:
1228:
228:Radical hydroxyl attacks can form baseless sites
67:
908:
1180:
1131:
506:
504:
281:Radical damage through radiomimetic compounds
79:, which are relatively stable precursors to
948:
946:
881:
510:
986:
984:
501:
219:
1206:
1157:
1026:
1016:
926:
754:
744:
647:
629:
565:
529:
1183:"How oxygen gave rise to eukaryotic sex"
952:
943:
877:
875:
873:
871:
777:
267:
223:
36:or to radiomimetic compounds. Damage to
1043:
981:
771:
471:
460:at combating certain types of tumors.
1229:
1181:Hörandl E, Speijer D (February 2018).
1132:Hörandl E, Hadacek F (December 2013).
839:
679:
677:
675:
536:Advances in Physical Organic Chemistry
389:Calicheamicin and the closely related
169:
868:
833:
532:"Reactivity of Nucleic Acid Radicals"
272:Route of deoxyribonolactone formation
32:can occur as a result of exposure to
1082:
902:
623:
588:
513:Ecological Chemistry and Engineering
443:
289:
712:
672:
636:The Journal of Biological Chemistry
582:
344:
13:
14:
1258:
630:Henle ES, Linn S (August 1997).
433:
415:
382:
357:
324:
309:
294:Many radiomimetic compounds are
21:DNA damage (naturally occurring)
1174:
1125:
263:
798:
523:
1:
599:10.1016/s0079-6603(08)60611-x
494:
68:Damage via radiation exposure
854:10.1016/0027-5107(96)00023-1
62:free radicals can easily add
7:
928:10.7164/antibiotics.42.1294
548:10.1016/bs.apoc.2016.02.001
10:
1263:
915:The Journal of Antibiotics
30:Free radical damage to DNA
18:
1150:10.1007/s00497-013-0234-7
1068:10.1182/blood.V93.11.3678
649:10.1074/jbc.272.31.19095
479:is a central feature of
1111:10.1126/science.3240341
1018:10.1073/pnas.87.10.3831
746:10.1073/pnas.95.17.9738
220:Radical hydroxyl attack
1199:10.1098/rspb.2017.2706
451:acute myeloid leukemia
302:reaction to produce a
273:
229:
83:. 60%–70% of cellular
542:. Elsevier: 119–202.
530:Greenberg MM (2016).
487:. The need to repair
426:enediyne bound to an
271:
227:
489:oxidative DNA damage
472:Evolution of Meiosis
304:1,4-didehydrobenzene
298:, which undergo the
246:base-excision repair
180:Haber–Weiss reaction
1103:1988Sci...240.1198Z
1009:1990PNAS...87.3831S
792:10.1021/cr00093a003
737:1998PNAS...95.9738B
481:sexual reproduction
300:Bergman cyclization
170:The Fenton reaction
46:indirect DNA damage
1247:Molecular genetics
1097:(4856): 1198–201.
967:10.1002/chir.20990
274:
230:
50:Malignant melanoma
34:ionizing radiation
896:10.1021/ja001017k
890:(34): 8245–8264.
698:10.1021/cr960437i
557:978-0-12-804716-3
444:Antitumor ability
376:-based products.
321:if not repaired.
290:General mechanism
184:Transition metals
81:hydroxyl radicals
44:attack is called
1254:
1221:
1220:
1210:
1178:
1172:
1171:
1161:
1129:
1123:
1122:
1086:
1080:
1079:
1047:
1041:
1040:
1030:
1020:
988:
979:
978:
950:
941:
940:
930:
906:
900:
899:
884:J. Am. Chem. Soc
879:
866:
865:
837:
831:
830:
802:
796:
795:
775:
769:
768:
758:
748:
716:
710:
709:
692:(3): 1089–1108.
686:Chemical Reviews
681:
670:
669:
651:
627:
621:
620:
589:Ward JF (1988).
586:
580:
579:
569:
527:
521:
520:
508:
437:
419:
386:
361:
345:Natural products
328:
313:
54:labile hydrogens
1262:
1261:
1257:
1256:
1255:
1253:
1252:
1251:
1227:
1226:
1225:
1224:
1179:
1175:
1130:
1126:
1087:
1083:
1062:(11): 3678–84.
1048:
1044:
989:
982:
951:
944:
907:
903:
880:
869:
838:
834:
803:
799:
776:
772:
731:(17): 9738–43.
717:
713:
682:
673:
642:(31): 19095–8.
628:
624:
609:
587:
583:
558:
528:
524:
509:
502:
497:
474:
446:
347:
292:
283:
266:
222:
202:
198:
176:Fenton reaction
172:
158:
154:
140:
130:
123:
119:
112:
104:
70:
40:as a result of
27:
17:
12:
11:
5:
1260:
1250:
1249:
1244:
1239:
1223:
1222:
1173:
1124:
1081:
1042:
1003:(10): 3831–5.
980:
942:
901:
867:
848:(1–2): 71–89.
832:
797:
786:(3): 503–529.
770:
711:
671:
622:
607:
581:
556:
522:
499:
498:
496:
493:
473:
470:
445:
442:
346:
343:
291:
288:
282:
279:
265:
262:
221:
218:
213:
212:
211:
210:
209:
208:
200:
196:
171:
168:
164:
163:
162:
161:
160:
159:
156:
152:
145:
138:
135:
128:
125:
121:
117:
114:
110:
102:
69:
66:
15:
9:
6:
4:
3:
2:
1259:
1248:
1245:
1243:
1240:
1238:
1235:
1234:
1232:
1218:
1214:
1209:
1204:
1200:
1196:
1192:
1188:
1187:Proc Biol Sci
1184:
1177:
1169:
1165:
1160:
1155:
1151:
1147:
1144:(4): 351–67.
1143:
1139:
1135:
1128:
1120:
1116:
1112:
1108:
1104:
1100:
1096:
1092:
1085:
1077:
1073:
1069:
1065:
1061:
1057:
1053:
1046:
1038:
1034:
1029:
1024:
1019:
1014:
1010:
1006:
1002:
998:
994:
987:
985:
976:
972:
968:
964:
961:(8): 660–71.
960:
956:
949:
947:
938:
934:
929:
924:
921:(8): 1294–8.
920:
916:
912:
905:
897:
893:
889:
885:
878:
876:
874:
872:
863:
859:
855:
851:
847:
843:
836:
828:
824:
820:
816:
812:
808:
801:
793:
789:
785:
781:
774:
766:
762:
757:
752:
747:
742:
738:
734:
730:
726:
722:
715:
707:
703:
699:
695:
691:
687:
680:
678:
676:
667:
663:
659:
655:
650:
645:
641:
637:
633:
626:
618:
614:
610:
608:9780125400350
604:
600:
596:
592:
585:
577:
573:
568:
563:
559:
553:
549:
545:
541:
537:
533:
526:
518:
514:
507:
505:
500:
492:
490:
486:
482:
478:
469:
466:
461:
459:
454:
452:
441:
438:
436:
431:
429:
425:
420:
418:
413:
411:
407:
403:
402:intercalation
399:
398:anthraquinone
394:
392:
387:
385:
380:
377:
375:
374:chromoprotein
371:
366:
362:
360:
355:
352:
351:Calicheamicin
342:
340:
335:
329:
327:
322:
320:
314:
312:
307:
305:
301:
297:
287:
278:
270:
261:
259:
256:and N7-C8 in
255:
250:
247:
243:
239:
235:
226:
217:
206:
194:
193:
192:
191:
190:
189:
188:
185:
181:
177:
167:
150:
146:
144:
136:
134:
126:
115:
108:
100:
99:
98:
97:
96:
95:
94:
91:
90:nucleic acids
86:
82:
78:
74:
65:
63:
59:
55:
51:
47:
43:
39:
35:
31:
26:
22:
1190:
1186:
1176:
1141:
1138:Plant Reprod
1137:
1127:
1094:
1090:
1084:
1059:
1055:
1045:
1000:
996:
958:
954:
918:
914:
904:
887:
883:
845:
841:
835:
813:(2): 65–83.
810:
806:
800:
783:
779:
773:
728:
724:
714:
689:
685:
639:
635:
625:
590:
584:
539:
535:
525:
516:
512:
475:
465:Tirapazamine
462:
455:
447:
439:
432:
421:
414:
395:
388:
381:
378:
367:
363:
356:
348:
330:
323:
315:
308:
293:
284:
275:
264:Abasic sites
251:
236:that can be
231:
214:
204:
173:
165:
148:
142:
132:
106:
71:
42:free radical
29:
28:
807:Biopolymers
424:chromophore
391:esperamicin
334:ring strain
254:pyrimidines
1242:DNA repair
1231:Categories
495:References
485:eukaryotes
458:adriamycin
428:apoprotein
354:molecule.
141:O → OH + H
131:O → H + OH
73:Radiolysis
25:DNA repair
19:See also:
955:Chirality
780:Chem. Rev
370:dynemicin
339:cytotoxic
319:apoptosis
296:enediynes
242:mutagenic
238:cytotoxic
203:→ Fe + OH
120:O + e → H
77:peroxides
58:DNA bases
1217:29436502
1193:(1872).
1168:23995700
1076:10339474
975:21800378
827:10898853
706:11848926
666:11016259
576:28529390
1208:5829205
1159:3825497
1119:3240341
1099:Bibcode
1091:Science
1037:2339123
1005:Bibcode
937:2759910
862:8781578
765:9707545
733:Bibcode
658:9235895
617:3065826
567:5435387
477:Meiosis
258:purines
234:lesions
1215:
1205:
1166:
1156:
1117:
1074:
1035:
1025:
973:
935:
860:
825:
763:
753:
704:
664:
656:
615:
605:
574:
564:
554:
372:, and
195:Fe + H
1056:Blood
1028:53997
756:21406
662:S2CID
410:thiol
406:NADPH
113:O + e
60:that
1213:PMID
1164:PMID
1115:PMID
1072:PMID
1033:PMID
971:PMID
933:PMID
858:PMID
823:PMID
761:PMID
702:PMID
654:PMID
613:PMID
603:ISBN
572:PMID
552:ISBN
519:(3).
408:and
207:+ OH
174:The
147:2 OH
105:O +
64:to.
23:and
1237:DNA
1203:PMC
1195:doi
1191:285
1154:PMC
1146:doi
1107:doi
1095:240
1064:doi
1023:PMC
1013:doi
963:doi
923:doi
892:doi
888:122
850:doi
846:355
815:doi
788:doi
751:PMC
741:doi
694:doi
644:doi
640:272
595:doi
562:PMC
544:doi
483:in
240:or
109:→ H
85:DNA
38:DNA
1233::
1211:.
1201:.
1189:.
1185:.
1162:.
1152:.
1142:26
1140:.
1136:.
1113:.
1105:.
1093:.
1070:.
1060:93
1058:.
1054:.
1031:.
1021:.
1011:.
1001:87
999:.
995:.
983:^
969:.
959:23
957:.
945:^
931:.
919:42
917:.
913:.
886:.
870:^
856:.
844:.
821:.
811:52
809:.
784:89
782:.
759:.
749:.
739:.
729:95
727:.
723:.
700:.
690:98
688:.
674:^
660:.
652:.
638:.
634:.
611:.
601:.
570:.
560:.
550:.
540:50
538:.
534:.
517:16
515:.
503:^
453:.
430:.
341:.
182:.
151:→H
107:hν
1219:.
1197::
1170:.
1148::
1121:.
1109::
1101::
1078:.
1066::
1039:.
1015::
1007::
977:.
965::
939:.
925::
898:.
894::
864:.
852::
829:.
817::
794:.
790::
767:.
743::
735::
708:.
696::
668:.
646::
619:.
597::
578:.
546::
205:·
201:2
199:O
197:2
157:2
155:O
153:2
149:·
143:·
139:2
137:H
133:·
129:2
127:H
124:O
122:2
118:2
116:H
111:2
103:2
101:H
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.