278:
189:
581:
394:
24:
793:, β-galactosidase may be used as a reporter to identify proteins that interact with each other. In this method, genome libraries may be screened for protein interaction using yeast or bacterial system. Where there is a successful interaction between proteins being screened, it will result to the binding of an activation domain to a promoter. If the promoter is linked to a
652:-1-thiogalactopyranoside). IPTG is a chemical structure analogue of lactose. However, IPTG cannot be hydrolyzed by β-galactosidase. IPTG is used as an inducer that binds to lac repressor releasing the DNA and allowing transcription. The presence of IPTG in the agar plate therefore increases the synthesis of β-galactosidase.
797:
gene, the production of β-galactosidase, which results in the formation of blue-pigmented colonies in the presence of X-gal, will therefore indicate a successful interaction between proteins. This technique may be limited to screening libraries of size of less than around 10. The successful cleavage
622:
cells, they form a functional β-galactosidase. The presence of an active β-galactosidase may be detected when cells are grown in plates containing X-gal, the blue-colored product precipitated within cells resulted in the characteristic blue colonies. However, the multiple cloning site, where a gene
631:
gene, α-complementation is therefore also disrupted and no functional β-galactosidase can form, resulting in white colonies. Cells containing successfully ligated insert can then be easily identified by its white coloration from the unsuccessful blue ones. Example of cloning vectors used for this
556:, an intensely blue product which is insoluble. X-gal itself is colorless, so the presence of blue-colored product may therefore be used as a test for the presence of active β-galactosidase. This also allows for bacterial β-galactosidase (so called
126:
453:
and collaborators in 1964. The formal chemical name is often shortened to less accurate but also less cumbersome phrases such as bromochloroindoxyl galactoside. The X from
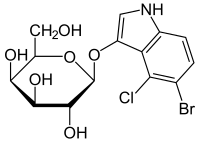
504:
927:"Development of a semi-quantitative plate-based alpha-galactosidase gene reporter for Schizosaccharomyces pombe and its use to isolate a constitutively active Mam2"
407:
614:
gene (lacZΔM15) in the cell. Both genes by themselves produce non-functional peptides, however, when expressed together, as when a plasmid containing
660:
X-gal has a number of variants, which are similar molecules with slight differences serving mainly to produce colors other than blue as a signal.
637:
301:
InChI=1S/C14H15BrClNO6/c15-5-1-2-6-9(10(5)16)7(3-17-6)22-14-13(21)12(20)11(19)8(4-18)23-14/h1-3,8,11-14,17-21H,4H2/t8-,11+,12+,13-,14-/m1/s1
327:
311:
InChI=1/C14H15BrClNO6/c15-5-1-2-6-9(10(5)16)7(3-17-6)22-14-13(21)12(20)11(19)8(4-18)23-14/h1-3,8,11-14,17-21H,4H2/t8-,11+,12+,13-,14-/m1/s1
606:
The blue/white screening method relies on the principle of α-complementation of the β-galactosidase gene, where a fragment of the
842:
Kiernan JA 2007. Indigogenic substrates for detection and localization of enzymes. Biotechnic & Histochemistry 82(2): 73-103.
603:. This method of screening is a convenient way of distinguishing a successful cloning product from other unsuccessful ones.
909:
829:
Horwitz JP et al., 1964. Substrates for cytochemical demonstration of enzyme activity. I. Some substituted 3-indolyl-β-
292:
1111:
414:
402:
1132:
1090:
1027:
54:
544:-lactose. X-gal, when cleaved by β-galactosidase, yields galactose and 5-bromo-4-chloro-3-hydroxyindole -
235:
184:
465:, in the place of its usual target, a β-galactoside. It is also used to detect activity of this enzyme in
256:
273:
1069:
1048:
473:. X-gal is one of many indoxyl glycosides and esters that yield insoluble blue compounds similar to
202:
1158:"A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions"
513:
196:
508:(Xαgal, X-alpha-gal), or 5-Bromo-4-chloro-3-indolyl-α-D-galactopyranoside, which is hydrolyzed by
1230:
1220:
489:
36:
1235:
1169:
244:
166:
876:
852:
548:. The latter then spontaneously dimerizes and is oxidized into 5,5'-dibromo-4,4'-dichloro-
102:
8:
790:
92:
1173:
277:
188:
146:
962:
596:
533:
462:
568:
509:
1197:
1192:
1157:
954:
946:
905:
600:
525:
458:
966:
1225:
1187:
1177:
938:
592:
438:
350:
224:
537:
595:, X-gal is used as a visual indication of whether a cell expresses a functional
466:
450:
385:
1214:
950:
561:
177:
457:
may be the source of the X in the X-gal contraction. X-gal is often used in
1201:
1182:
958:
926:
853:"Information on EC 3.2.1.22 - alpha-galactosidase - BRENDA Enzyme Database"
470:
877:"Information on EC 3.2.1.23 - beta-galactosidase - BRENDA Enzyme Database"
798:
of X-gal also creates a noticeably foul odor due to the volatilization of
623:
of interest may be ligated into the plasmid vector, is located within the
1004:
648:
gene. Often, the plate containing X-Gal also contains IPTG (isopropyl β-
549:
474:
373:
157:
942:
481:
442:
980:
384:
Except where otherwise noted, data are given for materials in their
1005:"Blue/White Cloning of a DNA Fragment and Assay of β-Galactosidase"
125:
580:
529:
454:
211:
811:
799:
446:
833:-glycopyranosides. Journal of Medicinal Chemistry 7: 574-575.
633:
137:
115:
261:
640:, pGem-T Vectors, and it also requires the use of specific
611:
607:
610:
gene (lacZα) in the plasmid can complement another mutant
925:
Goddard, Alan; Ladds, Graham; Davey, John (2005-01-15).
23:
492:
644:
host strains such as DH5α which carries the mutant
567:
Similarly, Xαgal is used as a reporter compound for
516:
3.2.1.22) instead of β-galactosidase (EC 3.2.1.23).
904:. I K International Publishing House. p. 116.
498:
924:
627:gene. Successful ligation therefore disrupts the
1212:
1091:"5-Bromo-6-chloro-3-indolyl-β-D-glucopyranoside"
223:
784:
101:
1155:
1133:"4-Methylumbelliferyl β-D-galactopyranoside"
477:as a result of enzyme-catalyzed hydrolysis.
726:Magenta glucoside, Magenta-GLC, Magenta gal
276:
187:
165:
1191:
1181:
1028:"5-Bromo-3-indolyl β-D-galactopyranoside"
532:, and therefore may be hydrolyzed by the
243:
1151:
1149:
461:to test for the presence of an enzyme,
272:
201:
79:)-2--6-(hydroxymethyl)oxane-3,4,5-triol
1213:
899:
178:
1146:
304:Key: OPIFSICVWOWJMJ-AEOCFKNESA-N
145:
692:Rose-Gal, Salmon-Gal, Y-Gal, Red-Gal
480:A less often used but very similar (
335:Brc3ccc2c(c(O1O((O)(O)1O)CO)c2)c3Cl
314:Key: OPIFSICVWOWJMJ-AEOCFKNEBZ
214:
13:
449:. The compound was synthesized by
14:
1247:
433:for 5-bromo-4-chloro-3-indolyl-β-
1156:Joung J, Ramm E, Pabo C (2000).
579:
392:
22:
1125:
1104:
1083:
1062:
1041:
388:(at 25 °C , 100 kPa).
1112:"Green-β-D-Gal - Biotium, Inc"
1020:
997:
973:
918:
893:
869:
845:
836:
823:
1:
1070:"Purple-beta-D-Gal - PubChem"
900:Sandhu, Sardul Singh (2010).
817:
729:5-Bromo-6-chloro-3-indolyl-β-
599:enzyme in a technique called
785:Protein-protein interactions
7:
931:Yeast (Chichester, England)
805:
655:
574:
536:enzyme which cleaves the β-
10:
1252:
902:Recombinant DNA Technology
586:
437:-galactopyranoside) is an
564:in various applications.
382:
343:
323:
288:
85:
53:
35:
30:
21:
445:linked to a substituted
763:4-Methylumbelliferyl β-
519:
499:{\displaystyle \alpha }
1183:10.1073/pnas.110149297
1162:Proc Natl Acad Sci USA
1049:"Salmon-Gal - PubChem"
881:www.brenda-enzymes.org
857:www.brenda-enzymes.org
618:is transformed into a
571:(e.g. Mel1 in yeast).
500:
695:6-Chloro-3-indolyl-β-
501:
378:408.629
55:Systematic IUPAC name
680:5-Bromo-3-indolyl β-
601:blue/white screening
490:
1174:2000PNAS...97.7382J
791:two-hybrid analysis
714:5-Iodo-3-indolyl-β-
18:
767:-galactopyranoside
752:-galactopyranoside
748:N-Methylindolyl-β-
718:-galactopyranoside
699:-galactopyranoside
684:-galactopyranoside
677:Blue-Gal, Bluo-Gal
560:) to be used as a
496:
429:(also abbreviated
415:Infobox references
48:-galactopyranoside
40:5-Bromo-4-chloro-1
16:
782:
781:
459:molecular biology
423:Chemical compound
421:
420:
257:CompTox Dashboard
127:Interactive image
1243:
1206:
1205:
1195:
1185:
1153:
1144:
1143:
1141:
1139:
1129:
1123:
1122:
1120:
1118:
1108:
1102:
1101:
1099:
1097:
1087:
1081:
1080:
1078:
1076:
1066:
1060:
1059:
1057:
1055:
1045:
1039:
1038:
1036:
1034:
1024:
1018:
1017:
1015:
1014:
1009:
1001:
995:
994:
992:
991:
981:"IPTG - Bioline"
977:
971:
970:
943:10.1002/yea.1190
922:
916:
915:
897:
891:
890:
888:
887:
873:
867:
866:
864:
863:
849:
843:
840:
834:
832:
827:
766:
751:
744:
733:-glucopyranoside
732:
717:
710:
698:
683:
663:
662:
651:
583:
543:
505:
503:
502:
497:
439:organic compound
436:
405:
399:
396:
395:
351:Chemical formula
281:
280:
265:
263:
247:
227:
216:
205:
191:
180:
169:
149:
129:
105:
47:
26:
19:
15:
1251:
1250:
1246:
1245:
1244:
1242:
1241:
1240:
1211:
1210:
1209:
1154:
1147:
1137:
1135:
1131:
1130:
1126:
1116:
1114:
1110:
1109:
1105:
1095:
1093:
1089:
1088:
1084:
1074:
1072:
1068:
1067:
1063:
1053:
1051:
1047:
1046:
1042:
1032:
1030:
1026:
1025:
1021:
1012:
1010:
1007:
1003:
1002:
998:
989:
987:
985:www.bioline.com
979:
978:
974:
923:
919:
912:
898:
894:
885:
883:
875:
874:
870:
861:
859:
851:
850:
846:
841:
837:
830:
828:
824:
820:
808:
787:
777:
773:
764:
749:
742:
730:
715:
708:
696:
681:
658:
649:
597:β-galactosidase
589:
577:
569:α-galactosidase
541:
538:glycosidic bond
534:β-galactosidase
522:
510:α-galactosidase
491:
488:
487:
463:β-galactosidase
434:
424:
417:
412:
411:
410: ?)
401:
397:
393:
389:
367:
363:
359:
353:
339:
336:
331:
330:
319:
316:
315:
312:
306:
305:
302:
296:
295:
284:
274:DTXSID901349452
266:
259:
250:
230:
217:
172:
152:
132:
119:
108:
95:
81:
80:
49:
45:
12:
11:
5:
1249:
1239:
1238:
1233:
1228:
1223:
1208:
1207:
1168:(13): 7382–7.
1145:
1124:
1103:
1082:
1061:
1040:
1019:
996:
972:
917:
911:978-9380578446
910:
892:
868:
844:
835:
821:
819:
816:
815:
814:
807:
804:
786:
783:
780:
779:
775:
771:
770:Fluorescent (λ
768:
761:
757:
756:
753:
746:
738:
737:
734:
727:
723:
722:
719:
712:
704:
703:
700:
693:
689:
688:
685:
678:
674:
673:
670:
667:
657:
654:
588:
585:
576:
573:
521:
518:
495:
484:) compound is
467:histochemistry
451:Jerome Horwitz
441:consisting of
422:
419:
418:
413:
391:
390:
386:standard state
383:
380:
379:
376:
370:
369:
365:
361:
357:
354:
349:
346:
345:
341:
340:
338:
337:
334:
326:
325:
324:
321:
320:
318:
317:
313:
310:
309:
307:
303:
300:
299:
291:
290:
289:
286:
285:
283:
282:
269:
267:
255:
252:
251:
249:
248:
240:
238:
232:
231:
229:
228:
220:
218:
210:
207:
206:
199:
193:
192:
182:
174:
173:
171:
170:
162:
160:
154:
153:
151:
150:
142:
140:
134:
133:
131:
130:
122:
120:
113:
110:
109:
107:
106:
98:
96:
91:
88:
87:
83:
82:
58:
57:
51:
50:
44:-indol-3-yl β-
39:
33:
32:
28:
27:
9:
6:
4:
3:
2:
1248:
1237:
1234:
1232:
1229:
1227:
1224:
1222:
1219:
1218:
1216:
1203:
1199:
1194:
1189:
1184:
1179:
1175:
1171:
1167:
1163:
1159:
1152:
1150:
1134:
1128:
1113:
1107:
1092:
1086:
1071:
1065:
1050:
1044:
1029:
1023:
1006:
1000:
986:
982:
976:
968:
964:
960:
956:
952:
948:
944:
940:
936:
932:
928:
921:
913:
907:
903:
896:
882:
878:
872:
858:
854:
848:
839:
826:
822:
813:
810:
809:
803:
801:
796:
792:
769:
762:
759:
758:
754:
747:
740:
739:
735:
728:
725:
724:
720:
713:
706:
705:
701:
694:
691:
690:
686:
679:
676:
675:
671:
668:
665:
664:
661:
653:
647:
643:
639:
635:
630:
626:
621:
617:
613:
609:
604:
602:
598:
594:
584:
582:
572:
570:
565:
563:
559:
555:
551:
547:
539:
535:
531:
527:
517:
515:
511:
507:
493:
483:
478:
476:
472:
468:
464:
460:
456:
452:
448:
444:
440:
432:
428:
416:
409:
404:
387:
381:
377:
375:
372:
371:
355:
352:
348:
347:
342:
333:
332:
329:
322:
308:
298:
297:
294:
287:
279:
275:
271:
270:
268:
258:
254:
253:
246:
242:
241:
239:
237:
234:
233:
226:
222:
221:
219:
213:
209:
208:
204:
200:
198:
195:
194:
190:
186:
183:
181:
179:ECHA InfoCard
176:
175:
168:
164:
163:
161:
159:
156:
155:
148:
144:
143:
141:
139:
136:
135:
128:
124:
123:
121:
117:
112:
111:
104:
100:
99:
97:
94:
90:
89:
84:
78:
74:
70:
66:
62:
56:
52:
43:
38:
34:
29:
25:
20:
1231:Chloroarenes
1221:Galactosides
1165:
1161:
1136:. Retrieved
1127:
1115:. Retrieved
1106:
1094:. Retrieved
1085:
1073:. Retrieved
1064:
1052:. Retrieved
1043:
1031:. Retrieved
1022:
1011:. Retrieved
999:
988:. Retrieved
984:
975:
937:(1): 31–41.
934:
930:
920:
901:
895:
884:. Retrieved
880:
871:
860:. Retrieved
856:
847:
838:
825:
794:
788:
659:
645:
641:
628:
624:
619:
615:
605:
593:gene cloning
590:
578:
566:
557:
553:
545:
524:X-gal is an
523:
485:
479:
471:bacteriology
430:
426:
425:
86:Identifiers
76:
72:
68:
64:
60:
41:
1236:Bromoarenes
638:pBluescript
344:Properties
185:100.027.855
147:CHEBI:75055
1215:Categories
1138:4 February
1117:4 February
1096:4 February
1075:4 February
1054:4 February
1033:4 February
1013:2023-09-10
990:2018-05-15
886:2023-07-31
862:2023-07-31
818:References
687:Dark blue
666:Short name
475:indigo dye
374:Molar mass
245:V595OG374W
158:ChemSpider
114:3D model (
93:CAS Number
37:IUPAC name
951:0749-503X
760:MUG, MUGA
707:Purple-β-
669:Long name
632:test are
494:α
443:galactose
103:7240-90-6
1202:10852947
967:33926866
959:15580593
806:See also
774:= 365, λ
741:Green-β-
736:Magenta
656:Variants
646:lacZΔM15
620:lacZΔM15
575:Reaction
562:reporter
1226:Indoles
1170:Bibcode
778:= 455)
721:Purple
642:E. coli
587:Cloning
530:lactose
455:indoxyl
408:what is
406: (
368:
212:PubChem
1200:
1190:
965:
957:
949:
908:
812:X-Gluc
800:indole
755:Green
672:Color
550:indigo
526:analog
482:chiral
447:indole
403:verify
400:
364:BrClNO
328:SMILES
31:Names
17:X-gal
1193:16554
1008:(PDF)
963:S2CID
702:Pink
634:pUC19
629:lacZα
625:lacZα
616:lacZα
558:lacZ
427:X-gal
293:InChI
225:65181
203:X-gal
167:58680
138:ChEBI
116:JSmol
1198:PMID
1140:2014
1119:2014
1098:2014
1077:2014
1056:2014
1035:2014
955:PMID
947:ISSN
906:ISBN
795:lacZ
745:-Gal
711:-Gal
612:lacZ
608:lacZ
520:Uses
506:-gal
469:and
431:BCIG
236:UNII
197:MeSH
1188:PMC
1178:doi
939:doi
789:In
591:In
540:in
528:of
262:EPA
215:CID
1217::
1196:.
1186:.
1176:.
1166:97
1164:.
1160:.
1148:^
983:.
961:.
953:.
945:.
935:22
933:.
929:.
879:.
855:.
802:.
776:em
772:ex
636:,
552:-
514:EC
486:X-
362:15
358:14
75:,6
71:,5
67:,4
63:,3
59:(2
1204:.
1180::
1172::
1142:.
1121:.
1100:.
1079:.
1058:.
1037:.
1016:.
993:.
969:.
941::
914:.
889:.
865:.
831:D
765:D
750:D
743:D
731:D
716:D
709:D
697:D
682:D
650:D
554:2
546:1
542:D
512:(
435:D
398:N
366:6
360:H
356:C
264:)
260:(
118:)
77:R
73:R
69:S
65:R
61:S
46:D
42:H
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.