82:, presents several advantages over traditional tissue biopsies. They are non-invasive, can be used repeatedly, and provide more useful information on metastatic risk, disease progression, and treatment effectiveness. For example, analysis of blood samples from cancer patients has found a propensity for increased CTC detection as the disease progresses. Blood tests are easy and safe to perform and multiple samples can be taken over time. By contrast, analysis of solid tumors necessitates invasive procedures that might limit patient compliance. The ability to monitor the disease progression over time could facilitate appropriate modification to a patient's therapy, potentially improving their prognosis and quality of life. The important aspect of the ability to prognose the future progression of the disease is elimination (at least temporarily) of the need for a surgery when the repeated CTC counts are low and not increasing; the obvious benefits of avoiding the surgery include avoiding the risk related to the innate tumor-genicity of cancer surgeries. To this end, technologies with the requisite sensitivity and reproducibility to detect CTCs in patients with metastatic disease have recently been developed. On the other hand, CTCs are very rare, often present as only a few cells per milliliter of blood, which makes their detection challenging. In addition, they often express a variety of markers which vary from patient to patient, which makes it difficult to develop techniques with high
369:
process, Maintrac uses just two steps towards the identification. In contrast to many other methods, maintrac does not purify the cells or enrich them, but identifies them within the context of the other blood compounds. To obtain vital cells and to reduce stress of those cells, blood cells are prepared by only one centrifugation step and erythrocyte lysis. Like CellSearch, maintrac uses an EpCAM antibody. It is, however, not used for enrichment but rather as a fluorescent marker to identify those cells. Together with the nuclear staining with propidium iodide the maintrac method can distinguish between dead and living cells. Only vital, propidium excluding EpCAM positive cells are counted as potential tumor cells. Only living cells can grow into tumors, therefore dying EpCAM positive cells can do no harm. The suspension is analysed by fluorescence microscopy, which automatically counts the events. Simultaneous event galleries are recorded to verify whether the software found a true living cell and to differentiate between skin epithelial cells for example. Close validation of the method showed that additional antibodies of cytokeratins or CD45 did not have any advantage.
338:
blood cells and white blood cells to pass through, while larger rare cells, including circulating tumor cells and fetal cells get caught. Trapped cells can either be automatically stained with antibodies for identification or can be released out of the cassette for subsequent analysis. These released / harvested cells are alive and can be analyzed by downstream cellular and molecular techniques, as well as cultured. The filtration cassette captures a plethora of different cancer cell types. In May 2022, the
Parsortix PC1 system was cleared by the FDA as a medical device for the capture and harvest of circulating tumor cells (CTCs) from metastatic breast cancer patient blood for subsequent analysis. In addition to the IVD application, the PC1 may be used with the MBC-01 Metastatic Breast Cancer Kit for use in research studies or Lab Developed Tests (LDTs) that have been created and validated in a clinical laboratory.
325:(epithelial markers). The sample is then scanned on an analyzer which takes images of the nuclear, cytokeratin, and CD45 stains. To be considered a CTC a cell must contain a nucleus, be positive for cytoplasmic expression of cytokeratin as well as negative for the expression of CD45 marker, and have a diameter larger than 5 μm. If the total number of tumor cells found to meet the criteria cited above is 5 or more, a blood sample is positive. In studies done on prostate, breast and colon cancer patients, median survival of metastatic patients with positive samples is about half the median survival of metastatic patients with negative samples. This system is characterized by a recovery capacity of 93% and a detection limit of one CTC per 7.5 mL of whole blood. For specific cancer types, alternative methods such as IsoFlux have shown greater
236:
total cells in circulating blood, 1–10 CTCs per mL of whole blood compared to a few million white blood cells and a billion red blood cells. Therefore, the major challenge for CTC researchers is the prevailing difficulty of CTC purification that allows the molecular characterization of CTCs. Several methods have been developed to isolate CTCs in the peripheral blood and essentially fall into two categories: biological methods and physical methods, as well as hybrid methods that combine both strategies. Techniques may also be classified based on whether they select CTCs for isolation (positive selection) or whether they exclude all blood cells (negative selection).
284:, as developed by GILUPI GmbH. An antibody coated metal wire is inserted into a peripheral vein and stays there for a defined period (30 min). During this time, CTCs from the blood can bind to the antibodies (currently anti-EpCAM). After the incubation time, the wire is removed, washed and the native CTCs, isolated from the blood of the patient, can be further analysed. Molecular genetics as well as immunofluorescent staining and several other methods are possible. Advantage of this method is the higher blood volume that can be analysed for CTCs (approx. 750 ml in 30 min compared to 7.5 ml of a drawn blood sample).
180:
cancerous cells, means that a key component of understanding CTCs biological properties require technologies and approaches capable of isolating 1 CTC per mL of blood, either by enrichment, or better yet with enrichment-free assays that identify all CTC subtypes in sufficiently high definition to satisfy diagnostic pathology image-quantity requirements in patients with a variety of cancer types. To date CTCs have been detected in several epithelial cancers (breast, prostate, lung, and colon) and clinical evidences indicate that patients with metastatic lesions are more likely to have CTCs isolated.
352:
next-generation sequencing. A hematopathology-trained algorithm incorporates numerous morphology measurements as well as expression from cytokeratin and CD45. The algorithm then proposes candidate CTCs that a trained reader confirms. Cells of interest are analyzed for relevant phenotypic and genotypic markers, with regional white blood cells included as negative controls. Epic's molecular assays measure protein expression and also interrogate genomic abnormalities in CTCs for more than 20 different cancer types.
20:
394:. ScreenCell is a filtration based device that allows sensitive and specific isolation of CTCs from human whole blood in a few minutes. Peripheral blood is drawn and processed within 4 hours with a ScreenCell isolation device to capture CTCs. The captured cells are ready for cell culture or for direct characterization using ViewRNA in situ hybridization assay. The Parsortix method separates CTCs based on their size and deformability.
503:(colloidal magnetic nanoparticles) and high gradient magnetic separators invented by Paul Liberti and motivated by theoretical calculations by Liberti and Leon Terstappen that indicated very small tumors shedding cells at less than 1.0% per day should result in detectable cells in blood. A variety of other technologies have been applied to CTC enumeration and identification since that time.
216:
151:
risk and poor prognosis. For example, one study involving prostate cancer showed an eight-fold longer mean survival rate for patients with only single CTCs versus those with CTC clusters, while other studies have shown similar correlations for colon cancer. In addition, enumerating CTC clusters can provide useful prognostic information for patients with already elevated CTC levels.
137:(programmed cell death). These may be used to monitor treatment response, as done experimentally by the Epic Sciences method, which identifies nuclear fragmentation or cytoplasmic blebbing associated with apoptosis. Measuring the ratio of traditional CTC to apoptotic CTCs—from baseline to therapy—provides clues to treatment efficacy in targeting and killing cancer cells.
373:
maintrac can be used to verify the success of a chemotherapy and to supervise the treatment during hormone or maintenance therapy
Maintrac has been used experimentally to monitor cancer recurrence. Studies using Maintrac have shown that EpCAM positive cells can be found in the blood in patient without cancer. Inflammatory conditions like
264:. Other techniques under research include microfluidic separation and combination of immunomagnetic assay and microfluidic separation. As the development of microfabrication technology, microscale magnetic structures are implemented to provide better control of the magnetic field and assist the CTCs detection. Oncolytic viruses such as
63:) in distant organs, a mechanism that is responsible for the vast majority of cancer-related deaths. The detection and analysis of CTCs can assist early patient prognoses and determine appropriate tailored treatments. Currently, there is one FDA-approved method for CTC detection, CellSearch, which is used to diagnose
368:
diagnostic methods to identify rare cells in body fluids and their molecular characteristics. It is based on positive selection using EpCAM-specific antibodies. Maintrac uses an approach based on microscopic identification of circulating tumor cells. To prevent damage and loss of the cells during the
305:
tube with an added preservative. Upon arrival in the lab, 7.5mL of blood is centrifuged and placed in a preparation system. This system first enriches the tumor cells immunomagnetically by means of ferrofluid nanoparticles and a magnet. Subsequently, recovered cells are permeabilized and stained with
150:
CTC clusters are two or more individual CTCs bound together. The CTC cluster may contain traditional, small or CK- CTCs. These clusters have cancer-specific biomarkers that identify them as CTCs. Several studies have reported that the presence of these clusters is associated with increased metastatic
494:
of the circulating cells to tumor cells from different lesions led
Ashworth to conclude that "One thing is certain, that if they came from an existing cancer structure, they must have passed through the greater part of the circulatory system to have arrived at the internal saphena vein of the sound
235:
CTCs are pivotal to understanding the biology of metastasis and promise potential as a biomarker to noninvasively evaluate tumor progression and response to treatment. However, isolation and characterization of CTCs represent a major technological challenge, since CTCs make up a minute number of the
168:
154:
However, one study has reported that contrary to existing consensus, at least a discrete population of these clusters are non-malignant, and derive instead from the tumor endothelium. These circulating tumor-endothelial clusters also show epithelial-mesenchymal markers but do not mirror the genetics
3489:
Lobodasch, Kurt; Fröhlich, Frank; Rengsberger, Matthias; Schubert, Rene; Dengler, Robert; Pachmann, Ulrich; Pachmann, Katharina (April 2007). "Quantification of circulating tumour cells for the monitoring of adjuvant therapy in breast cancer: An increase in cell number at completion of therapy is a
1523:
Cima, I.; Kong, S. L.; Sengupta, D.; Tan, I. B.; Phyo, W. M.; Lee, D.; Hu, M.; Iliescu, C.; Alexander, I.; Goh, W. L.; Rahmani, M.; Suhaimi, N.-A. M.; Vo, J. H.; Tai, J. A.; Tan, J. H.; Chua, C.; Ten, R.; Lim, W. J.; Chew, M. H.; Hauser, C. A. E.; van Dam, R. M.; Lim, W.-Y.; Prabhakar, S.; Lim, B.;
480:
Morphological appearance is judged by human operators and is therefore subject to large inter operator variation. Several CTC enumeration methods exist which use morphological appearance to identify CTC, which may also apply different morphological criteria. A recent study in prostate cancer showed
471:
showed they could generate bone, lung, ovary and brain metastases in mice, partially reflecting the secondary lesions as found in the corresponding patients. Remarkably, one CTC line—isolated long before the appearance of brain metastasis in patient—was highly competent to generate brain metastasis
346:
This method involves technology to separate nucleated cells from red blood cells, which lack a nucleus. All nucleated cells, including normal white blood cells and CTCs, are exposed to fluorescent-tagged antibodies specific for cancer biomarkers. In addition, Epic's imaging system captures pictures
3085:
Agerbæk, Mette Ø.; Bang-Christensen, Sara R.; Yang, Ming-Hsin; Clausen, Thomas M.; Pereira, Marina A.; Sharma, Shreya; Ditlev, Sisse B.; Nielsen, Morten A.; Choudhary, Swati; Gustavsson, Tobias; Sorensen, Poul H.; Meyer, Tim; Propper, David; Shamash, Jonathan; Theander, Thor G.; Aicher, Alexandra;
1373:
Ferraldeschi, Roberta; McDaniel, Andrew; Krupa, Rachel; Louw, Jessica; Tucker, Eric; Bales, Natalee; Marrinucci, Dena; Riisnaes, Ruth; Mateo, Joaquin; Dittamore, Ryan; De Bono, Johann
Sebastian; Tomlins, Scott A.; Attard, Gerhardt (February 2014). "CK- and small nuclear size circulating tumor cell
372:
Unlike other methods maintrac does not use the single cell count as a prognostic marker, rather
Maintrac utilizes the dynamics of the cell count. Rising tumor cell numbers are an important factor that tumor activity is ongoing. Decreasing cell counts are a sign for a successful therapy. Therefore,
337:
This automated method uses size filtration to enrich larger and less compressible circulating tumor cells from other blood components. The
Parsortix system can take in blood samples ranging from 1 mL to 40 mL. A disposable microfluidic cassette with a 6.5 micron gap allows the vast majority of red
198:
First evidence indicates that CTC markers applied in human medicine are conserved in other species. Five of the more common markers including CK19 are also useful to detect CTC in the blood of dogs with malignant mammary tumors. Newer approaches are able to identify more cells out 7.5 ml of blood,
3288:
Sánchez-Lorencio, M.I.; Ramirez, P.; Saenz, L.; Martínez Sánchez, M.V.; De La Orden, V.; Mediero-Valeros, B.; Veganzones-De-Castro, S.; Baroja-Mazo, A.; Revilla Nuin, B.; Gonzalez, M.R.; Cascales-Campos, P.A.; Noguera-Velasco, J.A.; Minguela, A.; Díaz-Rubio, E.; Pons, J.A.; Parrilla, P. (November
2129:
Pachmann K.; Camara O.; Kavallaris A.; Krauspe S.; Malarski N.; Gajda M.; Kroll T.; Jorke C.; Hammer U.; Altendorf-Hofmann A.; et al. (2008). "Monitoring the
Response of Circulating Epithelial Tumor Cells to Adjuvant Chemotherapy in Breast Cancer Allows Detection of Patients at Risk of Early
489:
CTCs were observed for the first time in 1869 in the blood of a man with metastatic cancer by Thomas
Ashworth, who postulated that "cells identical with those of the cancer itself being seen in the blood may tend to throw some light upon the mode of origin of multiple tumours existing in the same
140:
Small CTCs are cytokeratin-positive and CD45-negative, but with sizes and shapes similar to white blood cells. Importantly, small CTCs have cancer-specific biomarkers that identify them as CTCs. Small CTCs have been implicated in progressive disease and differentiation into small cell carcinomas,
4115:
Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R,
1451:
Divella R, Daniele A, Abbate I, Bellizzi A, Savino E, Simone G, Giannone G, Giuliani F, Fazio V, Gadaleta-Caldarola G, Gadaleta C, Lolli I, Sabbà C, Mazzocca A (2014). "The presence of clustered circulating tumor cells (CTCs) and circulating cytokines define an aggressive phenotype in metastatic
439:
positive, but much less effective in patients who are Her2 negative. Once the primary tumor is removed, biopsy of the current state of the cancer through traditional tissue typing is not possible anymore. Often tissue sections of the primary tumor, removed years prior, are used to do the typing.
402:
Hybrid methods combine physical separation (by gradients, magnetic fields, etc.) with antibody-mediated cell retrieval. An example of this is a sensitive double gradient centrifugation and magnetic cell sorting detection and enumeration method which has been used to detect circulating epithelial
179:
Circulating tumor cells are found in frequencies on the order of 1-10 CTC per mL of whole blood in patients with metastatic disease. For comparison, a mL of blood contains a few million white blood cells and a billion red blood cells. This low frequency, associated to difficulty of identifying
1315:
Marrinucci, Dena; Bethel, Kelly; Kolatkar, Anand; Luttgen, Madelyn; Malchiodi, Michael; Baehring, Franziska; Voigt, Katharina; Lazar, Daniel; Nieva, Jorge; Bazhenova, Lyudmilda; Ko, Andrew; Korn, W. Michael; Schram, Ethan; Coward, Michael; Yang, Xing; Metzner, Thomas; Lamy, Rachelle; Honnatti,
506:
Modern cancer research has demonstrated that CTCs derive from clones in the primary tumor, validating
Ashworth's remarks. The significant efforts put into understanding the CTCs biological properties have demonstrated the critical role circulating tumor cells play in the metastatic spread of
158:
Previously it was assumed that CTC clusters could not pass through narrow vessels, such as capillaries, due to their overall size. However, it has been shown that CTC clusters can "unwind" through "selective cleavage of intercellular adhesions" to traverse these constrictions single-file, then
4071:
Hodgkinson, Cassandra L; Morrow, Christopher J; Li, Yaoyong; Metcalf, Robert L; Rothwell, Dominic G; Trapani, Francesca; Polanski, Radoslaw; Burt, Deborah J; Simpson, Kathryn L; Morris, Karen; Pepper, Stuart D; Nonaka, Daisuke; Greystoke, Alastair; Kelly, Paul; Bola, Becky; Krebs, Matthew G;
219:
Kaplan Meier
Analysis of overall survival before starting a new line of therapy for patients with metastatic breast, colorectal and prostate cancer. Patients were divided into those with Favorable and Unfavorable CTC (Unfavorable: >5 CTC/7.5mL for breast and prostate, >3 CTC/7.5mL for
351:
and other staining techniques to look for abnormalities such as duplications, deletions, and rearrangements. The imaging and analysis technology also allows for the coordinates of every cell on a slide to be known so that a single cell can be retrieved from the slide for analysis using
347:
of all the cells on the slide (approximately 3 million), records the precise coordinates of each cell, and analyzes each cell for 90 different parameters, including the fluorescence intensity of the four fluorescent markers and 86 different morphological parameters. Epic can also use
2574:
Nagrath, Sunitha; Sequist, Lecia V.; Maheswaran, Shyamala; Bell, Daphne W.; Irimia, Daniel; Ulkus, Lindsey; Smith, Matthew R.; Kwak, Eunice L.; Digumarthy, Subba; Muzikansky, Alona; Ryan, Paula; Balis, Ulysses J.; Tompkins, Ronald G.; Haber, Daniel A.; Toner, Mehmet (December 2007).
129:
or EMT). These populations of CTCs may be the most resistant and most prone to metastasis. They are also more difficult to isolate because they express neither cytokeratins nor CD45. Otherwise, their morphology, gene expression and genomics are similar to those of other cancer
800:
175:
The detection of CTCs may have important prognostic and therapeutic implications but because their numbers can be very small, these cells are not easily detected. It is estimated that among the cells that have detached from the primary tumor, only 0.01% can form metastases.
511:. Furthermore, highly sensitive, single-cell analysis demonstrated a high level of heterogeneity seen at the single cell level for both protein expression and protein localization and the CTCs reflected both the primary biopsy and the changes seen in the metastatic sites.
4072:
Antonello, Jenny; Ayub, Mahmood; Faulkner, Suzanne; Priest, Lynsey; Carter, Louise; Tate, Catriona; Miller, Crispin J; Blackhall, Fiona; Brady, Ged; Dive, Caroline (1 June 2014). "Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer".
4657:
Fehm T, Sagalowsky A, Clifford E, Beitsch P, Saboorian H, Euhus D, Meng S, Morrison L, Tucker T, Lane N, Ghadimi BM, Heselmeyer-Haddad K, Ried T, Rao C, Uhr J (Jul 2002). "Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant".
754:
Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, Oommen NB, Hawche G, Jameson C, Thompson E, Sipkema R, Carden CP, Parker C, Dearnaley D, Kaye SB, Cooper CS, Molina A, Cox ME, Terstappen LW, de Bono JS (2009).
481:
that many different morphological definitions of circulating tumor cells have similar prognostic value, even though the absolute number of cells found in patients and normal donors varied by more than a decade between different morphological definitions.
838:
Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA (2012).
498:
The importance of CTCs in modern cancer research began in the mid 1990s with the demonstration that CTCs exist early on in the course of the disease. Those results were made possible by exquisitely sensitive magnetic separation technology employing
4165:
Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, Beitsch P, Khan A, Euhus D, Osborne C, Frenkel E, Hoover S, Leitch M, Clifford E, Vitetta E, Morrison L, Herlyn D, Terstappen LW, Fleming T, Fehm T, Tucker T, Lane N, Wang J, Uhr J (2004).
300:
analogues and conjugated with antibodies against EpCAM for the capture of CTCs. Isolation is coupled to an analyzer to take images of isolated cells upon their staining with specific fluorescent antibody conjugates. Blood is sampled in an
4747:
3210:
Luecke, Klaus, et al. "The GILUPI CellCollector as an in vivo tool for circulating tumor cell enumeration and molecular characterization in lung cancer patients." ASCO Annual Meeting Proceedings. Vol. 33. No. 15_suppl. 2015.
472:
in mice. This was the first predictive case for brain metastasis and a proof of concept that intrinsic molecular features of metastatic precursors amongst CTCs could provide novel insights into the mechanisms of metastasis.
377:
also show increased levels of EpCAM-positive cells. Patients with severe skin burns can also carry EpCAM positive cells in the blood. Therefore, the use of EpCAM-positive cells as a tool for early diagnosis is not optimal.
1495:
Ye Z, Mu Z, Wang C, Palazzo JP, Biederman L, Li B, Jaslow R, Avery T, Austin L, Yang H, Cristofanilli M (2016). "Prognostic values of circulating tumor cell (CTC) enumeration and their clusters in advanced breast cancer".
3988:
Polascik TJ, Wang ZP, Shue M, Di S, Gurganus RT, Hortopan SC, Ts'o PO, Partin AW (1999). "Influence of sextant prostate needle biopsy or surgery on the detection and harvest of intact circulating prostate cancer cells".
3025:
Wang, Huiqiang; Chen, Nanhai G.; Minev, Boris R.; Zimmermann, Martina; Aguilar, Richard J.; Zhang, Qian; Sturm, Julia B.; Fend, Falko; Yu, Yong A.; Cappello, Joseph; Lauer, Ulrich M.; Szalay, Aladar A. (September 2013).
4627:
1401:
Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S (2014).
3903:
Tkaczuk KH, Goloubeva O, Tait NS, Feldman F, Tan M, Lum ZP, Lesko SA, Van Echo DA, Ts'o PO (2008). "The significance of circulating epithelial cells in Breast Cancer patients by a novel negative selection method".
187:); however this approach is biased by the need for a sufficient expression of the selected protein on the cell surface, event necessary for the enrichment step. Moreover, since EpCAM and other proteins (e.g.
98:(cancers of epithelial origin, which are the most prevalent) can be classified according to the expression of epithelial markers, as well as their size and whether they are apoptotic. In general, CTCs are
228:(FDA) cleared methodology for enumeration of CTC in whole blood is the CellSearch system. Extensive clinical testing done using this method shows that presence of CTCs is a strong prognostic factor for
612:
Rack B, Schindlbeck C, Jückstock J, Andergassen U, Hepp P, Zwingers T, Friedl T, Lorenz R, Tesch H, Fasching P, Fehm T, Schneeweiss A, Lichtenegger W, Beckmann M, Friese K, Pantel K, Janni W (2014).
4391:
Klotz, Remi; Thomas, Amal; Teng, Teng; Han, Sung Min; Iriondo, Oihana; Li, Lin; Restrepo-Vassalli, Sara; Wang, Alan; Izadian, Negeen; MacKay, Matthew; Moon, Byoung-San (2019-01-01).
3725:"Increase in number of circulating disseminated epithelial cells after surgery for non-small cell lung cancer monitored by MAINTRAC is a predictor for relapse: A preliminary report"
3264:
3233:"50P * Enumeration and Molecular Characterization of Circulating Tumor Cells in Lung Cancer Patients Using the Gilupi Cellcollector , an Effective in Vivo Device for Capturing CTCS"
2005:
da Costa A, Oliveira JT, Gärtner F, Kohn B, Gruber AD, Klopfleisch R (2011). "Potential markers for detection of circulating canine mammary tumor cells in the peripheral blood".
4635:
3576:"Assessing the efficacy of targeted therapy using circulating epithelial tumor cells (CETC): the example of SERM therapy monitoring as a unique tool to individualize therapy"
1870:
Tanaka F, Yoneda K, Kondo N, Hashimoto M, Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T, Kuribayashi K, Fukuoka K, Nakano T, Hasegawa S (2009).
423:. The technique, called CTC-iChip, first removes cells too small to be CTCs, such as red blood cells, and then uses magnetic particles to remove white blood cells.
2681:
Peng, Chen; Yu-yen, Huang; Hoshino, Kazunori; Xiaojing, Zhang (2014). "Multiscale immunomagnetic enrichment of circulating tumor cells: from tubes to microchips".
799:
Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ (2008).
2724:
Huang, Yu-yen; Hoshino, Kazunori; Chen, Peng; Wu, Chun-hsien; Lane, Nancy; Huebschman, Michael; Liu, Huaying; Sokolov, Konstantin; Uhr, Jonathan W. (2012-10-30).
3164:"A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire"
2903:
Huang, Yu-Yen; Chen, Peng; Wu, Chun-Hsien; Hoshino, Kazunori; Sokolov, Konstantin; Lane, Nancy; Liu, Huaying; Huebschman, Michael; Frenkel, Eugene (2015-11-05).
3324:
Bethel, Kelly; Luttgen2, Madelyn; Damani, Samir; Kolatkar2, Anand; Lamy, Rachelle; Sabouri-Ghomi, Mohsen; Topol, Sarah; Topol2, Eric; Kuhn, Peter (9 Jan 2014).
1145:
Nieva, J; Wendel, M; Luttgen, MS; Marrinucci, D; Bazhenova, L; Kolatkar, A; Santala, R; Whittenberger, B; Burke, J; Torrey, M; Bethel, K; Kuhn, P (Feb 2012).
4225:
Hayes DF, Walker TM, Singh B, et al. (2002). "Monitoring Expression of HER-2 on Circulating Epithelial Cells in Patients with advanced Breast Cancer".
1589:
Au S, Storey B, Moore J, Tang Q, Chen Y, Javaid S, Sarioglu A, Sullivan R, Madden M, O'Keefe R, Haber D, Maheswaran S, Langenau D, Stott S, Toner M (2016).
801:"Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer"
3215:
3535:"The relevance of circulating epithelial tumor cells (CETC) for therapy monitoring during neoadjuvant (primary systemic) chemotherapy in breast cancer"
1107:
Esmaeilsabzali H, Beischlag TV, Cox ME, Parameswaran AM, Park EJ (2013). "Detection and isolation of circulating tumor cells: principles and methods".
440:
Further characterization of CTC may help determining the current tumor phenotype. FISH assays have been performed on CTC as well as determination of
3625:"Efficacy control of therapy using circulating epithelial tumor cells (CETC) as "Liquid Biopsy": trastuzumab in HER2/neu-positive breast carcinoma"
1147:"High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: a longitudinal analysis"
419:
patients. Similarly, researchers at Massachusetts General Hospital have developed a negative selection method which employs inertial focusing on a
3381:
2486:"Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the CellSearch System"
896:
Sleijfer S, Gratama JW, Sieuwerts AM, et al. (2007). "Circulating tumour cell detection on its way to routine diagnostic implementation?".
121:
Cytokeratin-negative CTCs are characterised by the lack of EpCAM or cytokeratins, which may indicate an undifferentiated phenotype (circulating
3440:"Quantification of the response of circulating epithelial cells to neodadjuvant treatment for breast cancer: a new tool for therapy monitoring"
1721:"Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer"
415:
has been employed to detect circulating prostate cancer cells and another technique known as RosetteStep has been used to isolate CTCs from
2445:"Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with non-malignant diseases"
3271:
3289:
2015). "Comparison of Two Types of Liquid Biopsies in Patients With Hepatocellular Carcinoma Awaiting Orthotopic Liver Transplantation".
183:
CTCs are usually (in 2011) captured from the vasculature by using specific antibodies able to recognize specific tumoral marker (usually
467:
The organ tropism of patient-derived CTC has been investigated in a mouse model. CTCs isolated from breast cancer patients and expanded
4344:"Pharmacokinetics and Pharmacodynamics of the IGF-IR Inhibitor Figitumumab (CP-751,871) in Combination with Paclitaxel and Carboplatin"
3395:
Pachmann, Katharina (5 April 2015). "Current and potential use of MAINTRAC method for cancer diagnosis and prediction of metastasis".
2632:
Hoshino, Kazunori; Huang, Yu-Yen; Lane, Nancy; Huebschman, Michael; Uhr, Jonathan W.; Frenkel, Eugene P.; Zhang, Xiaojing (Oct 2011).
757:"Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer"
260:
have been used. The most common technique is magnetic nanoparticle-based separation (immunomagnetic assay) as used in CellSearch or
4024:
Ali A, Furusato B, Ts'o PO, Lum ZP, Elsamanoudi S, Mohamed A, Srivastava S, Moul JW, Brassell SA, Sesterhenn IA, McLeod DG (2010).
2283:"Veridex LLC. CellSearch circulating tumor cell kit premarket notification—expanded indications for use—metastatic prostate cancer"
1648:
Ghossein RA, Bhattacharya S, Rosai J (1999). "Molecular detection of micrometastases and circulating tumor cells in solid tumors".
403:
cancer cells in breast cancer patients by negative selection. The principle of negative selection is based on the retrieval of all
3825:
Desitter I.; et al. (2011). "A New Device for Rapid Isolation by Size and Characterization of Rare Circulating Tumor Cells".
2257:
2222:
Paterlini-Brechot P, Benali NL.; Benali (2007). "Circulating tumor cells (CTC) detection: Clinical impact and future directions".
2040:
da Costa, A (2013). "Multiple RT-PCR markers for the detection of circulating tumour cells of metastatic canine mammary tumours".
159:
reverse the process once clear. This behavior could be a factor in why CTC clusters have such a significant metastatic potential.
3965:
3948:
3776:"Seeding of Epithelial Cells into Circulation During Surgery for Breast Cancer: The Fate of Malignant and Benign Mobilized Cells"
2404:"Circulating Tumor Cells (CTC) predict survival benefit from treatment in metastatic castration resistant prostate cancer (CRPC)"
114:, which demonstrate epithelial origin; the absence of CD45, indicating the cell is not of hematopoietic origin; and their larger
2175:"Carcinocythemia: A rare entity becoming more common? A 3-year, single institution series of seven cases and literature review"
4540:
Ashworth, T. R (1869). "A case of cancer in which cells similar to those in the tumours were seen in the blood after death".
441:
192:
704:"Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type"
3674:"Epithelial Cell Dissemination and Readhesion: Analysis of Factors Contributing to Metastasis Formation in Breast Cancer"
3533:
Camara O.; Rengsberger M.; Egbe A.; Koch A.; Gajda M.; Hammer U.; Jorke C.; Rabenstein C.; Untch M.; Pachmann K. (2007).
1911:
Negin, BP; Cohen, SJ (Jun 2010). "Circulating tumor cells in colorectal cancer: past, present, and future challenges".
348:
126:
1015:
Pantel K, Riethdorf S.; Riethdorf (2009). "Pathology: are circulating tumor cells predictive of overall survival?".
3774:
Camara Oumar; Kavallaris Andreas; Nöschel Helmut; Rengsberger Matthias; Jörke Cornelia; Pachmann Katharina (2006).
199:
like IsofFux or Maintrac. In very rare cases, CTCs are present in large enough quantities to be visible on routine
1060:"Detection of circulating tumor cells in prostate cancer patients: methodological pitfalls and clinical relevance"
3326:"Fluid phase biopsy for detection and characterization of circulating endothelial cells in myocardial infarction"
261:
4262:"Multigene Reverse Transcription-PCR Profiling of Circulating Tumor Cells in Hormone-Refractory Prostate Cancer"
23:
An illustration depicting primary tumor (in the form of tumor microenvironment) and the circulating tumor cells.
4685:
Fidler IJ (2003). "Timeline: The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited".
1954:
Mikolajczyk, SD; Millar, LS; Tsinberg, P; Coutts, SM; Zomorrodi, M; Pham, T; Bischoff, FZ; Pircher, TJ (2011).
528:
Riquet, M; Rivera, C; Gibault, L; Pricopi, C; Mordant, P; Badia, A; Arame, A; Le Pimpec Barthes, F (2014). "".
191:) are not expressed in some tumors and can be down regulated during the epithelial to mesenchymal transition (
3212:
1316:
Meghana; Yoshioka, Craig; Kunken, Joshua; Petrova, Yelena; Sok, Devin; Nelson, David; Kuhn, Peter (Feb 2012).
4771:
2840:"Microscale Magnetic Field Modulation for Enhanced Capture and Distribution of Rare Circulating Tumor Cells"
2484:
Riethdorf; Fritsche, H; Müller, V; Rau, T; Schindlbeck, C; Rack, B; Janni, W; Coith, C; et al. (2007).
2077:"Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay"
4628:"The tools that fueled the new world of Circulating Tumor Cells by Paul A. Liberti – BioMagnetic Solutions"
2290:
225:
3088:"The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner"
2783:"Computational Analysis of Microfluidic Immunomagnetic Rare Cell Separation from a Particulate Blood Flow"
4559:
Racila, E.; Euhus, D.; Weiss, A. J.; Rao, C.; McConnell, J.; Terstappen, L. W. M. M.; Uhr, J. W. (1998).
4501:"All circulating EpCAM1CK1CD452 objects predict overall survival in castration-resistant prostate cancer"
2970:"Inkjet-Print Micromagnet Array on Glass Slides for Immunomagnetic Enrichment of Circulating Tumor Cells"
326:
83:
4118:"Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells"
663:"Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes"
292:
CellSearch is the only FDA-approved platform for CTC isolation. This method is based on the use of iron
224:
To date, a variety of research methods have been developed to isolate and enumerate CTCs. The only U.S.
3028:"Optical Detection and Virotherapy of Live Metastatic Tumor Cells in Body Fluids with Vaccinia Strains"
2726:"Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system"
2173:
Ronen, Shira; Kroft, Steven H.; Olteanu, Horatiu; Hosking, Paul R.; Harrington, Alexandra M. (2019).
257:
4303:"Potential Applications for Circulating Tumor Cells expressing the Insulin Growth Factor-I Receptor"
3852:
Miller, M. Craig; Robinson, Peggy S.; Wagner, Christopher; O'Shannessy, Daniel J. (14 August 2018).
2075:
Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; and Ionescu-Zanetti, C. (2013).
431:
Some drugs are particularly effective against cancers which fit certain requirements. For example,
102:-resistant, which means that they can survive in the bloodstream without attaching to a substrate.
3574:
Pachmann K.; Camara O.; Kohlhase A.; Rabenstein C.; Kroll T.; Runnebaum I.B.; Hoeffken K. (2010).
2363:"Circulating Tumor Cells versus Imaging - Predicting Overall Survival in Metastatic Breast Cancer"
1956:"Detection of EpCAM-Negative and Cytokeratin-Negative Circulating Tumor Cells in Peripheral Blood"
4026:"Assessment of circulating tumor cells (CTCs) in prostate cancer patients with low-volume tumors"
3773:
3623:
Pachmann K.; Camara O.; Kroll T.; Gajda M.; Gellner A.K.; Wotschadlo J.; Runnebaum I.B. (2011).
841:"RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis"
614:"Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients"
416:
3375:
245:
2905:"Screening and Molecular Analysis of Single Circulating Tumor Cells Using Micromagnet Array"
244:
Biological methods isolate cells based on highly specific antigen binding, most commonly by
4572:
4179:
3337:
3099:
3039:
2916:
2851:
2588:
1602:
1525:
1329:
1264:
1158:
852:
491:
2968:
Chen, Peng; Huang, Yu-Yen; Bhave, Gauri; Hoshino, Kazunori; Zhang, Xiaojing (2015-08-20).
1251:
Racila, E; Euhus, D; Weiss, AJ; Rao, C; McConnell, J; Terstappen, LW; Uhr, JW (Apr 1998).
8:
420:
281:
280:
on the surface of CTCs. CTCs may also be retrieved directly from the blood by a modified
277:
269:
4576:
4393:"Circulating tumor cells exhibit metastatic tropism and reveal brain metastasis drivers"
4183:
3349:
3341:
3103:
3043:
2920:
2855:
2592:
1606:
1404:"Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis"
1333:
1268:
1162:
856:
4710:
4481:
4427:
4392:
4368:
4343:
4142:
4117:
4097:
4053:
3929:
3880:
3853:
3802:
3775:
3751:
3724:
3700:
3673:
3649:
3624:
3600:
3575:
3466:
3439:
3438:
Pachmann K.; Camara O.; Kavallaris A.; Schneider U.; Schünemann S.; Höffken K. (2005).
3420:
3358:
3325:
3302:
3188:
3163:
3120:
3087:
3062:
3027:
3002:
2969:
2945:
2904:
2880:
2839:
2815:
2782:
2758:
2725:
2706:
2658:
2633:
2609:
2577:"Isolation of rare circulating tumour cells in cancer patients by microchip technology"
2576:
2551:
2526:
2343:
2314:"Circulating Tumor Cells, Disease Progression and Survival in Metastatic Breast Cancer"
2155:
2101:
2076:
1982:
1955:
1936:
1847:
1822:
1798:
1771:
1747:
1720:
1693:
1676:
1625:
1590:
1571:
1509:
1477:
1428:
1403:
1350:
1341:
1317:
1228:
1203:
1179:
1170:
1146:
1084:
1059:
1040:
997:
873:
840:
728:
703:
679:
662:
638:
613:
594:
461:
374:
268:
viruses are developed to detect and identify CTCs. Alternative methods exist which use
48:
4202:
4167:
4776:
4702:
4667:
4608:
4603:
4590:
4560:
4522:
4473:
4432:
4414:
4373:
4324:
4283:
4242:
4207:
4147:
4101:
4089:
4057:
4045:
4041:
4006:
4002:
3970:
3921:
3885:
3834:
3807:
3756:
3705:
3654:
3605:
3556:
3515:
3507:
3471:
3412:
3363:
3306:
3193:
3125:
3067:
3007:
2989:
2950:
2932:
2885:
2867:
2820:
2802:
2763:
2745:
2710:
2698:
2663:
2614:
2556:
2507:
2466:
2425:
2384:
2335:
2239:
2204:
2196:
2147:
2106:
2057:
2022:
1987:
1928:
1893:
1852:
1803:
1752:
1698:
1657:
1630:
1563:
1555:
1469:
1433:
1355:
1292:
1287:
1252:
1233:
1184:
1124:
1120:
1089:
1032:
989:
954:
913:
878:
820:
778:
733:
684:
643:
586:
545:
457:
122:
68:
4728:
3933:
3424:
2159:
1940:
1575:
1481:
1387:
4714:
4694:
4598:
4580:
4512:
4485:
4463:
4422:
4404:
4363:
4355:
4314:
4273:
4234:
4197:
4187:
4137:
4129:
4081:
4037:
3998:
3960:
3913:
3875:
3865:
3797:
3787:
3746:
3736:
3695:
3685:
3644:
3636:
3595:
3587:
3546:
3499:
3461:
3451:
3404:
3353:
3345:
3298:
3244:
3183:
3175:
3115:
3107:
3057:
3047:
2997:
2981:
2940:
2924:
2875:
2859:
2810:
2794:
2753:
2737:
2690:
2653:
2645:
2604:
2596:
2546:
2538:
2497:
2456:
2415:
2374:
2347:
2325:
2261:
2231:
2186:
2139:
2096:
2088:
2049:
2014:
1977:
1967:
1920:
1883:
1842:
1834:
1793:
1783:
1742:
1732:
1688:
1620:
1610:
1545:
1537:
1505:
1461:
1423:
1415:
1383:
1345:
1337:
1282:
1272:
1223:
1215:
1174:
1166:
1116:
1079:
1071:
1044:
1024:
981:
944:
905:
868:
860:
812:
768:
723:
715:
674:
633:
625:
598:
576:
537:
407:
by using a panel of antibodies as well as traditional gradient centrifugation with
106:
Traditional CTCs are characterised by an intact, viable nucleus; the expression of
40:
4319:
4302:
3408:
2502:
2485:
2461:
2444:
2420:
2403:
2379:
2362:
1888:
1871:
1838:
1318:"Fluid Biopsy in Patients with Metastatic Prostate, Pancreatic and Breast Cancers"
1001:
949:
932:
773:
756:
464:. Single cell level qPCR can also be performed with the CTCs isolated from blood.
4409:
4359:
4278:
4261:
4133:
3503:
3219:
3052:
2235:
1541:
933:"Is There a Role for Circulating Tumor Cells in the Management of Breast Cancer?"
541:
361:
204:
72:
3671:
3249:
3232:
3111:
2053:
2018:
1419:
1374:(CTCs) phenotypes in metastatic castration-resistant prostate cancer (mCRPC)".
909:
581:
564:
248:
for positive selection. Antibodies against tumor specific biomarkers including
19:
3917:
3640:
3591:
2985:
2741:
1924:
1465:
1075:
1028:
985:
4765:
4594:
4418:
3949:"Identification and characterization of circulating prostate carcinoma cells"
3723:
Rolle A.; Günzel R.; Pachmann U.; Willen B.; Höffken K.; Pachmann K. (2005).
3511:
3437:
2993:
2936:
2871:
2838:
Chen, Peng; Huang, Yu-Yen; Hoshino, Kazunori; Zhang, John X.J. (2015-03-04).
2806:
2749:
2200:
2143:
1772:"Circulating tumor cells in breast cancer: a tool whose time has come of age"
1559:
816:
702:
Marrinucci, D; Bethel, K; Luttgen, M; Nieva, J; Kuhn, P; Kuhn, P (Sep 2009).
293:
229:
79:
64:
4517:
4500:
4192:
3573:
3551:
3534:
3287:
2781:
Hoshino, Kazunori; Chen, Peng; Huang, Yu-Yen; Zhang, Xiaojing (2012-05-15).
2282:
1615:
4706:
4671:
4585:
4526:
4477:
4436:
4377:
4328:
4287:
4246:
4211:
4151:
4093:
4049:
4010:
3974:
3947:
Wang ZP, Eisenberger MA, Carducci MA, Partin AW, Scher HI, Ts'o PO (2000).
3925:
3889:
3838:
3811:
3792:
3760:
3741:
3709:
3672:
Hekimian K.; Meisezahl S.; Trompelt K.; Rabenstein C.; Pachmann K. (2012).
3658:
3609:
3560:
3519:
3475:
3416:
3367:
3310:
3213:
http://hwmaint.meeting.ascopubs.org/cgi/content/abstract/33/15_suppl/e22035
3197:
3129:
3071:
3011:
2954:
2889:
2824:
2767:
2702:
2667:
2618:
2560:
2511:
2470:
2429:
2388:
2339:
2243:
2208:
2151:
2128:
2110:
2061:
2026:
1991:
1932:
1897:
1856:
1807:
1788:
1756:
1702:
1661:
1634:
1567:
1473:
1437:
1359:
1277:
1237:
1204:"Circulating tumor cell clusters: What we know and what we expect (Review)"
1188:
1128:
1093:
1036:
993:
972:
Pantel K, Alix-Panabières C, Riethdorf S (2009). "Cancer micrometastases".
958:
917:
882:
824:
782:
737:
688:
647:
590:
549:
111:
36:
4612:
4238:
3690:
3179:
2542:
1972:
1953:
1737:
1526:"Tumor-derived circulating endothelial cell clusters in colorectal cancer"
1296:
1219:
629:
4468:
4451:
3870:
3854:"The Parsortix Cell Separation System—A versatile liquid biopsy platform"
2330:
2313:
453:
432:
404:
386:
Physical methods are often filter-based, enabling the capture of CTCs by
311:
200:
188:
52:
4498:
3966:
10.1002/1097-0142(20000615)88:12<2787::aid-cncr18>3.0.co;2-2
2600:
1550:
1524:
Koh, P. K.; Robson, P.; Ying, J. Y.; Hillmer, A. M.; Tan, M.-H. (2016).
864:
167:
4452:"Statistical Considerations for Enumeration of Circulating Tumor Cells"
2694:
2649:
2527:"Circulating tumor cells: approaches to isolation and characterization"
2191:
2174:
719:
500:
449:
95:
60:
4168:"HER-2 gene amplification can be acquired as breast cancer progresses"
3086:
Daugaard, Mads; Heeschen, Christopher; Salanti, Ali (16 August 2018).
2928:
2863:
2798:
2092:
1872:"Circulating tumor cell as a diagnostic marker in primary lung cancer"
1591:"Clusters of circulating tumor cells traverse capillary-sized vessels"
1106:
3722:
3532:
2634:"Microchip-based immunomagnetic detection of circulating tumor cells"
2573:
2221:
1314:
508:
412:
387:
134:
115:
4698:
4085:
4025:
3622:
3488:
3456:
3323:
2631:
207:
or carcinoma cell leukemia and is associated with a poor prognosis.
3851:
2074:
611:
436:
265:
232:
in patients with metastatic breast, colorectal or prostate cancer.
44:
4499:
F. A. W. Coumans; C. J. M. Doggen; G. Attard; et al. (2010).
3084:
2524:
171:
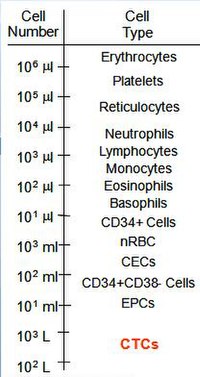
Number of various types of blood cells in whole blood versus CTCs.
4449:
3024:
391:
273:
215:
99:
4561:"Detection and characterization of carcinoma cells in the blood"
1253:"Detection and characterization of carcinoma cells in the blood"
1144:
1014:
701:
408:
297:
2401:
971:
4656:
3946:
1677:"Circulating tumor cells: finding the needle in the haystack"
1372:
445:
249:
184:
107:
1450:
1400:
3902:
2004:
930:
322:
318:
307:
302:
253:
4114:
2483:
1869:
1823:"Circulating tumor cells as biomarkers in prostate cancer"
1820:
1058:
Panteleakou Z, Lembessis P, Sourla A, et al. (2009).
1057:
753:
527:
306:
a nuclear stain, a fluorescent antibody conjugate against
125:) or the acquisition of a mesenchymal phenotype (known as
4070:
2680:
1647:
314:
4259:
2402:
JS DeBono; HI Scher; RB Montgomery; et al. (2008).
2312:
Cristofanilli M, Budd GT, Ellis MJ, et al. (2004).
2172:
1202:
Hong, Yupeng; Fang, Francia; Zhang, Qi (December 2016).
895:
798:
133:
Apoptotic CTCs are traditional CTCs that are undergoing
4260:
O'Hara SM, Moreno JG, Zweitzig DR, et al. (2004).
4164:
3144:
1674:
4023:
2361:
Budd G, Cristofanilli M, Ellis M, et al. (2006).
2360:
2311:
1250:
16:
Cell from a primary tumor carried by blood circulation
2967:
2837:
2780:
1769:
1718:
661:
Millner, LM; Linder, MW; Valdes R, Jr (Summer 2013).
660:
2443:
Allard WJ, Matera J, Miller MC, et al. (2004).
4558:
4301:de Bono JS, Attard G, Adjei A, et al. (2007).
1821:Danila, DC; Fleisher, M; Scher, HI (Jun 15, 2011).
1522:
141:
which often require a different therapeutic course.
4390:
4342:Karp DD, Pollak MN, Cohen RB, et al. (2009).
4300:
3987:
3161:
2902:
2723:
2442:
1140:
1138:
837:
1588:
4763:
4224:
562:
59:for the subsequent growth of additional tumors (
4565:Proceedings of the National Academy of Sciences
4552:
4341:
1714:
1712:
1595:Proceedings of the National Academy of Sciences
1494:
1257:Proceedings of the National Academy of Sciences
1135:
708:Archives of Pathology & Laboratory Medicine
4539:
4450:AGJ Tibbe; MC Miller; LWMM Terstappen (2007).
3145:"GILUPI - CellCollector in vivo CTC isolation"
2179:International Journal of Laboratory Hematology
2166:
1910:
1582:
794:
792:
749:
747:
1310:
1308:
1306:
1201:
4625:
4158:
4108:
4017:
3981:
3940:
3896:
1998:
1863:
1770:Swaby, RF; Cristofanilli, M (Apr 21, 2011).
1709:
1516:
1394:
1100:
965:
118:, irregular shape or subcellular morphology.
3824:
3380:: CS1 maint: numeric names: authors list (
3230:
2124:
2122:
2120:
1719:Miller MC, Doyle GV, Terstappen LW (2010).
831:
789:
744:
272:instead of antibodies, such as the malaria
195:), new enrichment strategies are required.
1303:
4684:
4602:
4584:
4516:
4467:
4426:
4408:
4367:
4318:
4277:
4201:
4191:
4141:
3964:
3879:
3869:
3801:
3791:
3750:
3740:
3699:
3689:
3648:
3599:
3550:
3465:
3455:
3357:
3248:
3187:
3119:
3061:
3051:
3001:
2944:
2879:
2814:
2757:
2657:
2608:
2550:
2501:
2460:
2419:
2378:
2329:
2190:
2100:
1981:
1971:
1887:
1846:
1797:
1787:
1746:
1736:
1692:
1624:
1614:
1549:
1427:
1349:
1286:
1276:
1227:
1178:
1083:
948:
872:
772:
727:
678:
667:Annals of Clinical and Laboratory Science
637:
580:
565:"Cancer metastasis: building a framework"
3394:
3162:Saucedo-Zeni Nadia; et al. (2012).
2117:
2039:
618:Journal of the National Cancer Institute
214:
166:
18:
1675:Zhe, X; Cher M.L.; Bonfil R.D. (2011).
1444:
563:Gupta, GP; Massagué, J (Nov 17, 2006).
490:person". A thorough comparison of the
426:
341:
4764:
3397:Expert Review of Molecular Diagnostics
931:Hayes DF, Smerage J.; Smerage (2008).
435:is very effective in patients who are
47:and is carried around the body in the
1913:Current Treatment Options in Oncology
296:coated with a polymer layer carrying
239:
287:
210:
203:examination. This is referred to as
4729:"Meeting Library - Meeting Library"
3265:"An Introduction to the CellSearch"
1488:
605:
381:
332:
35:) is a cell that has shed into the
13:
4745:
4626:Paul Liberti (November 22, 2013).
3780:World Journal of Surgical Oncology
3303:10.1016/j.transproceed.2015.10.003
3142:
1510:10.1158/1538-7445.SABCS15-P2-08-09
475:
276:protein, which binds to oncofetal
14:
4788:
3231:Scheumann N; et al. (2015).
3168:International Journal of Oncology
1208:International Journal of Oncology
397:
127:epithelial-mesenchymal transition
4042:10.1111/j.1440-1827.2010.02584.x
4003:10.1097/00005392-199909010-00034
2974:Annals of Biomedical Engineering
1121:10.1016/j.biotechadv.2013.08.016
1017:Nature Reviews Clinical Oncology
4739:
4721:
4678:
4650:
4619:
4533:
4492:
4443:
4384:
4335:
4294:
4253:
4218:
4064:
3845:
3818:
3767:
3716:
3665:
3616:
3567:
3526:
3482:
3431:
3388:
3317:
3281:
3257:
3224:
3204:
3155:
3136:
3078:
3018:
2961:
2896:
2831:
2774:
2717:
2674:
2625:
2567:
2518:
2477:
2436:
2395:
2354:
2305:
2275:
2250:
2215:
2068:
2033:
1947:
1904:
1814:
1763:
1668:
1641:
1388:10.1200/jco.2014.32.4_suppl.209
1366:
1244:
1195:
1051:
1008:
924:
145:
4122:Science Translational Medicine
1530:Science Translational Medicine
889:
695:
654:
556:
521:
364:platform applying microscopic
1:
4320:10.1158/1078-0432.CCR-07-0268
3490:predictor of early relapse".
3409:10.1586/14737159.2015.1032260
3350:10.1088/1478-3975/11/1/016002
2503:10.1158/1078-0432.CCR-06-1695
2462:10.1158/1078-0432.CCR-04-0378
2421:10.1158/1078-0432.CCR-08-0872
2380:10.1158/1078-0432.CCR-05-1769
1889:10.1158/1078-0432.CCR-09-1095
1839:10.1158/1078-0432.CCR-10-2650
950:10.1158/1078-0432.CCR-07-4481
774:10.1158/0008-5472.CAN-08-3667
530:Revue de Pneumologie Clinique
514:
4410:10.1158/2159-8290.CD-19-0384
4360:10.1097/JTO.0b013e3181ba2f1d
4348:Journal of Thoracic Oncology
4279:10.1373/clinchem.2003.028563
4172:Proc. Natl. Acad. Sci. U.S.A
4134:10.1126/scitranslmed.3005616
3504:10.1016/j.breast.2006.12.005
3053:10.1371/journal.pone.0071105
2291:Food and Drug Administration
2260:. March 2010. Archived from
2258:"Veridex CellSearch Website"
2236:10.1016/j.canlet.2006.12.014
1542:10.1126/scitranslmed.aad7369
1376:Journal of Clinical Oncology
1342:10.1088/1478-3975/9/1/016003
1171:10.1088/1478-3975/9/1/016004
542:10.1016/j.pneumo.2013.07.001
411:. A similar method known as
226:Food and Drug Administration
162:
7:
3291:Transplantation Proceedings
2531:The Journal of Cell Biology
2525:Yu M.; et al. (2011).
355:
84:sensitivity and specificity
10:
4793:
4542:Australian Medical Journal
4116:Haber DA, Toner M (2013).
3629:J. Cancer Res. Clin. Oncol
3580:J. Cancer Res. Clin. Oncol
3112:10.1038/s41467-018-05793-2
2054:10.1016/j.tvjl.2012.08.021
2019:10.1016/j.tvjl.2010.09.027
1504:(4 Supplement): P2–08–09.
1420:10.1016/j.cell.2014.07.013
910:10.1016/j.ejca.2007.09.016
582:10.1016/j.cell.2006.11.001
484:
78:The detection of CTCs, or
4632:biomargneticsolutions.com
3918:10.1007/s10549-007-9771-9
3641:10.1007/s00432-011-1000-6
3592:10.1007/s00432-010-0942-4
2986:10.1007/s10439-015-1427-z
2742:10.1007/s10544-012-9718-8
1925:10.1007/s11864-010-0115-3
1466:10.1007/s10552-014-0457-4
1076:10.2119/molmed.2008.00116
1029:10.1038/nrclinonc.2009.23
986:10.1038/nrclinonc.2009.44
94:CTCs that originate from
4660:Clinical Cancer Research
3906:Breast Cancer Res. Treat
3250:10.1093/annonc/mdv045.14
2144:10.1200/JCO.2007.13.6523
1827:Clinical Cancer Research
817:10.1200/JCO.2007.15.8923
390:rather than by specific
89:
4752:www.abstractsonline.com
4733:meetinglibrary.asco.org
4193:10.1073/pnas.0402993101
2730:Biomedical Microdevices
1616:10.1073/pnas.1524448113
310:(leukocyte marker) and
4586:10.1073/pnas.95.8.4589
3793:10.1186/1477-7819-4-67
3742:10.1186/1477-7819-3-18
1789:10.1186/1741-7015-9-43
1278:10.1073/pnas.95.8.4589
417:small-cell lung cancer
221:
172:
155:of the primary tumor.
29:circulating tumor cell
24:
4687:Nature Reviews Cancer
4638:on September 26, 2017
4518:10.1093/annonc/mdq030
4239:10.3892/ijo.21.5.1111
3552:10.1093/annonc/mdm206
3180:10.3892/ijo.2012.1557
3092:Nature Communications
2543:10.1083/jcb.201010021
1454:Cancer Causes Control
1220:10.3892/ijo.2016.3747
362:diagnostic blood test
246:monoclonal antibodies
218:
170:
22:
4772:Anatomical pathology
4469:10.1002/cyto.a.20369
3871:10.1002/cyto.a.23571
3729:World J. Surg. Oncol
2787:Analytical Chemistry
2331:10.1056/NEJMoa040766
1452:colorectal cancer".
427:CTC characterization
342:Epic Sciences method
4577:1998PNAS...95.4589R
4184:2004PNAS..101.9393M
3827:Anticancer Research
3691:10.5402/2012/601810
3342:2014PhBio..11a6002B
3104:2018NatCo...9.3279A
3044:2013PLoSO...871105W
2921:2015NatSR...516047H
2856:2015NatSR...5E8745C
2601:10.1038/nature06385
2593:2007Natur.450.1235N
2587:(7173): 1235–1239.
1973:10.1155/2011/252361
1960:Journal of Oncology
1738:10.1155/2010/617421
1607:2016PNAS..113.4947A
1334:2012PhBio...9a6003M
1269:1998PNAS...95.4589R
1163:2012PhBio...9a6004N
865:10.1038/nature11217
857:2012Natur.487..510Y
630:10.1093/jnci/dju066
421:microfluidic device
282:Seldinger technique
278:chondroitin sulfate
270:engineered proteins
4505:Annals of Oncology
3997:(3 Pt 1): 749–52.
3237:Annals of Oncology
3218:2016-03-10 at the
2909:Scientific Reports
2844:Scientific Reports
2695:10.1039/C3LC51107C
2650:10.1039/c1lc20270g
2192:10.1111/ijlh.12924
2042:Veterinary Journal
974:Nat Rev Clin Oncol
720:10.5858/133.9.1468
462:immunofluorescence
240:Biological methods
222:
173:
25:
4403:(1): CD–19–0384.
4354:(11): 1397–1403.
3864:(12): 1234–1239.
3444:Breast Cancer Res
2929:10.1038/srep16047
2864:10.1038/srep08745
2799:10.1021/ac2032386
2793:(10): 4292–4299.
2644:(20): 3449–3457.
2455:(20): 6897–6904.
2093:10.1593/tlo.13367
288:CellSearch method
211:Detection methods
123:cancer stem cells
49:blood circulation
4784:
4756:
4755:
4743:
4737:
4736:
4725:
4719:
4718:
4682:
4676:
4675:
4654:
4648:
4647:
4645:
4643:
4634:. Archived from
4623:
4617:
4616:
4606:
4588:
4571:(8): 4589–4594.
4556:
4550:
4549:
4537:
4531:
4530:
4520:
4496:
4490:
4489:
4471:
4456:Cytometry Part A
4447:
4441:
4440:
4430:
4412:
4397:Cancer Discovery
4388:
4382:
4381:
4371:
4339:
4333:
4332:
4322:
4298:
4292:
4291:
4281:
4257:
4251:
4250:
4222:
4216:
4215:
4205:
4195:
4162:
4156:
4155:
4145:
4112:
4106:
4105:
4068:
4062:
4061:
4021:
4015:
4014:
3985:
3979:
3978:
3968:
3944:
3938:
3937:
3900:
3894:
3893:
3883:
3873:
3858:Cytometry Part A
3849:
3843:
3842:
3822:
3816:
3815:
3805:
3795:
3771:
3765:
3764:
3754:
3744:
3720:
3714:
3713:
3703:
3693:
3669:
3663:
3662:
3652:
3635:(9): 1317–1327.
3620:
3614:
3613:
3603:
3571:
3565:
3564:
3554:
3545:(9): 1484–1492.
3530:
3524:
3523:
3486:
3480:
3479:
3469:
3459:
3435:
3429:
3428:
3392:
3386:
3385:
3379:
3371:
3361:
3330:Physical Biology
3321:
3315:
3314:
3297:(9): 2639–2642.
3285:
3279:
3278:
3276:
3270:. Archived from
3269:
3261:
3255:
3254:
3252:
3228:
3222:
3208:
3202:
3201:
3191:
3174:(4): 1241–1250.
3159:
3153:
3152:
3140:
3134:
3133:
3123:
3082:
3076:
3075:
3065:
3055:
3022:
3016:
3015:
3005:
2980:(5): 1710–1720.
2965:
2959:
2958:
2948:
2900:
2894:
2893:
2883:
2835:
2829:
2828:
2818:
2778:
2772:
2771:
2761:
2721:
2715:
2714:
2678:
2672:
2671:
2661:
2629:
2623:
2622:
2612:
2571:
2565:
2564:
2554:
2522:
2516:
2515:
2505:
2481:
2475:
2474:
2464:
2440:
2434:
2433:
2423:
2399:
2393:
2392:
2382:
2358:
2352:
2351:
2333:
2309:
2303:
2302:
2300:
2299:
2287:
2279:
2273:
2272:
2270:
2269:
2254:
2248:
2247:
2219:
2213:
2212:
2194:
2170:
2164:
2163:
2138:(8): 1208–1215.
2126:
2115:
2114:
2104:
2072:
2066:
2065:
2037:
2031:
2030:
2002:
1996:
1995:
1985:
1975:
1951:
1945:
1944:
1908:
1902:
1901:
1891:
1876:Clin. Cancer Res
1867:
1861:
1860:
1850:
1818:
1812:
1811:
1801:
1791:
1767:
1761:
1760:
1750:
1740:
1716:
1707:
1706:
1696:
1672:
1666:
1665:
1650:Clin. Cancer Res
1645:
1639:
1638:
1628:
1618:
1586:
1580:
1579:
1553:
1536:(345): 345ra89.
1520:
1514:
1513:
1492:
1486:
1485:
1448:
1442:
1441:
1431:
1398:
1392:
1391:
1382:(4_suppl): 209.
1370:
1364:
1363:
1353:
1322:Physical Biology
1312:
1301:
1300:
1290:
1280:
1263:(8): 4589–4594.
1248:
1242:
1241:
1231:
1214:(6): 2206–2216.
1199:
1193:
1192:
1182:
1151:Physical Biology
1142:
1133:
1132:
1104:
1098:
1097:
1087:
1055:
1049:
1048:
1012:
1006:
1005:
969:
963:
962:
952:
928:
922:
921:
893:
887:
886:
876:
835:
829:
828:
796:
787:
786:
776:
751:
742:
741:
731:
699:
693:
692:
682:
658:
652:
651:
641:
609:
603:
602:
584:
560:
554:
553:
525:
382:Physical methods
333:Parsortix method
230:overall survival
4792:
4791:
4787:
4786:
4785:
4783:
4782:
4781:
4762:
4761:
4760:
4759:
4746:Design, ISITE.
4744:
4740:
4727:
4726:
4722:
4699:10.1038/nrc1098
4683:
4679:
4655:
4651:
4641:
4639:
4624:
4620:
4557:
4553:
4538:
4534:
4497:
4493:
4448:
4444:
4389:
4385:
4340:
4336:
4307:Clin Cancer Res
4299:
4295:
4258:
4254:
4223:
4219:
4163:
4159:
4113:
4109:
4086:10.1038/nm.3600
4074:Nature Medicine
4069:
4065:
4022:
4018:
3986:
3982:
3959:(12): 2787–95.
3945:
3941:
3901:
3897:
3850:
3846:
3823:
3819:
3772:
3768:
3721:
3717:
3670:
3666:
3621:
3617:
3572:
3568:
3531:
3527:
3487:
3483:
3457:10.1186/bcr1328
3450:(6): R975–979.
3436:
3432:
3393:
3389:
3373:
3372:
3322:
3318:
3286:
3282:
3274:
3267:
3263:
3262:
3258:
3229:
3225:
3220:Wayback Machine
3209:
3205:
3160:
3156:
3141:
3137:
3083:
3079:
3023:
3019:
2966:
2962:
2901:
2897:
2836:
2832:
2779:
2775:
2722:
2718:
2679:
2675:
2630:
2626:
2572:
2568:
2523:
2519:
2490:Clin Cancer Res
2482:
2478:
2449:Clin Cancer Res
2441:
2437:
2408:Clin Cancer Res
2400:
2396:
2373:(21): 6404–09.
2367:Clin Cancer Res
2359:
2355:
2310:
2306:
2297:
2295:
2285:
2281:
2280:
2276:
2267:
2265:
2256:
2255:
2251:
2220:
2216:
2171:
2167:
2127:
2118:
2073:
2069:
2038:
2034:
2003:
1999:
1952:
1948:
1909:
1905:
1868:
1864:
1833:(12): 3903–12.
1819:
1815:
1768:
1764:
1717:
1710:
1681:Am J Cancer Res
1673:
1669:
1646:
1642:
1601:(18): 4937–52.
1587:
1583:
1521:
1517:
1498:Cancer Research
1493:
1489:
1460:(11): 1531–41.
1449:
1445:
1399:
1395:
1371:
1367:
1313:
1304:
1249:
1245:
1200:
1196:
1143:
1136:
1109:Biotechnol. Adv
1105:
1101:
1070:(3–4): 101–14.
1056:
1052:
1013:
1009:
970:
966:
943:(12): 3646–50.
937:Clin Cancer Res
929:
925:
904:(18): 2645–50.
894:
890:
851:(7408): 510–3.
836:
832:
811:(19): 3213–21.
797:
790:
752:
745:
700:
696:
659:
655:
610:
606:
561:
557:
526:
522:
517:
487:
478:
476:Cell morphology
429:
400:
384:
375:Crohn's disease
358:
344:
335:
290:
242:
213:
205:carcinocythemia
165:
148:
92:
43:from a primary
17:
12:
11:
5:
4790:
4780:
4779:
4774:
4758:
4757:
4738:
4720:
4677:
4666:(7): 2073–84.
4649:
4618:
4551:
4532:
4491:
4462:(3): 132–142.
4442:
4383:
4334:
4313:(12): 3611–6.
4293:
4272:(5): 826–835.
4252:
4217:
4178:(25): 9393–8.
4157:
4107:
4080:(8): 897–903.
4063:
4036:(10): 667–72.
4016:
3980:
3939:
3895:
3844:
3833:(2): 427–442.
3817:
3766:
3715:
3664:
3615:
3586:(5): 821–828.
3566:
3525:
3498:(2): 211–218.
3481:
3430:
3403:(5): 597–605.
3387:
3316:
3280:
3277:on 2014-01-23.
3256:
3223:
3203:
3154:
3135:
3077:
3017:
2960:
2895:
2830:
2773:
2736:(4): 673–681.
2716:
2689:(3): 446–458.
2673:
2624:
2566:
2537:(3): 373–382.
2517:
2476:
2435:
2414:(19): 6302–9.
2394:
2353:
2304:
2274:
2249:
2230:(2): 180–204.
2214:
2165:
2132:J. Clin. Oncol
2116:
2087:(5): 528–538.
2067:
2032:
1997:
1946:
1903:
1882:(22): 6980–6.
1862:
1813:
1762:
1708:
1687:(6): 740–751.
1667:
1656:(8): 1950–60.
1640:
1581:
1515:
1487:
1443:
1414:(5): 1110–22.
1393:
1365:
1302:
1243:
1194:
1134:
1115:(7): 1063–84.
1099:
1050:
1007:
964:
923:
888:
830:
805:J. Clin. Oncol
788:
743:
714:(9): 1468–71.
694:
673:(3): 295–304.
653:
604:
555:
536:(1–2): 16–25.
519:
518:
516:
513:
486:
483:
477:
474:
428:
425:
399:
398:Hybrid methods
396:
383:
380:
360:Maintrac is a
357:
354:
343:
340:
334:
331:
289:
286:
241:
238:
212:
209:
164:
161:
147:
144:
143:
142:
138:
131:
119:
91:
88:
15:
9:
6:
4:
3:
2:
4789:
4778:
4775:
4773:
4770:
4769:
4767:
4753:
4749:
4742:
4734:
4730:
4724:
4716:
4712:
4708:
4704:
4700:
4696:
4692:
4688:
4681:
4673:
4669:
4665:
4661:
4653:
4642:September 25,
4637:
4633:
4629:
4622:
4614:
4610:
4605:
4600:
4596:
4592:
4587:
4582:
4578:
4574:
4570:
4566:
4562:
4555:
4547:
4543:
4536:
4528:
4524:
4519:
4514:
4511:(9): 1851–7.
4510:
4506:
4502:
4495:
4487:
4483:
4479:
4475:
4470:
4465:
4461:
4457:
4453:
4446:
4438:
4434:
4429:
4424:
4420:
4416:
4411:
4406:
4402:
4398:
4394:
4387:
4379:
4375:
4370:
4365:
4361:
4357:
4353:
4349:
4345:
4338:
4330:
4326:
4321:
4316:
4312:
4308:
4304:
4297:
4289:
4285:
4280:
4275:
4271:
4267:
4263:
4256:
4248:
4244:
4240:
4236:
4233:(5): 1111–8.
4232:
4228:
4221:
4213:
4209:
4204:
4199:
4194:
4189:
4185:
4181:
4177:
4173:
4169:
4161:
4153:
4149:
4144:
4139:
4135:
4131:
4127:
4123:
4119:
4111:
4103:
4099:
4095:
4091:
4087:
4083:
4079:
4075:
4067:
4059:
4055:
4051:
4047:
4043:
4039:
4035:
4031:
4027:
4020:
4012:
4008:
4004:
4000:
3996:
3992:
3984:
3976:
3972:
3967:
3962:
3958:
3954:
3950:
3943:
3935:
3931:
3927:
3923:
3919:
3915:
3912:(2): 355–64.
3911:
3907:
3899:
3891:
3887:
3882:
3877:
3872:
3867:
3863:
3859:
3855:
3848:
3840:
3836:
3832:
3828:
3821:
3813:
3809:
3804:
3799:
3794:
3789:
3785:
3781:
3777:
3770:
3762:
3758:
3753:
3748:
3743:
3738:
3734:
3730:
3726:
3719:
3711:
3707:
3702:
3697:
3692:
3687:
3683:
3679:
3675:
3668:
3660:
3656:
3651:
3646:
3642:
3638:
3634:
3630:
3626:
3619:
3611:
3607:
3602:
3597:
3593:
3589:
3585:
3581:
3577:
3570:
3562:
3558:
3553:
3548:
3544:
3540:
3536:
3529:
3521:
3517:
3513:
3509:
3505:
3501:
3497:
3493:
3485:
3477:
3473:
3468:
3463:
3458:
3453:
3449:
3445:
3441:
3434:
3426:
3422:
3418:
3414:
3410:
3406:
3402:
3398:
3391:
3383:
3377:
3369:
3365:
3360:
3355:
3351:
3347:
3343:
3339:
3336:(1): 016002.
3335:
3331:
3327:
3320:
3312:
3308:
3304:
3300:
3296:
3292:
3284:
3273:
3266:
3260:
3251:
3246:
3242:
3238:
3234:
3227:
3221:
3217:
3214:
3207:
3199:
3195:
3190:
3185:
3181:
3177:
3173:
3169:
3165:
3158:
3150:
3149:www.gilupi.de
3146:
3139:
3131:
3127:
3122:
3117:
3113:
3109:
3105:
3101:
3097:
3093:
3089:
3081:
3073:
3069:
3064:
3059:
3054:
3049:
3045:
3041:
3038:(9): e71105.
3037:
3033:
3029:
3021:
3013:
3009:
3004:
2999:
2995:
2991:
2987:
2983:
2979:
2975:
2971:
2964:
2956:
2952:
2947:
2942:
2938:
2934:
2930:
2926:
2922:
2918:
2914:
2910:
2906:
2899:
2891:
2887:
2882:
2877:
2873:
2869:
2865:
2861:
2857:
2853:
2849:
2845:
2841:
2834:
2826:
2822:
2817:
2812:
2808:
2804:
2800:
2796:
2792:
2788:
2784:
2777:
2769:
2765:
2760:
2755:
2751:
2747:
2743:
2739:
2735:
2731:
2727:
2720:
2712:
2708:
2704:
2700:
2696:
2692:
2688:
2684:
2683:Lab on a Chip
2677:
2669:
2665:
2660:
2655:
2651:
2647:
2643:
2639:
2638:Lab on a Chip
2635:
2628:
2620:
2616:
2611:
2606:
2602:
2598:
2594:
2590:
2586:
2582:
2578:
2570:
2562:
2558:
2553:
2548:
2544:
2540:
2536:
2532:
2528:
2521:
2513:
2509:
2504:
2499:
2495:
2491:
2487:
2480:
2472:
2468:
2463:
2458:
2454:
2450:
2446:
2439:
2431:
2427:
2422:
2417:
2413:
2409:
2405:
2398:
2390:
2386:
2381:
2376:
2372:
2368:
2364:
2357:
2349:
2345:
2341:
2337:
2332:
2327:
2324:(8): 781–91.
2323:
2319:
2315:
2308:
2293:
2292:
2284:
2278:
2264:on 2008-06-05
2263:
2259:
2253:
2245:
2241:
2237:
2233:
2229:
2225:
2218:
2210:
2206:
2202:
2198:
2193:
2188:
2184:
2180:
2176:
2169:
2161:
2157:
2153:
2149:
2145:
2141:
2137:
2133:
2125:
2123:
2121:
2112:
2108:
2103:
2098:
2094:
2090:
2086:
2082:
2081:Transl. Oncol
2078:
2071:
2063:
2059:
2055:
2051:
2047:
2043:
2036:
2028:
2024:
2020:
2016:
2012:
2008:
2001:
1993:
1989:
1984:
1979:
1974:
1969:
1965:
1961:
1957:
1950:
1942:
1938:
1934:
1930:
1926:
1922:
1919:(1–2): 1–13.
1918:
1914:
1907:
1899:
1895:
1890:
1885:
1881:
1877:
1873:
1866:
1858:
1854:
1849:
1844:
1840:
1836:
1832:
1828:
1824:
1817:
1809:
1805:
1800:
1795:
1790:
1785:
1781:
1777:
1773:
1766:
1758:
1754:
1749:
1744:
1739:
1734:
1730:
1726:
1722:
1715:
1713:
1704:
1700:
1695:
1690:
1686:
1682:
1678:
1671:
1663:
1659:
1655:
1651:
1644:
1636:
1632:
1627:
1622:
1617:
1612:
1608:
1604:
1600:
1596:
1592:
1585:
1577:
1573:
1569:
1565:
1561:
1557:
1552:
1547:
1543:
1539:
1535:
1531:
1527:
1519:
1511:
1507:
1503:
1499:
1491:
1483:
1479:
1475:
1471:
1467:
1463:
1459:
1455:
1447:
1439:
1435:
1430:
1425:
1421:
1417:
1413:
1409:
1405:
1397:
1389:
1385:
1381:
1377:
1369:
1361:
1357:
1352:
1347:
1343:
1339:
1335:
1331:
1328:(1): 016003.
1327:
1323:
1319:
1311:
1309:
1307:
1298:
1294:
1289:
1284:
1279:
1274:
1270:
1266:
1262:
1258:
1254:
1247:
1239:
1235:
1230:
1225:
1221:
1217:
1213:
1209:
1205:
1198:
1190:
1186:
1181:
1176:
1172:
1168:
1164:
1160:
1157:(1): 016004.
1156:
1152:
1148:
1141:
1139:
1130:
1126:
1122:
1118:
1114:
1110:
1103:
1095:
1091:
1086:
1081:
1077:
1073:
1069:
1065:
1061:
1054:
1046:
1042:
1038:
1034:
1030:
1026:
1022:
1018:
1011:
1003:
999:
995:
991:
987:
983:
980:(6): 339–51.
979:
975:
968:
960:
956:
951:
946:
942:
938:
934:
927:
919:
915:
911:
907:
903:
899:
892:
884:
880:
875:
870:
866:
862:
858:
854:
850:
846:
842:
834:
826:
822:
818:
814:
810:
806:
802:
795:
793:
784:
780:
775:
770:
767:(7): 2912–8.
766:
762:
758:
750:
748:
739:
735:
730:
725:
721:
717:
713:
709:
705:
698:
690:
686:
681:
676:
672:
668:
664:
657:
649:
645:
640:
635:
631:
627:
623:
619:
615:
608:
600:
596:
592:
588:
583:
578:
575:(4): 679–95.
574:
570:
566:
559:
551:
547:
543:
539:
535:
531:
524:
520:
512:
510:
504:
502:
496:
493:
482:
473:
470:
465:
463:
460:status using
459:
455:
451:
447:
443:
438:
434:
424:
422:
418:
414:
410:
406:
395:
393:
389:
379:
376:
370:
367:
363:
353:
350:
339:
330:
328:
324:
320:
316:
313:
309:
304:
299:
295:
294:nanoparticles
285:
283:
279:
275:
271:
267:
263:
259:
255:
251:
247:
237:
233:
231:
227:
217:
208:
206:
202:
196:
194:
190:
186:
181:
177:
169:
160:
156:
152:
139:
136:
132:
128:
124:
120:
117:
113:
109:
105:
104:
103:
101:
97:
87:
85:
81:
80:liquid biopsy
76:
74:
70:
66:
62:
58:
54:
50:
46:
42:
38:
34:
30:
21:
4751:
4741:
4732:
4723:
4693:(6): 453–8.
4690:
4686:
4680:
4663:
4659:
4652:
4640:. Retrieved
4636:the original
4631:
4621:
4568:
4564:
4554:
4545:
4541:
4535:
4508:
4504:
4494:
4459:
4455:
4445:
4400:
4396:
4386:
4351:
4347:
4337:
4310:
4306:
4296:
4269:
4265:
4255:
4230:
4226:
4220:
4175:
4171:
4160:
4128:(179): 179.
4125:
4121:
4110:
4077:
4073:
4066:
4033:
4029:
4019:
3994:
3990:
3983:
3956:
3952:
3942:
3909:
3905:
3898:
3861:
3857:
3847:
3830:
3826:
3820:
3783:
3779:
3769:
3732:
3728:
3718:
3681:
3677:
3667:
3632:
3628:
3618:
3583:
3579:
3569:
3542:
3538:
3528:
3495:
3491:
3484:
3447:
3443:
3433:
3400:
3396:
3390:
3376:cite journal
3333:
3329:
3319:
3294:
3290:
3283:
3272:the original
3259:
3240:
3236:
3226:
3206:
3171:
3167:
3157:
3148:
3138:
3095:
3091:
3080:
3035:
3031:
3020:
2977:
2973:
2963:
2912:
2908:
2898:
2847:
2843:
2833:
2790:
2786:
2776:
2733:
2729:
2719:
2686:
2682:
2676:
2641:
2637:
2627:
2584:
2580:
2569:
2534:
2530:
2520:
2496:(3): 920–8.
2493:
2489:
2479:
2452:
2448:
2438:
2411:
2407:
2397:
2370:
2366:
2356:
2321:
2317:
2307:
2296:. Retrieved
2294:. March 2010
2289:
2277:
2266:. Retrieved
2262:the original
2252:
2227:
2223:
2217:
2185:(1): 69–79.
2182:
2178:
2168:
2135:
2131:
2084:
2080:
2070:
2048:(1): 34–39.
2045:
2041:
2035:
2013:(1): 165–8.
2010:
2006:
2000:
1963:
1959:
1949:
1916:
1912:
1906:
1879:
1875:
1865:
1830:
1826:
1816:
1779:
1776:BMC Medicine
1775:
1765:
1728:
1724:
1684:
1680:
1670:
1653:
1649:
1643:
1598:
1594:
1584:
1551:10754/615874
1533:
1529:
1518:
1501:
1497:
1490:
1457:
1453:
1446:
1411:
1407:
1396:
1379:
1375:
1368:
1325:
1321:
1260:
1256:
1246:
1211:
1207:
1197:
1154:
1150:
1112:
1108:
1102:
1067:
1063:
1053:
1023:(4): 190–1.
1020:
1016:
1010:
977:
973:
967:
940:
936:
926:
901:
898:Eur J Cancer
897:
891:
848:
844:
833:
808:
804:
764:
760:
711:
707:
697:
670:
666:
656:
621:
617:
607:
572:
568:
558:
533:
529:
523:
505:
497:
488:
479:
468:
466:
430:
401:
385:
371:
365:
359:
345:
336:
312:cytokeratins
291:
243:
234:
223:
197:
189:cytokeratins
182:
178:
174:
157:
153:
149:
146:CTC clusters
112:cytokeratins
93:
77:
56:
32:
28:
26:
4227:Int J Oncol
4030:Pathol. Int
3098:(1): 3279.
2224:Cancer Lett
501:ferrofluids
405:blood cells
327:sensitivity
201:blood smear
55:and become
53:extravasate
51:. CTCs can
37:vasculature
4766:Categories
3678:ISRN Oncol
3539:Ann. Oncol
3492:The Breast
2298:2010-03-14
2268:2010-03-14
2130:Relapse".
761:Cancer Res
515:References
492:morphology
96:carcinomas
69:colorectal
61:metastases
41:lymphatics
4595:0027-8424
4419:2159-8274
4266:Clin Chem
4102:205393324
4058:205478411
3735:(1): 18.
3512:0960-9776
2994:0090-6964
2937:2045-2322
2915:: 16047.
2872:2045-2322
2807:0003-2700
2750:1387-2176
2711:205863853
2201:1751-5521
1560:1946-6234
509:carcinoma
433:Herceptin
413:ISET Test
163:Frequency
135:apoptosis
4777:Oncology
4707:12778135
4672:12114406
4548:: 146–7.
4527:20147742
4478:17200956
4437:31601552
4378:19745765
4329:17575225
4288:14988224
4247:12370762
4212:15194824
4152:23552373
4094:24880617
4050:20846264
4011:10458358
3975:10870062
3934:25370612
3926:18064568
3890:30107082
3839:21378321
3812:17002789
3761:15801980
3710:22530147
3659:21739182
3610:20694797
3561:17761704
3520:17291754
3476:16280045
3425:34030968
3417:25843106
3368:24406475
3311:26680058
3216:Archived
3198:22825490
3143:GILUPI.
3130:30115931
3072:24019862
3032:PLOS ONE
3012:26289942
2955:26538094
2890:25735563
2850:: 8745.
2825:22510236
2768:23109037
2703:24292816
2668:21863182
2619:18097410
2561:21300848
2512:17289886
2471:15501967
2430:18829513
2389:17085652
2340:15317891
2244:17314005
2209:30216684
2160:20074388
2152:18323545
2111:24151533
2062:23036177
2027:21051248
1992:21577258
1966:: 1–10.
1941:11881681
1933:20143276
1898:19887487
1857:21680546
1808:21510857
1757:20016752
1703:22016824
1662:10473071
1635:27091969
1576:26085239
1568:27358499
1482:16377917
1474:25135616
1438:25171411
1360:22306768
1238:27779656
1189:22306961
1129:23999357
1094:19081770
1037:19333222
994:19399023
959:18559576
918:17977713
883:22763454
825:18591556
783:19339269
738:19722757
689:23884225
648:24832787
591:17110329
550:24566031
469:in vitro
444:, Her2,
392:epitopes
366:in vitro
356:Maintrac
266:vaccinia
75:cancer.
73:prostate
4748:"OASIS"
4715:9195161
4613:9539782
4573:Bibcode
4486:6648226
4428:6954305
4369:2941876
4180:Bibcode
4143:3760275
3991:J. Urol
3881:6586069
3803:1599731
3752:1087511
3701:3317055
3684:: 1–8.
3650:3155034
3601:3074080
3467:1410761
3359:4143170
3338:Bibcode
3243:: i14.
3189:3583719
3121:6095877
3100:Bibcode
3063:3760980
3040:Bibcode
3003:4761332
2946:4633592
2917:Bibcode
2881:4348664
2852:Bibcode
2816:3359653
2759:3584207
2659:3379551
2610:3090667
2589:Bibcode
2552:3101098
2348:7445998
2102:3799195
1983:3090615
1848:3743247
1799:3107794
1748:2793426
1731:: 1–8.
1725:J Oncol
1694:3195935
1626:4983862
1603:Bibcode
1429:4149753
1351:3387996
1330:Bibcode
1297:9539782
1265:Bibcode
1229:5117994
1180:3388002
1159:Bibcode
1085:2600498
1064:Mol Med
1045:8904131
874:3408856
853:Bibcode
729:4422331
680:5060940
639:4112925
599:7362869
485:History
274:VAR2CSA
100:anoikis
4713:
4705:
4670:
4611:
4601:
4593:
4525:
4484:
4476:
4435:
4425:
4417:
4376:
4366:
4327:
4286:
4245:
4210:
4203:438987
4200:
4150:
4140:
4100:
4092:
4056:
4048:
4009:
3973:
3953:Cancer
3932:
3924:
3888:
3878:
3837:
3810:
3800:
3786:: 67.
3759:
3749:
3708:
3698:
3657:
3647:
3608:
3598:
3559:
3518:
3510:
3474:
3464:
3423:
3415:
3366:
3356:
3309:
3196:
3186:
3128:
3118:
3070:
3060:
3010:
3000:
2992:
2953:
2943:
2935:
2888:
2878:
2870:
2823:
2813:
2805:
2766:
2756:
2748:
2709:
2701:
2666:
2656:
2617:
2607:
2581:Nature
2559:
2549:
2510:
2469:
2428:
2387:
2346:
2338:
2242:
2207:
2199:
2158:
2150:
2109:
2099:
2060:
2025:
2007:Vet. J
1990:
1980:
1939:
1931:
1896:
1855:
1845:
1806:
1796:
1782:: 43.
1755:
1745:
1701:
1691:
1660:
1633:
1623:
1574:
1566:
1558:
1480:
1472:
1436:
1426:
1358:
1348:
1295:
1285:
1236:
1226:
1187:
1177:
1127:
1092:
1082:
1043:
1035:
1002:890927
1000:
992:
957:
916:
881:
871:
845:Nature
823:
781:
736:
726:
687:
677:
646:
636:
597:
589:
548:
495:leg".
442:IGF-1R
409:Ficoll
298:biotin
220:colon)
130:cells.
65:breast
4711:S2CID
4604:22534
4482:S2CID
4098:S2CID
4054:S2CID
3930:S2CID
3421:S2CID
3275:(PDF)
3268:(PDF)
2707:S2CID
2344:S2CID
2286:(PDF)
2156:S2CID
1937:S2CID
1572:S2CID
1478:S2CID
1288:22534
1041:S2CID
998:S2CID
624:(5).
595:S2CID
446:Bcl-2
250:EpCAM
185:EpCAM
108:EpCAM
90:Types
57:seeds
45:tumor
4703:PMID
4668:PMID
4644:2017
4609:PMID
4591:ISSN
4523:PMID
4474:PMID
4433:PMID
4415:ISSN
4374:PMID
4325:PMID
4284:PMID
4243:PMID
4208:PMID
4148:PMID
4090:PMID
4046:PMID
4007:PMID
3971:PMID
3922:PMID
3886:PMID
3835:PMID
3808:PMID
3757:PMID
3706:PMID
3682:2012
3655:PMID
3606:PMID
3557:PMID
3516:PMID
3508:ISSN
3472:PMID
3413:PMID
3382:link
3364:PMID
3307:PMID
3194:PMID
3126:PMID
3068:PMID
3008:PMID
2990:ISSN
2951:PMID
2933:ISSN
2886:PMID
2868:ISSN
2821:PMID
2803:ISSN
2764:PMID
2746:ISSN
2699:PMID
2664:PMID
2615:PMID
2557:PMID
2508:PMID
2467:PMID
2426:PMID
2385:PMID
2336:PMID
2318:NEJM
2240:PMID
2205:PMID
2197:ISSN
2148:PMID
2107:PMID
2058:PMID
2023:PMID
1988:PMID
1964:2011
1929:PMID
1894:PMID
1853:PMID
1804:PMID
1753:PMID
1729:2010
1699:PMID
1658:PMID
1631:PMID
1564:PMID
1556:ISSN
1470:PMID
1434:PMID
1408:Cell
1356:PMID
1293:PMID
1234:PMID
1185:PMID
1125:PMID
1090:PMID
1033:PMID
990:PMID
955:PMID
914:PMID
879:PMID
821:PMID
779:PMID
734:PMID
685:PMID
644:PMID
587:PMID
569:Cell
546:PMID
454:PTEN
437:Her2
388:size
349:FISH
321:and
308:CD45
303:EDTA
262:MACS
256:and
254:HER2
116:size
110:and
71:and
4695:doi
4599:PMC
4581:doi
4513:doi
4464:doi
4460:71A
4423:PMC
4405:doi
4364:PMC
4356:doi
4315:doi
4274:doi
4235:doi
4198:PMC
4188:doi
4176:101
4138:PMC
4130:doi
4082:doi
4038:doi
3999:doi
3995:162
3961:doi
3914:doi
3910:111
3876:PMC
3866:doi
3798:PMC
3788:doi
3747:PMC
3737:doi
3696:PMC
3686:doi
3645:PMC
3637:doi
3633:137
3596:PMC
3588:doi
3584:137
3547:doi
3500:doi
3462:PMC
3452:doi
3405:doi
3354:PMC
3346:doi
3299:doi
3245:doi
3184:PMC
3176:doi
3116:PMC
3108:doi
3058:PMC
3048:doi
2998:PMC
2982:doi
2941:PMC
2925:doi
2876:PMC
2860:doi
2811:PMC
2795:doi
2754:PMC
2738:doi
2691:doi
2654:PMC
2646:doi
2605:PMC
2597:doi
2585:450
2547:PMC
2539:doi
2535:192
2498:doi
2457:doi
2416:doi
2375:doi
2326:doi
2322:351
2232:doi
2228:253
2187:doi
2140:doi
2097:PMC
2089:doi
2050:doi
2046:196
2015:doi
2011:190
1978:PMC
1968:doi
1921:doi
1884:doi
1843:PMC
1835:doi
1794:PMC
1784:doi
1743:PMC
1733:doi
1689:PMC
1621:PMC
1611:doi
1599:113
1546:hdl
1538:doi
1506:doi
1462:doi
1424:PMC
1416:doi
1412:158
1384:doi
1346:PMC
1338:doi
1283:PMC
1273:doi
1224:PMC
1216:doi
1175:PMC
1167:doi
1117:doi
1080:PMC
1072:doi
1025:doi
982:doi
945:doi
906:doi
869:PMC
861:doi
849:487
813:doi
769:doi
724:PMC
716:doi
712:133
675:PMC
634:PMC
626:doi
622:106
577:doi
573:127
538:doi
450:ERG
258:PSA
193:EMT
39:or
33:CTC
4768::
4750:.
4731:.
4709:.
4701:.
4689:.
4662:.
4630:.
4607:.
4597:.
4589:.
4579:.
4569:95
4567:.
4563:.
4546:14
4544:.
4521:.
4509:21
4507:.
4503:.
4480:.
4472:.
4458:.
4454:.
4431:.
4421:.
4413:.
4401:10
4399:.
4395:.
4372:.
4362:.
4350:.
4346:.
4323:.
4311:13
4309:.
4305:.
4282:.
4270:50
4268:.
4264:.
4241:.
4231:21
4229:.
4206:.
4196:.
4186:.
4174:.
4170:.
4146:.
4136:.
4124:.
4120:.
4096:.
4088:.
4078:20
4076:.
4052:.
4044:.
4034:60
4032:.
4028:.
4005:.
3993:.
3969:.
3957:88
3955:.
3951:.
3928:.
3920:.
3908:.
3884:.
3874:.
3862:93
3860:.
3856:.
3831:31
3829:.
3806:.
3796:.
3782:.
3778:.
3755:.
3745:.
3731:.
3727:.
3704:.
3694:.
3680:.
3676:.
3653:.
3643:.
3631:.
3627:.
3604:.
3594:.
3582:.
3578:.
3555:.
3543:18
3541:.
3537:.
3514:.
3506:.
3496:16
3494:.
3470:.
3460:.
3446:.
3442:.
3419:.
3411:.
3401:15
3399:.
3378:}}
3374:{{
3362:.
3352:.
3344:.
3334:11
3332:.
3328:.
3305:.
3295:47
3293:.
3241:26
3239:.
3235:.
3192:.
3182:.
3172:41
3170:.
3166:.
3147:.
3124:.
3114:.
3106:.
3094:.
3090:.
3066:.
3056:.
3046:.
3034:.
3030:.
3006:.
2996:.
2988:.
2978:44
2976:.
2972:.
2949:.
2939:.
2931:.
2923:.
2911:.
2907:.
2884:.
2874:.
2866:.
2858:.
2846:.
2842:.
2819:.
2809:.
2801:.
2791:84
2789:.
2785:.
2762:.
2752:.
2744:.
2734:15
2732:.
2728:.
2705:.
2697:.
2687:14
2685:.
2662:.
2652:.
2642:11
2640:.
2636:.
2613:.
2603:.
2595:.
2583:.
2579:.
2555:.
2545:.
2533:.
2529:.
2506:.
2494:13
2492:.
2488:.
2465:.
2453:10
2451:.
2447:.
2424:.
2412:14
2410:.
2406:.
2383:.
2371:12
2369:.
2365:.
2342:.
2334:.
2320:.
2316:.
2288:.
2238:.
2226:.
2203:.
2195:.
2183:41
2181:.
2177:.
2154:.
2146:.
2136:26
2134:.
2119:^
2105:.
2095:.
2083:.
2079:.
2056:.
2044:.
2021:.
2009:.
1986:.
1976:.
1962:.
1958:.
1935:.
1927:.
1917:11
1915:.
1892:.
1880:15
1878:.
1874:.
1851:.
1841:.
1831:17
1829:.
1825:.
1802:.
1792:.
1778:.
1774:.
1751:.
1741:.
1727:.
1723:.
1711:^
1697:.
1683:.
1679:.
1652:.
1629:.
1619:.
1609:.
1597:.
1593:.
1570:.
1562:.
1554:.
1544:.
1532:.
1528:.
1502:76
1500:.
1476:.
1468:.
1458:25
1456:.
1432:.
1422:.
1410:.
1406:.
1380:32
1378:.
1354:.
1344:.
1336:.
1324:.
1320:.
1305:^
1291:.
1281:.
1271:.
1261:95
1259:.
1255:.
1232:.
1222:.
1212:49
1210:.
1206:.
1183:.
1173:.
1165:.
1153:.
1149:.
1137:^
1123:.
1113:31
1111:.
1088:.
1078:.
1068:15
1066:.
1062:.
1039:.
1031:.
1019:.
996:.
988:.
976:.
953:.
941:14
939:.
935:.
912:.
902:43
900:.
877:.
867:.
859:.
847:.
843:.
819:.
809:26
807:.
803:.
791:^
777:.
765:69
763:.
759:.
746:^
732:.
722:.
710:.
706:.
683:.
671:43
669:.
665:.
642:.
632:.
620:.
616:.
593:.
585:.
571:.
567:.
544:.
534:70
532:.
458:AR
456:,
452:,
448:,
329:.
323:19
319:18
317:,
252:,
86:.
67:,
27:A
4754:.
4735:.
4717:.
4697::
4691:3
4674:.
4664:8
4646:.
4615:.
4583::
4575::
4529:.
4515::
4488:.
4466::
4439:.
4407::
4380:.
4358::
4352:4
4331:.
4317::
4290:.
4276::
4249:.
4237::
4214:.
4190::
4182::
4154:.
4132::
4126:5
4104:.
4084::
4060:.
4040::
4013:.
4001::
3977:.
3963::
3936:.
3916::
3892:.
3868::
3841:.
3814:.
3790::
3784:4
3763:.
3739::
3733:3
3712:.
3688::
3661:.
3639::
3612:.
3590::
3563:.
3549::
3522:.
3502::
3478:.
3454::
3448:7
3427:.
3407::
3384:)
3370:.
3348::
3340::
3313:.
3301::
3253:.
3247::
3200:.
3178::
3151:.
3132:.
3110::
3102::
3096:9
3074:.
3050::
3042::
3036:8
3014:.
2984::
2957:.
2927::
2919::
2913:5
2892:.
2862::
2854::
2848:5
2827:.
2797::
2770:.
2740::
2713:.
2693::
2670:.
2648::
2621:.
2599::
2591::
2563:.
2541::
2514:.
2500::
2473:.
2459::
2432:.
2418::
2391:.
2377::
2350:.
2328::
2301:.
2271:.
2246:.
2234::
2211:.
2189::
2162:.
2142::
2113:.
2091::
2085:6
2064:.
2052::
2029:.
2017::
1994:.
1970::
1943:.
1923::
1900:.
1886::
1859:.
1837::
1810:.
1786::
1780:9
1759:.
1735::
1705:.
1685:1
1664:.
1654:5
1637:.
1613::
1605::
1578:.
1548::
1540::
1534:8
1512:.
1508::
1484:.
1464::
1440:.
1418::
1390:.
1386::
1362:.
1340::
1332::
1326:9
1299:.
1275::
1267::
1240:.
1218::
1191:.
1169::
1161::
1155:9
1131:.
1119::
1096:.
1074::
1047:.
1027::
1021:6
1004:.
984::
978:6
961:.
947::
920:.
908::
885:.
863::
855::
827:.
815::
785:.
771::
740:.
718::
691:.
650:.
628::
601:.
579::
552:.
540::
315:8
31:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.