318:(BEAMing) is a technique that builds upon Droplet Digital PCR in order to identify mutations in ctDNA using flow cytometry. After ctDNA is extracted from blood, PCR is performed with primers designed to target the regions of interest. These primers also contain specific DNA sequences, or tags. The amplified DNA is mixed with streptavidin-coated magnetic beads and emulsified into droplets. Biotinylated primers designed to bind to the tags are used to amplify the DNA. Biotinylation allows the amplified DNA to bind to the magnetic beads, which are coated with streptavidin. After the PCR is complete, the DNA-bound beads are separated using a magnet. The DNA on the beads are then denatured and allowed to hybridize with fluorescent oligonucleotides specific to each DNA template. The resulting bead-DNA complexes are then analyzed using flow cytometry. This technique is able to capture allele and mutation frequencies due to coupling with ddPCR. However, unlike with ddPCR, a larger number of DNA sequences can be interrogated due to the flexibility of using fluorescently bound probes. Another advantage of this system is that the DNA isolated can also be used for downstream sequencing. Sensitivity is 1.6 in 10 to 4.3 in 10.
530:
separation of cells within the tumor. For example, since a biopsy only samples a small part of the tumor, clones that resides in a different location may go unnoticed. This can mislead research that focuses on studying the role of tumor heterogeneity in cancer progression and relapse. The use of ctDNA in research can alleviate these concerns because it could provide a more representative 'screenshot' of the genetic diversity of cancer at both primary and metastatic sites. For example, ctDNA has been shown to be useful in studying the clonal evolution of a patient's cancer before and after treatment regimens. Early detection of cancer is still challenging but recent progress in the analysis of the epigenetic features of cfDNA, or the fragmentation pattern unlock improve the sensitivity of liquid biopsy. Furthermore, ctDNA analysis is an emerging tool for understanding the clonal composition of metastatic tumors, detecting different mutations on a genomic scale, studying subclonal diversity that affects the prognosis of the disease as different resistant phenotypes can be found and the appearance of new mechanisms of genomic and transcriptomic resistance to treatment.
307:. Droplet Digital PCR utilizes a droplet generator to partition single pieces of DNA into droplets using an oil/water emulsion. Then individual polymerase chain reactions occur in each droplet using selected primers against regions of ctDNA and proceeds to endpoint. The presence of the sequences of interest is measured by fluorescent probes, which bind to the amplified region. ddPCR allows for highly quantitative assessment of allele and mutant frequencies in ctDNA but is limited by the number of fluorescent probes that can be used in one assay (up to 5). The sensitivity of the assay can vary depending on the amount of DNA analyzed and is around 1 in 10,000. Specificity should be augmented through the use of either minor groove binding (MGB) modified probes or of an alternative such as locked nucleic acids (LNAs).
362:. Safe-Seq decreases the error rate of massively parallel sequencing in order to increase the sensitivity to rare mutants. It achieves this by addition of a unique identifier (UID) sequence to each DNA template. The DNA is then amplified using the added UIDs and sequenced. All DNA molecules with the same UID (a UID family) should have the same reported DNA sequence since they were amplified from one molecule. However, mutations can be introduced through amplification, or incorrect base assignments may be called in the sequencing and analysis steps. The presence of the UID allows these methodology errors to be separated from true mutations of the ctDNA. A mutation is considered a ‘supermutant’ if 95% of the sequenced reads are in agreement. The sensitivity of this approach is 9 in 1 million.
504:(MRD), and thus the possibility of tumor recurrence, in cases where bulk tumors are absent by conventional imaging methods. A comparison of MRD detection by CT imaging compared to ctDNA has been previously done in individuals with stage II colon cancer; in this study, researchers were able to detect ctDNA in individuals who showed no sign of clinical malignancy by a CT scan, suggesting that ctDNA detection has greater sensitivity to assess MRD. However, the authors acknowledge that ctDNA analysis is not without limitations; plasma samples collected post-operatively were only able to predict recurrence at 36 months in 48% of cases. Subsequently, ctDNA assays have been developed for both
337:. This technique uses biotinylated oligonucleotide selector probes to target sequences of DNA relevant to ctDNA detection. Publicly available cancer databases were used to construct a library of probes against recurrent mutations in cancer by calculating their recurrence index. The protocol was optimized for the low DNA levels observed in ctDNA collection. Then the isolated DNA undergoes deep sequencing for increased sensitivity. This technique allows for the interrogation of hundreds of DNA regions. The ctDNA detection sensitivity of CAPP-Seq is reported to be 2.5 molecules in 1,000,000.
374:
strand with an α tag on the 5’ end and a β tag on the 3’ end and the other strand with a β tag on the 5’ end and an α tag on the 3’ end. These DNA fragments are then amplified with primers against the invariant sequences of the tags. The amplified DNA is sequenced and analyzed. DNA with the duplex adaptors are compared and mutations are only accepted if there is a consensus between both strands. This method takes into account both errors from sequencing and errors from early stage PCR amplification. The sensitivity of the approach to discovering mutants is 1 in 10^7.
539:
validation must be established before ctDNA analysis can become a routine clinical assay. Furthermore, creation of a panel of ‘standard’ tumor-associated biomarkers may be necessary given the resolution of current ctDNA sequencing and detection methods. Sequencing tumor-specific aberrations from plasma samples may also help exclude contaminating cfDNA from analysis; elevated levels of cfDNA from normal cells may be attributed to non-cancer related causes. These sequencing techniques can also determine the clonal evolution of cancer,
287:. This is especially important when analyzing ctDNA not only because there are relatively low levels of DNA circulating in the bloodstream, but also because ctDNA makes up a small proportion of the total cell-free DNA extracted. Therefore, amplification of regions of interest can drastically improve sensitivity of ctDNA detection. However, amplification through PCR can introduce errors given the inherent error rate of DNA polymerases. Errors introduced during sequencing can also decrease the sensitivity of detecting ctDNA mutations.
272:
analysis of ctDNA to each patient is also possible by combining liquid biopsies with standard primary tissue biopsies. Whole genome or whole exome sequencing of the primary tumor biopsy allows for discovery of genetic mutations specific to a patient's tumor, and can be used for subsequent targeted sequencing of the patient's ctDNA. The highest sensitivity of ctDNA detection is accomplished through targeted sequencing of specific
193:. Limiting the sequencing to only the whole exome instead can decrease expense and increase speed, but at the cost of losing information about mutations in the non-coding regulatory regions of DNA. While simply looking at DNA polymorphisms through sequencing does not differentiate DNA from tumor or normal cells, this problem can be resolved by comparing against a control sample of normal DNA (for example, DNA obtained through a
263:. Bisulfite treatment chemically converts unmethylated cytosines into a uracil while leaving methylated cytosines unmodified. DNA is subsequently sequenced, and any alterations to the DNA methylation pattern can be identified. DNA hydroxymethylation is a similarly associated mark that has been shown to be a predictive marker of healthy versus diseased conditions in cfDNA, including cancer.)
186:
untargeted methods may be necessary in certain applications, it is more expensive and has lower resolution. This makes it difficult to detect rare mutations, or in situations where low ctDNA levels are present (such as minimal residual disease). Furthermore, there can be problems distinguishing between DNA from tumor cells and DNA from normal cells using a whole genome approach.
107:
infiltration to tumor sites, which reduces effective clearance of ctDNA from the bloodstream. Comparison of mutations in ctDNA and DNA extracted from primary tumors of the same patients revealed the presence of identical cancer-relevant genetic changes. This led to the possibility of using ctDNA for earlier cancer detection and treatment follow up.
276:(SNPs). Commonly mutated genes, such as oncogenes, which typically have hotspot mutations, are good candidates for targeted sequencing approaches. Conversely, most tumor suppressor genes have a wide array of possible loss of function mutations throughout the gene, and as such are not suitable for targeted sequencing.
429:
elementary to identify clinically relevant differences in the cancer phenotype and to see how therapy is affecting patients. Furthermore, the relative homogeneity in driver gene alterations among metastases justifies that genomic and functional alterations in prostate cancer are shared between ctDNA and tissue.
168:
this can decrease the sensitivity of ctDNA detection. Therefore, the majority of studies use plasma for ctDNA isolation. Plasma is then processed again by centrifugation to remove residual intact blood cells. The supernatant is used for DNA extraction, which can be performed using commercially available kits.
20:
237:
After the whole genome is sequenced using a high throughput sequencing method, such as
Illumina HiSeq, personalized analysis of rearranged ends (PARE) is applied to the data to analyze chromosomal rearrangements and translocations. This technique was originally designed to analyze solid tumor DNA but
345:
Tagged amplicon deep sequencing (TAM-Seq) allows targeted sequencing of entire genes to detect mutations in ctDNA. First a general amplification step is performed using primers that span the entire gene of interest in 150-200bp sections. Then, a microfluidics system is used to attached adaptors with
200:
Whole genome sequencing enables to recover the structural properties of cfDNA, the size of fragments and their fragmentation patterns. These unique patterns can be an important source of information to improve the detection of ctDNA or localize the tissue of origin of these fragments. Size-selection
176:
The analysis of ctDNA after extraction requires the use of various amplification and sequencing methods. These methods can be separated into two main groups based on whether the goal is to interrogate all genes in an untargeted approach, or if the goal is to monitor specific genes and mutations in a
373:
is an improvement on the single UIDs added in the Safe-Seq technique. In duplex sequencing, randomized double-stranded DNA act as unique tags and are attached to an invariant spacer. Tags are attached to both ends of a DNA fragment (α and β tags), which results in two unique templates for PCR - one
167:
fractions of blood can be separated through a centrifugation step. ctDNA or cfDNA can be subsequently extracted from these fractions. Although serum tends to have greater levels of cfDNA, this is primarily attributed to DNA from lymphocytes. High levels of contaminating cfDNA is sub-optimal because
432:
This makes ctDNA a powerful emerging tool for the detection of genetic mutations at the genomic scale in patients suffering from metastatic cancer to observe the clinical relevance of the clonal composition of these tumors to understand better cancer control. This subclonal reconstruction based on
428:
patterns (taking into account nucleosomes present in transcription start sites (TSSs) and AR-binding sites (ARBs). In this way, the genomic and transcriptomic evolution of ctDNA can be observed, performed in living patients who are developing resistance to treatment, therefore, ctDNA sequencing is
185:
A whole genome or whole exome sequencing approaches may be necessary to discover new mutations in tumor DNA while monitoring disease burden or tracking drug resistance. Untargeted approaches are also useful in research to observe tumor heterogeneity or to discover new drug targets. However, while
520:
The question of whether measurement of the amount or qualities of ctDNA could be used to determine outcomes in people with cancer has been a subject of study. As of 2015 this was very uncertain. Although some studies have shown a trend of higher ctDNA levels in people with high stage metastatic
271:
In a targeted approach, sequencing of ctDNA can be directed towards a genetic panel constructed based on mutational hotspots for the cancer of interest. This is especially important for informing treatment in situations where mutations are identified in druggable targets. Personalizing targeted
529:
The emergence of drug-resistant tumors due to intra- and inter-tumoral heterogeneity an issue in treatment efficacy. A minor genetic clone within the tumor can expand after treatment if it carries a drug-resistant mutation. Initial biopsies can miss these clones due to low frequency or spatial
120:
When blood is collected in EDTA tubes and stored, the white blood cells begin to lyse and release genomic wild type DNA in to the sample in quantities typically many fold higher than the ctDNA is present in. This makes detection of mutations or other ctDNA biomarkers more difficult. The use of
538:
Implementation of ctDNA in clinical practice is largely hindered by the lack of standardized methods for ctDNA processing and analysis. Standardization of methods for sample collection (including time of collection), downstream processing (DNA extraction and amplification), quantification and
106:
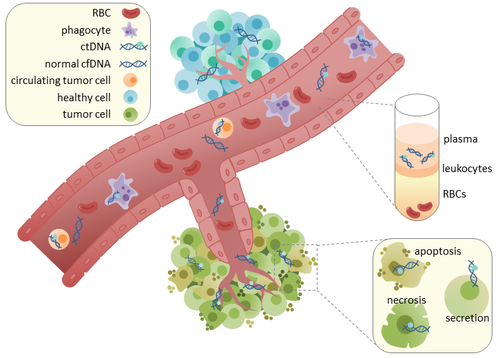
are responsible for clearance of apoptotic or necrotic cellular debris, which includes cfDNA. ctDNA in healthy patients is only present at low levels but higher levels of ctDNA in cancer patients can be detected with increasing tumor sizes. This possibly occurs due to inefficient immune cell
73:
Recent studies have laid the foundation for inferring gene expression from cfDNA (and ctDNA), with EPIC-seq emerging as a notable advancement. This method has substantially raised the bar for the noninvasive inference of expression levels of individual genes, thereby augmenting the assay's
382:
Integrated digital error suppression (iDES) improves CAPP-Seq analysis of ctDNA in order to decrease error and therefore increase sensitivity of detection. Reported in 2016, iDES combines CAPP-Seq with duplex barcoding sequencing technology and with a computational algorithm that removes
2345:
Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, Hadfield J, May AP, Caldas C, Brenton JD, Rosenfeld N (May 2012). "Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA".
346:
a unique identifier to each amplicon to further amplify the DNA in parallel singleplex reactions. This technique was shown to successfully identify mutations scattered in the TP53 tumor suppressor gene in advanced ovarian cancer patients. The sensitivity of this technique is 1 in 50.
383:
stereotypical errors associated with the CAPP-Seq hybridization step. The method also integrates duplex sequencing where possible, and includes methods for more efficient duplex recovery from cell free DNA. The sensitivity of this improved version of CAPP-Seq is 4 in 100,000 copies.
433:
ctDNA thanks to Whole-genome sequencing poses a unique set of challenges and opportunities for scientific research in oncology. Furthermore, serial ctDNA reveals treatment-driven selection for androgen receptor augmentation because it increases the dimensionality of the data.
2613:
Fitzgerald, Sandra; Blenkiron, Cherie; Stephens, Rosalie; Mathy, Jon A.; Somers-Edgar, Tiffany; Rolfe, Gill; Martin, Richard; Jackson, Christopher; Eccles, Michael; Robb, Tamsin; Rodger, Euan; Lawrence, Ben; Guilford, Parry; Lasham, Annette; Print, Cristin G. (2023).
2502:
Herberts, Cameron; Annala, Matti; Sipola, Joonatan; Ng, Sarah W. S.; Chen, Xinyi E.; Nurminen, Anssi; Korhonen, Olga V.; Munzur, Aslı D.; Beja, Kevin; Schönlau, Elena; Bernales, Cecily Q.; Ritch, Elie; Bacon, Jack V. W.; Lack, Nathan A.; Nykter, Matti (August 2022).
2771:
Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin SF, Tsui DW, Marass F, Gale D, Ali HR, Shah P, Contente-Cuomo T, Farahani H, Shumansky K, Kingsbury Z, Humphray S, Bentley D, Shah SP, Wallis M, Rosenfeld N, Caldas C (November 2015).
93:
from dying cells, or active release from viable tumor cells. Studies in both human (healthy and cancer patients) and xenografted mice show that the size of fragmented cfDNA is predominantly 166bp long, which corresponds to the length of DNA wrapped around a
1071:
Heitzer E, Auer M, Hoffmann EM, Pichler M, Gasch C, Ulz P, Lax S, Waldispuehl-Geigl J, Mauermann O, Mohan S, Pristauz G, Lackner C, Höfler G, Eisner F, Petru E, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl JB, Speicher MR (July 2013).
1662:
Vallée A, Marcq M, Bizieux A, Kouri CE, Lacroix H, Bennouna J, Douillard JY, Denis MG (November 2013). "Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients".
453:
One of the challenges in using ctDNA as a cancer biomarker is whether ctDNA can be distinguished with cfDNA from normal cells. cfDNA is released by non-malignant cells during normal cellular turnover, but also during procedures such as
197:.) Importantly, whole genome and whole exome sequencing are useful for initial mutation discovery. This provides information for the use of more sensitive targeted techniques, which can then be used for disease monitoring purposes.
229:. Copy number variations are common in cancers and describe situations where loss of heterozygosity of a gene may lead to decreased function due to lower expression, or duplication of a gene, which leads to overexpression.
479:
The clinical utility of ctDNA for the detection of primary disease is in part limited by the sensitivity of current technology to detect small tumors with low levels of ctDNA present and a priori unknown somatic mutations.
250:
marking is essential for normal gene expression and cell function and aberrant alterations in epigenetic patterns is a hallmark of cancer. A normal epigenetic status is maintained in a cell at least in part through
1399:
Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M (April 1994). "Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia".
142:
Perform a double centrifugation step (centrifuge the blood to extract plasma, then repeat on the plasma to remove from debris in the bottom of the tube) to remove more cellular debris prior to DNA extraction.
154:
The main appeal of ctDNA analysis is that it is extracted in a non-invasive manner through blood collection. Acquisition of cfDNA or ctDNA typically requires collection of approximately 3mL of blood into
125:
mutations in matched samples collected in both EDTA K3 and Streck BCT tubes. The advantages of cell stabilisation tubes can be realised in situation where blood cannot be processed to plasma immediately.
2566:
Zou, Donghui; Day, Robert; Cocadiz, Judy A; Parackal, Sarah; Mitchell, Wilson; Black, Michael A; Lawrence, Ben; Fitzgerald, Sandra; Print, Cristin; Jackson, Christopher; Guilford, Parry (2020-11-13).
819:
Akca H, Demiray A, Yaren A, Bir F, Koseler A, Iwakawa R, Bagci G, Yokota J (March 2013). "Utility of serum DNA and pyrosequencing for the detection of EGFR mutations in non-small cell lung cancer".
121:
commercially available cell stabilisation tubes can prevent or delay the lysis of white cells thereby reducing the dilution effect of the ctDNA. Sherwood et al. demonstrated superior detection of
521:
cancer, ctDNA burden does not always correlate with traditional cancer staging. As of 2013 it appeared unlikely that ctDNA would be of clinical utility as a sole predictor of prognosis.
2047:
Song CX, Yin S, Ma L, Wheeler A, Chen Y, Zhang Y, Liu B, Xiong J, Zhang W, Hu J, Zhou Z, Dong B, Tian Z, Jeffrey SS, Chua MS, So S, Li W, Wei Y, Diao J, Xie D, Quake SR (October 2017).
1698:
Lee TH, Montalvo L, Chrebtow V, Busch MP (February 2001). "Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma".
225:
where a dye is used to stain chromosomal bands in order to visualize the chromosomes, digital karyotyping uses DNA sequences of loci throughout the genome in order to calculate
102:, suggesting that apoptosis may be the primary method of ctDNA release. The fragmentation of cfDNA is altered in the plasma of cancer patients. In healthy tissue, infiltrating
62:), a broader term which describes DNA that is freely circulating in the bloodstream, but is not necessarily of tumor origin. Because ctDNA may reflect the entire tumor
946:
Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P (November 2001). "About the possible origin and mechanism of circulating DNA apoptosis and active DNA release".
1895:
Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O'Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, Diaz LA, Velculescu VE (November 2012).
1559:"A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR"
2244:
Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW, Alizadeh AA, Diehn M (May 2014).
1605:"Optimised Pre-Analytical Methods Improve KRAS Mutation Detection in Circulating Tumour DNA (ctDNA) from Patients with Non-Small Cell Lung Cancer (NSCLC)"
2201:
Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D (July 2006). "BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions".
330:
440:
burden, influence circulating tumor DNA and the choice of new techniques to select other lesions that reflect clinically dominant disease.
201:
of short fragments (<150bp) with in vitro or in silico methods could improve the recovery of mutations and copy number aberrations.
85:
system. The precise mechanism of ctDNA release is unclear. The biological processes postulated to be involved in ctDNA release include
43:, and active secretion from tumor cells have been hypothesized. Once ctDNA is isolated, it can be sequenced for mutational analysis.
70:" in the form of blood draws may be taken at various time points to monitor tumor progression throughout the treatment regimen.
2448:
Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, Prindle MJ, Kuong KJ, Shen JC, Risques RA, Loeb LA (November 2014).
406:
and control patients who did not present this DNA, including somatic mutations and structural rearrangements in their genomes.
255:. Measuring aberrant methylation patterns in ctDNA is possible due to stable methylation of regions of DNA referred to as “
3801:
2144:"Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations"
2975:
2568:"Circulating tumor DNA is a sensitive marker for routine monitoring of treatment response in advanced colorectal cancer"
129:
Other procedures can also reduce the amount of "contaminating" wild type DNA and make detection of ctDNA more feasible:
3286:
1284:
Pisetsky DS, Fairhurst AM (June 2007). "The origin of extracellular DNA during the clearance of dead and dying cells".
296:
284:
3399:
3254:
2107:
Butler TM, Spellman PT, Gray J (February 2017). "Circulating-tumor DNA as an early detection and diagnostic tool".
1332:
Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, Alizadeh AA, Diehn M, Reiter JG (December 2020).
1851:"Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA"
1445:
K-ras point mutations in the blood plasma DNA of patients with colorectal tumors in
Challenges of Modern Medicine
577:
Wan J, Massie C, Garcia-Corbacho J, Mouliere F, Brenton J, Caldas C, Pacey S, Baird R, Rosenfeld N (April 2017).
2895:
273:
2774:"Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer"
982:
81:(CTCs), which describes viable, intact tumor cells that shed from primary tumors and enter the bloodstream or
3786:
3714:
2838:
552:
2000:"5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers"
854:
Schwarzenbach H, Hoon DS, Pantel K (June 2011). "Cell-free nucleic acids as biomarkers in cancer patients".
493:
74:
applicability in disease characterization, histological classification, and monitoring treatment efficacy.
1897:"Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing"
99:
3745:
735:
Mutter, Jurik A; Shahrokh
Esfahani, Mohammad; Schroers-Martin, Joseph; et al. (28 November 2023).
497:
190:
58:
in the bloodstream that is not associated with cells. ctDNA should not be confused with cell-free DNA (
2049:"5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages"
3539:
1232:
Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. (November 2018).
280:
1971:
Beaumont G, Dobbins S, Latta D, McMillin WP (May 1990). "Mequitazine in the treatment of hayfever".
1170:
Mouliere F, Robert B, Arnau
Peyrotte E, Del Rio M, Ychou M, Molina F, Gongora C, Thierry AR (2011).
1121:
Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Del Rio M, Molina F (October 2010).
3574:
3532:
501:
394:
investigations have been performed on ctDNA present in different patients with treatment-resistant
359:
304:
218:
500:
may be absent after tumor resection. Therefore, ctDNA analysis poses a potential avenue to detect
3667:
3631:
3569:
3361:
2968:
1074:"Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer"
1014:"Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen"
421:
391:
737:"Inferred Gene Expression By Cell-Free DNA Profiling Allows Noninvasive Lymphoma Classification"
684:
Esfahani, Mohammad
Shahrokh; Hamilton, Emily G.; Mehrmohamadi, Mahya; et al. (April 2022).
3791:
3672:
3626:
768:
Alig, Stefan K.; Shahrokh
Esfahani, Mohammad; Garofalo, Andrea; et al. (25 January 2024).
78:
2916:"Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time?"
1123:"Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts"
3724:
3705:
3149:
3117:
2567:
2246:"An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage"
226:
1516:
409:
This novel and promising technique has provided information on resistance to treatment with
3677:
3522:
3512:
3392:
3319:
3314:
3271:
3221:
3112:
3091:
2785:
2516:
2402:
2155:
1616:
1471:
1345:
1183:
1025:
637:
260:
139:
Never use heparinised tubes, heparin inhibits PCR by mimicking the helical structure of DNA
3377:
8:
3564:
3496:
3276:
3244:
3086:
3054:
2296:
Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. (May 2016).
649:
540:
334:
2949:
NucPosDB: a database of nucleosome positioning in vivo and nucleosomics of cell-free DNA
2868:
2789:
2648:
2615:
2520:
2406:
2159:
1711:
1620:
1475:
1349:
1187:
1029:
796:
769:
641:
3806:
3657:
3304:
3296:
3281:
3249:
3229:
3137:
2961:
2877:
2852:
2806:
2773:
2753:
2697:
2672:
2616:"Dynamic ctDNA Mutational Complexity in Patients with Melanoma Receiving Immunotherapy"
2548:
2474:
2449:
2425:
2390:
2371:
2322:
2297:
2270:
2245:
2226:
2073:
2048:
2024:
1999:
1921:
1896:
1769:
1742:
1723:
1639:
1604:
1497:
1425:
1413:
1381:
1368:
1333:
1309:
1258:
1233:
1206:
1171:
1147:
1122:
1098:
1073:
923:
898:
879:
712:
685:
658:
625:
606:
2298:"Integrated digital error suppression for improved detection of circulating tumor DNA"
2178:
2143:
1575:
1558:
1048:
1013:
983:"Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system"
959:
578:
436:
Further work is needed to understand how metastatic location and size, in relation to
3796:
3760:
3750:
3662:
3476:
3341:
2937:
2882:
2811:
2745:
2702:
2653:
2635:
2595:
2587:
2552:
2540:
2532:
2504:
2479:
2430:
2363:
2327:
2275:
2218:
2183:
2124:
2078:
2029:
1980:
1953:
1926:
1872:
1828:
1774:
1715:
1680:
1644:
1580:
1539:
1517:"Optimizing the yield and utility of circulating cell-free DNA from plasma and serum"
1489:
1417:
1385:
1373:
1301:
1263:
1211:
1152:
1103:
1053:
994:
963:
928:
871:
836:
801:
717:
663:
598:
505:
459:
411:
370:
214:
2757:
2375:
1727:
1429:
1313:
832:
3649:
3527:
3261:
3127:
3013:
2927:
2872:
2864:
2801:
2793:
2737:
2721:
2692:
2684:
2643:
2627:
2579:
2524:
2469:
2461:
2420:
2410:
2391:"Detection and quantification of rare mutations with massively parallel sequencing"
2355:
2317:
2309:
2265:
2257:
2230:
2210:
2173:
2163:
2116:
2068:
2060:
2019:
2011:
1916:
1908:
1862:
1818:
1764:
1754:
1707:
1672:
1634:
1624:
1570:
1531:
1501:
1479:
1409:
1363:
1353:
1293:
1253:
1245:
1201:
1191:
1142:
1134:
1093:
1085:
1043:
1033:
955:
918:
910:
883:
863:
828:
791:
781:
748:
707:
697:
653:
645:
610:
590:
1676:
3765:
3740:
3385:
3329:
3309:
2932:
2915:
2359:
1912:
1823:
1806:
1629:
1249:
1196:
395:
355:
300:
252:
210:
1867:
1850:
466:. It is thought that leukocytes are the primary contributors to cfDNA in serum.
159:-coated tubes. The use of EDTA is important to reduce coagulation of blood. The
3687:
3645:
3351:
3346:
3266:
2741:
2688:
2631:
2528:
2395:
Proceedings of the
National Academy of Sciences of the United States of America
2148:
Proceedings of the
National Academy of Sciences of the United States of America
1018:
Proceedings of the
National Academy of Sciences of the United States of America
786:
702:
579:"Liquid biopsies come of age: towards implementation of circulating tumour DNA"
425:
403:
2120:
1759:
1535:
1443:
Vasioukhin V, Stroun M, Maurice P, Lyautey J, Lederrey C, Anker P (May 1994).
1297:
753:
736:
3780:
3491:
3234:
3047:
2850:
2844:
2639:
2591:
2583:
2536:
770:"Distinct Hodgkin lymphoma subtypes defined by noninvasive genomic profiling"
557:
164:
67:
24:
2415:
2168:
1603:
Sherwood JL, Corcoran C, Brown H, Sharpe AD, Musilova M, Kohlmann A (2016).
3811:
3432:
3336:
3166:
3142:
3132:
3037:
3025:
3018:
2941:
2886:
2851:
Karachaliou N, Mayo-de-Las-Casas C, Molina-Vila MA, Rosell R (March 2015).
2815:
2749:
2706:
2657:
2599:
2544:
2505:"Deep whole-genome ctDNA chronology of treatment-resistant prostate cancer"
2483:
2465:
2434:
2367:
2331:
2279:
2222:
2187:
2128:
2082:
2033:
1930:
1876:
1832:
1778:
1719:
1684:
1648:
1584:
1543:
1493:
1377:
1358:
1305:
1267:
1215:
1156:
1107:
1038:
967:
875:
840:
805:
721:
667:
602:
463:
417:
160:
133:
Never freeze a blood sample before extracting the plasma for ctDNA analysis
28:
2833:
Applying circulating tumor DNA methylation in the diagnosis of lung cancer
1984:
1957:
1421:
1057:
998:
932:
734:
3755:
3695:
3554:
3517:
3440:
3324:
3190:
3071:
3066:
3042:
1138:
377:
256:
247:
194:
136:
Process the sample to plasma within 2–4 hours (if collected in EDTA tube)
2832:
2064:
2015:
914:
3700:
3549:
3486:
3450:
3356:
3199:
3195:
3171:
3100:
3081:
2948:
2797:
1998:
Li W, Zhang X, Lu X, You L, Song Y, Luo Z, et al. (October 2017).
1234:"Enhanced detection of circulating tumor DNA by fragment size analysis"
594:
399:
95:
19:
2719:
1334:"A mathematical model of ctDNA shedding predicts tumor detection size"
1089:
310:
3719:
3605:
3590:
3471:
3455:
3211:
3154:
3076:
3003:
2984:
2389:
Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B (June 2011).
2313:
2214:
2142:
Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B (July 2003).
686:"Inferring gene expression from cell-free DNA fragmentation profiles"
222:
103:
86:
82:
36:
2725:
2673:"Circulating biomarkers to monitor cancer progression and treatment"
2261:
1484:
1459:
867:
3709:
3600:
3559:
3544:
3417:
3409:
3366:
3161:
3122:
3107:
3030:
3008:
1169:
509:
326:
321:
279:
Targeted approaches have the advantage of amplifying ctDNA through
98:
plus a linker. Fragmentation of this length might be indicative of
90:
40:
1012:
Rogers JC, Boldt D, Kornfeld S, Skinner A, Valeri CR (July 1972).
3610:
3595:
3239:
3204:
3184:
3061:
2998:
1524:
Clinica
Chimica Acta; International Journal of Clinical Chemistry
1172:"High fragmentation characterizes tumour-derived circulating DNA"
948:
Clinica Chimica Acta; International Journal of Clinical Chemistry
683:
489:
455:
315:
2853:"Real-time liquid biopsies become a reality in cancer treatment"
2612:
1231:
576:
3481:
3413:
1557:
Norton SE, Lechner JM, Williams T, Fernando MR (October 2013).
945:
767:
63:
1442:
899:"Nucleic acids spontaneously released by living frog auricles"
66:, it has gained traction for its potential clinical utility; "
2450:"Detecting ultralow-frequency mutations by Duplex Sequencing"
1556:
1398:
437:
59:
51:
32:
2953:
2839:
Circulating tumor DNA: A new generation of cancer biomarkers
2770:
1970:
1894:
1120:
488:
Evidence of disease by traditional imaging methods, such as
290:
3445:
3407:
2344:
1602:
232:
156:
2388:
2141:
1741:
Qin Z, Ljubimov VA, Zhou C, Tong Y, Liang J (April 2016).
1070:
1011:
209:
This method was originally developed by the laboratory of
2670:
1331:
241:
55:
2726:"Liquid biopsy: monitoring cancer-genetics in the blood"
1697:
2295:
1661:
1515:
Xue X, Teare MD, Holen I, Zhu YM, Woll PJ (June 2009).
853:
354:
Safe-sequencing (Safe-Seq) was originally described by
2501:
2447:
378:
Integrated digital error suppression-enhanced CAPP-Seq
2845:
ctDNA 'Liquid Biopsy' Could Revolutionize Cancer Care
2565:
1807:"Circulating tumor DNA as a liquid biopsy for cancer"
424:, contribution of ctDNA to metastasis through global
2243:
35:. The mechanism of ctDNA release is unknown, though
1848:
1740:
818:
543:and drug resistance mechanisms involved in cancer.
316:
Beads, emulsification, amplification, and magnetics
311:
Beads, emulsification, amplification, and magnetics
2677:Computational and Structural Biotechnology Journal
2106:
980:
515:
448:
340:
1283:
77:ctDNA originates directly from the tumor or from
3778:
2200:
2046:
1514:
981:Anker P, Stroun M, Maurice PA (September 1975).
728:
533:
327:Cancer personalized profiling by deep sequencing
322:Cancer Personalized Profiling by deep Sequencing
295:Droplet digital PCR (ddPCR) is derived from the
259:”. Methylation of ctDNA can be detected through
115:
2497:
2495:
2493:
1943:
1844:
1842:
1804:
1598:
1596:
1594:
1327:
1325:
1323:
1227:
1225:
679:
677:
189:Whole genome or exome sequencing typically use
16:Tumor-derived fragmented DNA in the bloodstream
2291:
2289:
145:Plasma is better than serum for ctDNA recovery
3393:
2969:
2671:Rapisuwon S, Vietsch EE, Wellstein A (2016).
2109:Current Opinion in Genetics & Development
1451:
483:
2490:
1997:
1839:
1591:
1392:
1320:
1222:
1163:
761:
674:
474:
2286:
1805:Heitzer E, Ulz P, Geigl JB (January 2015).
1743:"Cell-free circulating tumor DNA in cancer"
896:
570:
191:high throughput DNA sequencing technologies
3400:
3386:
2976:
2962:
2720:Crowley E, Di Nicolantonio F, Loupakis F,
1849:van der Pol Y, Mouliere F (October 2019).
623:
386:
23:Circulating tumor DNA (ctDNA) is found in
2931:
2876:
2805:
2696:
2647:
2473:
2424:
2414:
2321:
2269:
2177:
2167:
2072:
2023:
1920:
1866:
1822:
1768:
1758:
1638:
1628:
1574:
1483:
1367:
1357:
1257:
1205:
1195:
1146:
1097:
1047:
1037:
922:
795:
785:
752:
711:
701:
657:
291:Droplet digital polymerase chain reaction
2893:
1973:The British Journal of Clinical Practice
416:, intratumoral heterogeneity (thanks to
233:Personalized analysis of rearranged ends
180:
18:
2913:
398:(the vast majority, and in some cases,
329:(CAPP-Seq) was originally described by
3779:
266:
242:DNA methylation and hydroxymethylation
204:
149:
3381:
2957:
2102:
2100:
2098:
2096:
2094:
2092:
1890:
1888:
1886:
1800:
1798:
1796:
1794:
1792:
1790:
1788:
1460:"Cancer biomarkers: Written in blood"
1279:
1277:
630:Annual Review of Analytical Chemistry
624:Nonaka, T; Wong, DTW (13 June 2022).
617:
420:evolution and molecular chronology),
238:was modified for ctDNA applications.
1457:
650:10.1146/annurev-anchem-061020-123959
365:
171:
2896:"Teasing Out Circulating Tumor DNA"
2869:10.3978/j.issn.2305-5839.2015.01.16
1712:10.1046/j.1537-2995.2001.41020276.x
13:
3287:Fluorescence in situ hybridization
2894:Marusina, Kate (8 February 2018).
2826:
2713:
2089:
1883:
1785:
1414:10.1111/j.1365-2141.1994.tb04828.x
1274:
1064:
524:
349:
14:
3823:
2730:Nature Reviews. Clinical Oncology
2620:Molecular Diagnosis & Therapy
1576:10.1016/j.clinbiochem.2013.06.002
443:
333:and Maximilian Diehn's groups at
297:digital polymerase chain reaction
3255:Oral and maxillofacial pathology
2857:Annals of Translational Medicine
1447:. Vol. 5. pp. 141–150.
2764:
2664:
2606:
2559:
2441:
2382:
2338:
2237:
2194:
2135:
2040:
1991:
1964:
1944:Preobrazhenskiĭ BS (1966). "".
1937:
1734:
1691:
1655:
1550:
1508:
1436:
1114:
1078:International Journal of Cancer
1005:
974:
897:Stroun M, Anker P (July 1972).
833:10.1016/j.cancergen.2013.01.005
516:ctDNA as a prognostic biomarker
449:“Normal” vs tumor DNA detection
341:Tagged amplicon deep sequencing
274:single nucleotide polymorphisms
2348:Science Translational Medicine
1901:Science Translational Medicine
1402:British Journal of Haematology
939:
890:
847:
812:
1:
3715:Clonally transmissible cancer
2983:
1677:10.1016/j.lungcan.2013.08.014
960:10.1016/S0009-8981(01)00665-9
563:
553:Circulating mitochondrial DNA
534:Challenges for implementation
116:Pre-analytical considerations
2933:10.1016/j.canlet.2019.10.014
2360:10.1126/scitranslmed.3003726
1913:10.1126/scitranslmed.3004742
1824:10.1373/clinchem.2014.222679
1630:10.1371/journal.pone.0150197
1250:10.1126/scitranslmed.aat4921
1197:10.1371/journal.pone.0023418
7:
3802:Surgical removal procedures
1946:Vestnik Otorinolaringologii
1868:10.1016/j.ccell.2019.09.003
546:
469:
100:apoptotic DNA fragmentation
10:
3828:
3746:Index of oncology articles
2742:10.1038/nrclinonc.2013.110
2689:10.1016/j.csbj.2016.05.004
2632:10.1007/s40291-023-00651-4
2529:10.1038/s41586-022-04975-9
787:10.1038/s41586-023-06903-x
703:10.1038/s41587-022-01222-4
484:ctDNA in cancer monitoring
281:polymerase chain reactions
110:
3733:
3686:
3644:
3619:
3583:
3505:
3464:
3431:
3424:
3295:
3220:
2991:
2121:10.1016/j.gde.2016.12.003
1760:10.1186/s40880-016-0092-4
1747:Chinese Journal of Cancer
1536:10.1016/j.cca.2009.02.018
1298:10.1080/08916930701358826
754:10.1182/blood-2023-186853
747:(Supplement 1): 245–245.
475:ctDNA in cancer screening
2914:Du-Bois, Asante (2019).
502:minimal residual disease
360:Johns Hopkins University
305:Johns Hopkins University
219:Johns Hopkins University
3668:Prostate cancer staging
3632:Paraneoplastic syndrome
3362:Microbiological culture
2992:Principles of pathology
2416:10.1073/pnas.1105422108
2169:10.1073/pnas.1133470100
903:The Biochemical Journal
422:chromosomal instability
392:Whole-genome sequencing
387:Whole-genome sequencing
79:circulating tumor cells
3706:Tumor suppressor genes
3673:Gleason grading system
3627:Precancerous condition
2584:10.1093/carcin/bgaa102
2466:10.1038/nprot.2014.170
1359:10.1126/sciadv.abc4308
1127:Nucleic Acids Research
1039:10.1073/pnas.69.7.1685
856:Nature Reviews. Cancer
299:, originally named by
44:
3725:Carcinogenic bacteria
3465:Malignant progression
3325:Diagnostic immunology
3150:Programmed cell death
3118:Liquefactive necrosis
2778:Nature Communications
1563:Clinical Biochemistry
583:Nature Reviews Cancer
227:copy number variation
181:Untargeted approaches
48:Circulating tumor DNA
22:
3787:Anatomical pathology
3678:Dukes classification
3320:Medical microbiology
3315:Transfusion medicine
3272:Immunohistochemistry
3222:Anatomical pathology
3113:Coagulative necrosis
2302:Nature Biotechnology
1458:Yong E (July 2014).
690:Nature Biotechnology
626:"Saliva Diagnostics"
414:signaling inhibitors
54:-derived fragmented
3497:Sentinel lymph node
3277:Electron microscopy
3245:Molecular pathology
3123:Gangrenous necrosis
3055:Cellular adaptation
2790:2015NatCo...6.8760M
2521:2022Natur.608..199H
2407:2011PNAS..108.9530K
2160:2003PNAS..100.8817D
2065:10.1038/cr.2017.106
2016:10.1038/cr.2017.121
1621:2016PLoSO..1150197S
1476:2014Natur.511..524Y
1350:2020SciA....6.4308A
1188:2011PLoSO...623418M
1030:1972PNAS...69.1685R
915:10.1042/bj1280100pb
642:2022ARAC...15..107N
541:tumor heterogeneity
335:Stanford University
267:Targeted approaches
261:bisulfite treatment
205:Digital karyotyping
177:targeted approach.
150:Extraction of ctDNA
3545:Respiratory system
3305:Clinical chemistry
3297:Clinical pathology
3282:Immunofluorescence
3250:Forensic pathology
3230:Surgical pathology
3138:Fibrinoid necrosis
2798:10.1038/ncomms9760
1811:Clinical Chemistry
1139:10.1093/nar/gkq421
595:10.1038/nrc.2017.7
45:
3774:
3773:
3761:Cancer and nausea
3640:
3639:
3477:Carcinoma in situ
3375:
3374:
3342:Mass spectrometry
2578:(11): 1507–1517.
2515:(7921): 199–208.
2059:(10): 1231–1242.
2010:(10): 1243–1257.
1907:(162): 162ra154.
1470:(7511): 524–526.
1090:10.1002/ijc.28030
780:(7996): 778–787.
506:colorectal cancer
412:androgen receptor
371:Duplex sequencing
366:Duplex sequencing
358:and his group at
215:Victor Velculescu
213:, Luis Diaz, and
172:Analysis of ctDNA
3819:
3575:Endocrine system
3540:Digestive system
3429:
3428:
3402:
3395:
3388:
3379:
3378:
3262:Gross processing
3128:Caseous necrosis
2978:
2971:
2964:
2955:
2954:
2945:
2935:
2910:
2908:
2906:
2890:
2880:
2820:
2819:
2809:
2768:
2762:
2761:
2717:
2711:
2710:
2700:
2668:
2662:
2661:
2651:
2610:
2604:
2603:
2563:
2557:
2556:
2499:
2488:
2487:
2477:
2460:(11): 2586–606.
2454:Nature Protocols
2445:
2439:
2438:
2428:
2418:
2386:
2380:
2379:
2354:(136): 136ra68.
2342:
2336:
2335:
2325:
2314:10.1038/nbt.3520
2293:
2284:
2283:
2273:
2241:
2235:
2234:
2215:10.1038/nmeth898
2198:
2192:
2191:
2181:
2171:
2139:
2133:
2132:
2104:
2087:
2086:
2076:
2044:
2038:
2037:
2027:
1995:
1989:
1988:
1968:
1962:
1961:
1941:
1935:
1934:
1924:
1892:
1881:
1880:
1870:
1846:
1837:
1836:
1826:
1802:
1783:
1782:
1772:
1762:
1738:
1732:
1731:
1695:
1689:
1688:
1659:
1653:
1652:
1642:
1632:
1600:
1589:
1588:
1578:
1554:
1548:
1547:
1521:
1512:
1506:
1505:
1487:
1455:
1449:
1448:
1440:
1434:
1433:
1396:
1390:
1389:
1371:
1361:
1344:(50): eabc4308.
1338:Science Advances
1329:
1318:
1317:
1281:
1272:
1271:
1261:
1229:
1220:
1219:
1209:
1199:
1167:
1161:
1160:
1150:
1118:
1112:
1111:
1101:
1068:
1062:
1061:
1051:
1041:
1009:
1003:
1002:
978:
972:
971:
943:
937:
936:
926:
909:(3): 100P–101P.
894:
888:
887:
851:
845:
844:
816:
810:
809:
799:
789:
765:
759:
758:
756:
732:
726:
725:
715:
705:
681:
672:
671:
661:
621:
615:
614:
574:
221:. Unlike normal
3827:
3826:
3822:
3821:
3820:
3818:
3817:
3816:
3777:
3776:
3775:
3770:
3729:
3682:
3636:
3615:
3579:
3501:
3460:
3420:
3406:
3376:
3371:
3330:Immunopathology
3310:Hematopathology
3291:
3216:
2987:
2982:
2904:
2902:
2829:
2827:Further reading
2824:
2823:
2769:
2765:
2724:(August 2013).
2718:
2714:
2669:
2665:
2611:
2607:
2564:
2560:
2500:
2491:
2446:
2442:
2387:
2383:
2343:
2339:
2294:
2287:
2262:10.1038/nm.3519
2250:Nature Medicine
2242:
2238:
2199:
2195:
2154:(15): 8817–22.
2140:
2136:
2105:
2090:
2045:
2041:
1996:
1992:
1969:
1965:
1942:
1938:
1893:
1884:
1847:
1840:
1803:
1786:
1739:
1735:
1696:
1692:
1660:
1656:
1615:(2): e0150197.
1601:
1592:
1555:
1551:
1519:
1513:
1509:
1485:10.1038/511524a
1456:
1452:
1441:
1437:
1397:
1393:
1330:
1321:
1282:
1275:
1230:
1223:
1168:
1164:
1133:(18): 6159–75.
1119:
1115:
1069:
1065:
1010:
1006:
987:Cancer Research
979:
975:
954:(1–2): 139–42.
944:
940:
895:
891:
868:10.1038/nrc3066
852:
848:
821:Cancer Genetics
817:
813:
766:
762:
733:
729:
682:
675:
622:
618:
575:
571:
566:
549:
536:
527:
525:Cancer research
518:
486:
477:
472:
451:
446:
396:prostate cancer
389:
380:
368:
356:Bert Vogelstein
352:
350:Safe-sequencing
343:
324:
313:
301:Bert Vogelstein
293:
269:
253:DNA methylation
244:
235:
211:Bert Vogelstein
207:
183:
174:
152:
118:
113:
68:liquid biopsies
31:fractions from
17:
12:
11:
5:
3825:
3815:
3814:
3809:
3804:
3799:
3794:
3789:
3772:
3771:
3769:
3768:
3763:
3758:
3753:
3748:
3743:
3737:
3735:
3731:
3730:
3728:
3727:
3722:
3717:
3712:
3703:
3698:
3692:
3690:
3688:Carcinogenesis
3684:
3683:
3681:
3680:
3675:
3670:
3665:
3660:
3654:
3652:
3642:
3641:
3638:
3637:
3635:
3634:
3629:
3623:
3621:
3617:
3616:
3614:
3613:
3608:
3603:
3598:
3593:
3587:
3585:
3581:
3580:
3578:
3577:
3572:
3570:Nervous system
3567:
3562:
3557:
3552:
3547:
3542:
3537:
3536:
3535:
3533:nasopharyngeal
3530:
3525:
3520:
3509:
3507:
3503:
3502:
3500:
3499:
3494:
3489:
3484:
3479:
3474:
3468:
3466:
3462:
3461:
3459:
3458:
3453:
3448:
3443:
3437:
3435:
3426:
3422:
3421:
3405:
3404:
3397:
3390:
3382:
3373:
3372:
3370:
3369:
3364:
3359:
3354:
3352:Flow cytometry
3349:
3347:Chromatography
3344:
3339:
3333:
3332:
3327:
3322:
3317:
3312:
3307:
3301:
3299:
3293:
3292:
3290:
3289:
3284:
3279:
3274:
3269:
3267:Histopathology
3264:
3258:
3257:
3252:
3247:
3242:
3237:
3232:
3226:
3224:
3218:
3217:
3215:
3214:
3209:
3208:
3207:
3202:
3193:
3181:
3175:
3174:
3169:
3164:
3159:
3158:
3157:
3147:
3146:
3145:
3140:
3135:
3130:
3125:
3120:
3115:
3105:
3103:
3097:
3096:
3095:
3094:
3089:
3079:
3074:
3069:
3064:
3059:
3057:
3051:
3050:
3045:
3040:
3035:
3034:
3033:
3023:
3022:
3021:
3016:
3011:
3006:
2995:
2993:
2989:
2988:
2981:
2980:
2973:
2966:
2958:
2952:
2951:
2946:
2920:Cancer Letters
2911:
2891:
2848:
2842:
2836:
2828:
2825:
2822:
2821:
2763:
2712:
2663:
2626:(4): 537–550.
2605:
2572:Carcinogenesis
2558:
2489:
2440:
2401:(23): 9530–5.
2381:
2337:
2308:(5): 547–555.
2285:
2236:
2203:Nature Methods
2193:
2134:
2088:
2039:
1990:
1963:
1948:(in Russian).
1936:
1882:
1861:(4): 350–368.
1838:
1784:
1733:
1690:
1654:
1590:
1569:(15): 1561–5.
1549:
1507:
1450:
1435:
1408:(4): 774–779.
1391:
1319:
1273:
1238:Sci Transl Med
1221:
1162:
1113:
1063:
1004:
993:(9): 2375–82.
973:
938:
889:
846:
811:
760:
727:
696:(4): 585–597.
673:
636:(1): 107–121.
616:
589:(4): 223–238.
568:
567:
565:
562:
561:
560:
555:
548:
545:
535:
532:
526:
523:
517:
514:
485:
482:
476:
473:
471:
468:
450:
447:
445:
444:Considerations
442:
426:transcriptomic
404:bladder cancer
388:
385:
379:
376:
367:
364:
351:
348:
342:
339:
323:
320:
312:
309:
292:
289:
268:
265:
243:
240:
234:
231:
206:
203:
182:
179:
173:
170:
151:
148:
147:
146:
143:
140:
137:
134:
117:
114:
112:
109:
15:
9:
6:
4:
3:
2:
3824:
3813:
3810:
3808:
3805:
3803:
3800:
3798:
3795:
3793:
3792:Medical signs
3790:
3788:
3785:
3784:
3782:
3767:
3764:
3762:
3759:
3757:
3754:
3752:
3749:
3747:
3744:
3742:
3739:
3738:
3736:
3732:
3726:
3723:
3721:
3718:
3716:
3713:
3711:
3707:
3704:
3702:
3699:
3697:
3694:
3693:
3691:
3689:
3685:
3679:
3676:
3674:
3671:
3669:
3666:
3664:
3661:
3659:
3656:
3655:
3653:
3651:
3647:
3643:
3633:
3630:
3628:
3625:
3624:
3622:
3618:
3612:
3609:
3607:
3604:
3602:
3599:
3597:
3594:
3592:
3589:
3588:
3586:
3582:
3576:
3573:
3571:
3568:
3566:
3563:
3561:
3558:
3556:
3553:
3551:
3548:
3546:
3543:
3541:
3538:
3534:
3531:
3529:
3526:
3524:
3523:oropharyngeal
3521:
3519:
3516:
3515:
3514:
3513:Head and neck
3511:
3510:
3508:
3504:
3498:
3495:
3493:
3492:Primary tumor
3490:
3488:
3485:
3483:
3480:
3478:
3475:
3473:
3470:
3469:
3467:
3463:
3457:
3454:
3452:
3449:
3447:
3444:
3442:
3439:
3438:
3436:
3434:
3433:Benign tumors
3430:
3427:
3423:
3419:
3415:
3411:
3403:
3398:
3396:
3391:
3389:
3384:
3383:
3380:
3368:
3365:
3363:
3360:
3358:
3355:
3353:
3350:
3348:
3345:
3343:
3340:
3338:
3335:
3334:
3331:
3328:
3326:
3323:
3321:
3318:
3316:
3313:
3311:
3308:
3306:
3303:
3302:
3300:
3298:
3294:
3288:
3285:
3283:
3280:
3278:
3275:
3273:
3270:
3268:
3265:
3263:
3260:
3259:
3256:
3253:
3251:
3248:
3246:
3243:
3241:
3238:
3236:
3235:Cytopathology
3233:
3231:
3228:
3227:
3225:
3223:
3219:
3213:
3210:
3206:
3203:
3201:
3197:
3194:
3192:
3189:
3188:
3187:
3186:
3182:
3180:
3179:Accumulations
3177:
3176:
3173:
3170:
3168:
3165:
3163:
3160:
3156:
3153:
3152:
3151:
3148:
3144:
3141:
3139:
3136:
3134:
3131:
3129:
3126:
3124:
3121:
3119:
3116:
3114:
3111:
3110:
3109:
3106:
3104:
3102:
3099:
3098:
3093:
3090:
3088:
3085:
3084:
3083:
3080:
3078:
3075:
3073:
3070:
3068:
3065:
3063:
3060:
3058:
3056:
3053:
3052:
3049:
3048:Wound healing
3046:
3044:
3041:
3039:
3036:
3032:
3029:
3028:
3027:
3024:
3020:
3017:
3015:
3012:
3010:
3007:
3005:
3002:
3001:
3000:
2997:
2996:
2994:
2990:
2986:
2979:
2974:
2972:
2967:
2965:
2960:
2959:
2956:
2950:
2947:
2943:
2939:
2934:
2929:
2925:
2921:
2917:
2912:
2901:
2900:ClinicalOMICs
2897:
2892:
2888:
2884:
2879:
2874:
2870:
2866:
2862:
2858:
2854:
2849:
2846:
2843:
2840:
2837:
2834:
2831:
2830:
2817:
2813:
2808:
2803:
2799:
2795:
2791:
2787:
2783:
2779:
2775:
2767:
2759:
2755:
2751:
2747:
2743:
2739:
2736:(8): 472–84.
2735:
2731:
2727:
2723:
2716:
2708:
2704:
2699:
2694:
2690:
2686:
2682:
2678:
2674:
2667:
2659:
2655:
2650:
2645:
2641:
2637:
2633:
2629:
2625:
2621:
2617:
2609:
2601:
2597:
2593:
2589:
2585:
2581:
2577:
2573:
2569:
2562:
2554:
2550:
2546:
2542:
2538:
2534:
2530:
2526:
2522:
2518:
2514:
2510:
2506:
2498:
2496:
2494:
2485:
2481:
2476:
2471:
2467:
2463:
2459:
2455:
2451:
2444:
2436:
2432:
2427:
2422:
2417:
2412:
2408:
2404:
2400:
2396:
2392:
2385:
2377:
2373:
2369:
2365:
2361:
2357:
2353:
2349:
2341:
2333:
2329:
2324:
2319:
2315:
2311:
2307:
2303:
2299:
2292:
2290:
2281:
2277:
2272:
2267:
2263:
2259:
2256:(5): 548–54.
2255:
2251:
2247:
2240:
2232:
2228:
2224:
2220:
2216:
2212:
2208:
2204:
2197:
2189:
2185:
2180:
2175:
2170:
2165:
2161:
2157:
2153:
2149:
2145:
2138:
2130:
2126:
2122:
2118:
2114:
2110:
2103:
2101:
2099:
2097:
2095:
2093:
2084:
2080:
2075:
2070:
2066:
2062:
2058:
2054:
2053:Cell Research
2050:
2043:
2035:
2031:
2026:
2021:
2017:
2013:
2009:
2005:
2004:Cell Research
2001:
1994:
1986:
1982:
1978:
1974:
1967:
1959:
1955:
1951:
1947:
1940:
1932:
1928:
1923:
1918:
1914:
1910:
1906:
1902:
1898:
1891:
1889:
1887:
1878:
1874:
1869:
1864:
1860:
1856:
1852:
1845:
1843:
1834:
1830:
1825:
1820:
1817:(1): 112–23.
1816:
1812:
1808:
1801:
1799:
1797:
1795:
1793:
1791:
1789:
1780:
1776:
1771:
1766:
1761:
1756:
1752:
1748:
1744:
1737:
1729:
1725:
1721:
1717:
1713:
1709:
1706:(2): 276–82.
1705:
1701:
1694:
1686:
1682:
1678:
1674:
1670:
1666:
1658:
1650:
1646:
1641:
1636:
1631:
1626:
1622:
1618:
1614:
1610:
1606:
1599:
1597:
1595:
1586:
1582:
1577:
1572:
1568:
1564:
1560:
1553:
1545:
1541:
1537:
1533:
1529:
1525:
1518:
1511:
1503:
1499:
1495:
1491:
1486:
1481:
1477:
1473:
1469:
1465:
1461:
1454:
1446:
1439:
1431:
1427:
1423:
1419:
1415:
1411:
1407:
1403:
1395:
1387:
1383:
1379:
1375:
1370:
1365:
1360:
1355:
1351:
1347:
1343:
1339:
1335:
1328:
1326:
1324:
1315:
1311:
1307:
1303:
1299:
1295:
1291:
1287:
1280:
1278:
1269:
1265:
1260:
1255:
1251:
1247:
1243:
1239:
1235:
1228:
1226:
1217:
1213:
1208:
1203:
1198:
1193:
1189:
1185:
1182:(9): e23418.
1181:
1177:
1173:
1166:
1158:
1154:
1149:
1144:
1140:
1136:
1132:
1128:
1124:
1117:
1109:
1105:
1100:
1095:
1091:
1087:
1084:(2): 346–56.
1083:
1079:
1075:
1067:
1059:
1055:
1050:
1045:
1040:
1035:
1031:
1027:
1024:(7): 1685–9.
1023:
1019:
1015:
1008:
1000:
996:
992:
988:
984:
977:
969:
965:
961:
957:
953:
949:
942:
934:
930:
925:
920:
916:
912:
908:
904:
900:
893:
885:
881:
877:
873:
869:
865:
862:(6): 426–37.
861:
857:
850:
842:
838:
834:
830:
826:
822:
815:
807:
803:
798:
793:
788:
783:
779:
775:
771:
764:
755:
750:
746:
742:
738:
731:
723:
719:
714:
709:
704:
699:
695:
691:
687:
680:
678:
669:
665:
660:
655:
651:
647:
643:
639:
635:
631:
627:
620:
612:
608:
604:
600:
596:
592:
588:
584:
580:
573:
569:
559:
558:Liquid biopsy
556:
554:
551:
550:
544:
542:
531:
522:
513:
511:
507:
503:
499:
495:
491:
481:
467:
465:
461:
457:
441:
439:
434:
430:
427:
423:
419:
415:
413:
407:
405:
401:
397:
393:
384:
375:
372:
363:
361:
357:
347:
338:
336:
332:
328:
319:
317:
308:
306:
302:
298:
288:
286:
282:
277:
275:
264:
262:
258:
254:
249:
239:
230:
228:
224:
220:
216:
212:
202:
198:
196:
192:
187:
178:
169:
166:
162:
158:
144:
141:
138:
135:
132:
131:
130:
127:
124:
108:
105:
101:
97:
92:
88:
84:
80:
75:
71:
69:
65:
61:
57:
53:
49:
42:
38:
34:
30:
26:
21:
3408:Overview of
3337:Enzyme assay
3183:
3178:
3167:Karyorrhexis
3143:Myocytolysis
3133:Fat necrosis
3038:Inflammation
3026:Hemodynamics
3019:Pathogenesis
2923:
2919:
2903:. Retrieved
2899:
2860:
2856:
2781:
2777:
2766:
2733:
2729:
2715:
2680:
2676:
2666:
2623:
2619:
2608:
2575:
2571:
2561:
2512:
2508:
2457:
2453:
2443:
2398:
2394:
2384:
2351:
2347:
2340:
2305:
2301:
2253:
2249:
2239:
2209:(7): 551–9.
2206:
2202:
2196:
2151:
2147:
2137:
2112:
2108:
2056:
2052:
2042:
2007:
2003:
1993:
1979:(5): 183–8.
1976:
1972:
1966:
1949:
1945:
1939:
1904:
1900:
1858:
1854:
1814:
1810:
1750:
1746:
1736:
1703:
1699:
1693:
1671:(2): 373–4.
1668:
1664:
1657:
1612:
1608:
1566:
1562:
1552:
1530:(2): 100–4.
1527:
1523:
1510:
1467:
1463:
1453:
1444:
1438:
1405:
1401:
1394:
1341:
1337:
1292:(4): 281–4.
1289:
1286:Autoimmunity
1285:
1241:
1237:
1179:
1175:
1165:
1130:
1126:
1116:
1081:
1077:
1066:
1021:
1017:
1007:
990:
986:
976:
951:
947:
941:
906:
902:
892:
859:
855:
849:
827:(3): 73–80.
824:
820:
814:
777:
773:
763:
744:
740:
730:
693:
689:
633:
629:
619:
586:
582:
572:
537:
528:
519:
487:
478:
464:chemotherapy
460:radiotherapy
452:
435:
431:
418:phylogenetic
410:
408:
390:
381:
369:
353:
344:
331:Ash Alizadeh
325:
314:
303:’s group at
294:
278:
270:
245:
236:
208:
199:
188:
184:
175:
153:
128:
122:
119:
76:
72:
47:
46:
3756:Cancer pain
3696:Cancer cell
3441:Hyperplasia
3191:Hemosiderin
3072:Hyperplasia
3067:Hypertrophy
3043:Cell damage
2784:(1): 8760.
1952:(1): 3–11.
1855:Cancer Cell
1700:Transfusion
1665:Lung Cancer
285:digital PCR
257:CpG islands
223:karyotyping
195:buccal swab
50:(ctDNA) is
3781:Categories
3701:Carcinogen
3565:Urogenital
3506:Topography
3487:Metastasis
3451:Pseudocyst
3425:Conditions
3357:Blood bank
3200:Lipofuscin
3196:Lipochrome
3172:Karyolysis
3101:Cell death
3082:Metaplasia
2722:Bardelli A
2683:: 211–22.
564:References
400:metastatic
248:epigenetic
104:phagocytes
96:nucleosome
3807:Neoplasms
3720:Oncovirus
3710:oncogenes
3663:Ann Arbor
3606:Papilloma
3591:Carcinoma
3584:Histology
3528:laryngeal
3472:Dysplasia
3456:Hamartoma
3212:Steatosis
3155:Apoptosis
3092:Glandular
3077:Dysplasia
3009:Neoplasia
3004:Infection
2985:Pathology
2926:: 59–71.
2863:(3): 36.
2640:1177-1062
2592:0143-3334
2553:250730778
2537:1476-4687
2115:: 14–21.
1386:228096858
283:(PCR) or
87:apoptosis
83:lymphatic
37:apoptosis
3797:Oncology
3741:Research
3601:Blastoma
3418:oncology
3367:Serology
3162:Pyknosis
3108:Necrosis
3087:Squamous
3031:Ischemia
2942:31610267
2887:25815297
2847:Nov 2014
2841:Feb 2014
2835:May 2019
2816:26530965
2758:25537784
2750:23836314
2707:27358717
2658:37099071
2649:10131510
2600:32955091
2545:35859180
2484:25299156
2435:21586637
2376:34723244
2368:22649089
2332:27018799
2280:24705333
2223:16791214
2188:12857956
2129:28126649
2083:28820176
2034:28925386
1931:23197571
1877:31614115
1833:25388429
1779:27056366
1728:45714834
1720:11239235
1685:24007628
1649:26918901
1609:PLOS ONE
1585:23769817
1544:19281804
1494:25079538
1430:26365875
1378:33310847
1314:11499768
1306:17516210
1268:30404863
1216:21909401
1176:PLOS ONE
1157:20494973
1108:23319339
968:11694251
876:21562580
841:23491080
806:38081297
797:11293530
722:35361996
668:35696523
603:28233803
547:See also
510:melanoma
470:Research
91:necrosis
41:necrosis
3751:History
3650:grading
3646:Staging
3611:Adenoma
3596:Sarcoma
3240:Autopsy
3205:Melanin
3185:pigment
3062:Atrophy
2999:Disease
2905:5 March
2878:4356857
2807:4659935
2786:Bibcode
2698:4913179
2517:Bibcode
2475:4271547
2426:3111315
2403:Bibcode
2323:4907374
2271:4016134
2231:7059151
2156:Bibcode
2074:5630676
2025:5630683
1985:1975200
1958:5988180
1922:3641759
1770:4823888
1640:4769175
1617:Bibcode
1502:4445938
1472:Bibcode
1422:7918071
1369:7732186
1346:Bibcode
1259:6483061
1244:(466).
1207:3167805
1184:Bibcode
1148:2952865
1099:3708119
1058:4505646
1026:Bibcode
999:1149042
933:4634816
924:1173871
884:6061607
713:9337986
659:9348814
638:Bibcode
611:4561229
456:surgery
246:Proper
111:Methods
3482:Cancer
3414:cancer
3410:tumors
2940:
2885:
2875:
2814:
2804:
2756:
2748:
2705:
2695:
2656:
2646:
2638:
2598:
2590:
2551:
2543:
2535:
2509:Nature
2482:
2472:
2433:
2423:
2374:
2366:
2330:
2320:
2278:
2268:
2229:
2221:
2186:
2179:166396
2176:
2127:
2081:
2071:
2032:
2022:
1983:
1956:
1929:
1919:
1875:
1831:
1777:
1767:
1753:: 36.
1726:
1718:
1683:
1647:
1637:
1583:
1542:
1500:
1492:
1464:Nature
1428:
1420:
1384:
1376:
1366:
1312:
1304:
1266:
1256:
1214:
1204:
1155:
1145:
1106:
1096:
1056:
1049:426778
1046:
997:
966:
931:
921:
882:
874:
839:
804:
794:
774:Nature
720:
710:
666:
656:
609:
601:
161:plasma
64:genome
29:plasma
3734:Misc.
3620:Other
3560:Blood
3014:Cause
2754:S2CID
2549:S2CID
2372:S2CID
2227:S2CID
1724:S2CID
1520:(PDF)
1498:S2CID
1426:S2CID
1382:S2CID
1310:S2CID
880:S2CID
741:Blood
607:S2CID
462:, or
438:tumor
165:serum
60:cfDNA
52:tumor
33:blood
25:serum
3766:Diet
3555:Skin
3550:Bone
3518:oral
3446:Cyst
3416:and
2938:PMID
2907:2018
2883:PMID
2812:PMID
2746:PMID
2703:PMID
2654:PMID
2636:ISSN
2596:PMID
2588:ISSN
2541:PMID
2533:ISSN
2480:PMID
2431:PMID
2364:PMID
2328:PMID
2276:PMID
2219:PMID
2184:PMID
2125:PMID
2079:PMID
2030:PMID
1981:PMID
1954:PMID
1927:PMID
1873:PMID
1829:PMID
1775:PMID
1716:PMID
1681:PMID
1645:PMID
1581:PMID
1540:PMID
1490:PMID
1418:PMID
1374:PMID
1302:PMID
1264:PMID
1212:PMID
1153:PMID
1104:PMID
1054:PMID
995:PMID
964:PMID
929:PMID
872:PMID
837:PMID
802:PMID
718:PMID
664:PMID
599:PMID
508:and
163:and
157:EDTA
123:KRAS
89:and
27:and
3812:DNA
3658:TNM
2928:doi
2924:468
2873:PMC
2865:doi
2802:PMC
2794:doi
2738:doi
2693:PMC
2685:doi
2644:PMC
2628:doi
2580:doi
2525:doi
2513:608
2470:PMC
2462:doi
2421:PMC
2411:doi
2399:108
2356:doi
2318:PMC
2310:doi
2266:PMC
2258:doi
2211:doi
2174:PMC
2164:doi
2152:100
2117:doi
2069:PMC
2061:doi
2020:PMC
2012:doi
1917:PMC
1909:doi
1863:doi
1819:doi
1765:PMC
1755:doi
1708:doi
1673:doi
1635:PMC
1625:doi
1571:doi
1532:doi
1528:404
1480:doi
1468:511
1410:doi
1364:PMC
1354:doi
1294:doi
1254:PMC
1246:doi
1202:PMC
1192:doi
1143:PMC
1135:doi
1094:PMC
1086:doi
1082:133
1044:PMC
1034:doi
956:doi
952:313
919:PMC
911:doi
907:128
864:doi
829:doi
825:206
792:PMC
782:doi
778:625
749:doi
745:142
708:PMC
698:doi
654:PMC
646:doi
591:doi
498:MRI
496:or
494:PET
402:),
217:at
56:DNA
3783::
3412:,
2936:.
2922:.
2918:.
2898:.
2881:.
2871:.
2859:.
2855:.
2810:.
2800:.
2792:.
2780:.
2776:.
2752:.
2744:.
2734:10
2732:.
2728:.
2701:.
2691:.
2681:14
2679:.
2675:.
2652:.
2642:.
2634:.
2624:27
2622:.
2618:.
2594:.
2586:.
2576:41
2574:.
2570:.
2547:.
2539:.
2531:.
2523:.
2511:.
2507:.
2492:^
2478:.
2468:.
2456:.
2452:.
2429:.
2419:.
2409:.
2397:.
2393:.
2370:.
2362:.
2350:.
2326:.
2316:.
2306:34
2304:.
2300:.
2288:^
2274:.
2264:.
2254:20
2252:.
2248:.
2225:.
2217:.
2205:.
2182:.
2172:.
2162:.
2150:.
2146:.
2123:.
2113:42
2111:.
2091:^
2077:.
2067:.
2057:27
2055:.
2051:.
2028:.
2018:.
2008:27
2006:.
2002:.
1977:44
1975:.
1950:28
1925:.
1915:.
1903:.
1899:.
1885:^
1871:.
1859:36
1857:.
1853:.
1841:^
1827:.
1815:61
1813:.
1809:.
1787:^
1773:.
1763:.
1751:35
1749:.
1745:.
1722:.
1714:.
1704:41
1702:.
1679:.
1669:82
1667:.
1643:.
1633:.
1623:.
1613:11
1611:.
1607:.
1593:^
1579:.
1567:46
1565:.
1561:.
1538:.
1526:.
1522:.
1496:.
1488:.
1478:.
1466:.
1462:.
1424:.
1416:.
1406:86
1404:.
1380:.
1372:.
1362:.
1352:.
1340:.
1336:.
1322:^
1308:.
1300:.
1290:40
1288:.
1276:^
1262:.
1252:.
1242:10
1240:.
1236:.
1224:^
1210:.
1200:.
1190:.
1178:.
1174:.
1151:.
1141:.
1131:38
1129:.
1125:.
1102:.
1092:.
1080:.
1076:.
1052:.
1042:.
1032:.
1022:69
1020:.
1016:.
991:35
989:.
985:.
962:.
950:.
927:.
917:.
905:.
901:.
878:.
870:.
860:11
858:.
835:.
823:.
800:.
790:.
776:.
772:.
743:.
739:.
716:.
706:.
694:40
692:.
688:.
676:^
662:.
652:.
644:.
634:15
632:.
628:.
605:.
597:.
587:17
585:.
581:.
512:.
492:,
490:CT
458:,
39:,
3708:/
3648:/
3401:e
3394:t
3387:v
3198:/
2977:e
2970:t
2963:v
2944:.
2930::
2909:.
2889:.
2867::
2861:3
2818:.
2796::
2788::
2782:6
2760:.
2740::
2709:.
2687::
2660:.
2630::
2602:.
2582::
2555:.
2527::
2519::
2486:.
2464::
2458:9
2437:.
2413::
2405::
2378:.
2358::
2352:4
2334:.
2312::
2282:.
2260::
2233:.
2213::
2207:3
2190:.
2166::
2158::
2131:.
2119::
2085:.
2063::
2036:.
2014::
1987:.
1960:.
1933:.
1911::
1905:4
1879:.
1865::
1835:.
1821::
1781:.
1757::
1730:.
1710::
1687:.
1675::
1651:.
1627::
1619::
1587:.
1573::
1546:.
1534::
1504:.
1482::
1474::
1432:.
1412::
1388:.
1356::
1348::
1342:6
1316:.
1296::
1270:.
1248::
1218:.
1194::
1186::
1180:6
1159:.
1137::
1110:.
1088::
1060:.
1036::
1028::
1001:.
970:.
958::
935:.
913::
886:.
866::
843:.
831::
808:.
784::
757:.
751::
724:.
700::
670:.
648::
640::
613:.
593::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.