1776:
1817:
1644:
1608:
1089:
1165:
1940:
1716:
1540:
945:
1214:
1845:
1680:
1752:
1504:
1576:
1464:
1881:
81:
280:, and double bond geometry. These are structural features that cannot be specified by connectivity alone, and therefore SMILES which encode this information are termed isomeric SMILES. A notable feature of these rules is that they allow rigorous partial specification of chirality. The term isomeric SMILES is also applied to SMILES in which
1971:
The SMILES notation is described extensively in the SMILES theory manual provided by
Daylight Chemical Information Systems and a number of illustrative examples are presented. Daylight's depict utility provides users with the means to check their own examples of SMILES and is a valuable educational
241:
algorithm used to generate it, and is termed the canonical SMILES. These algorithms first convert the SMILES to an internal representation of the molecular structure; an algorithm then examines that structure and produces a unique SMILES string. Various algorithms for generating canonical SMILES
217:
The term SMILES refers to a line notation for encoding molecular structures and specific instances should strictly be called SMILES strings. However, the term SMILES is also commonly used to refer to both a single SMILES string and a number of SMILES strings; the exact meaning is usually apparent
334:) based on the main principle of chemoinformatics that similar molecules have similar properties. The predictive models implemented a syntactic pattern recognition approach (which involved defining a molecular distance) as well as a more robust scheme based on statistical pattern recognition.
2025:
SMILES can be converted back to two-dimensional representations using structure diagram generation (SDG) algorithms. This conversion is sometimes ambiguous. Conversion to three-dimensional representation is achieved by energy-minimization approaches. There are many downloadable and web-based
2016:
SMILES corresponds to discrete molecular structures. However many materials are macromolecules, which are too large (and often stochastic) to conveniently generate SMILES for. BigSMILES is an extension of SMILES that aims to provide an efficient representation system for macromolecules.
715:
SMILES does not require that ring numbers be used in any particular order, and permits ring number zero, although this is rarely used. Also, it is permitted to reuse ring numbers after the first ring has closed, although this usually makes formulae harder to read. For example,
1198:. Consider the four bonds in the order in which they appear, left to right, in the SMILES form. Looking toward the central carbon from the perspective of the first bond, the other three are either clockwise or counter-clockwise. These cases are indicated with
1991:
searching. One common misconception is that SMARTS-based substructural searching involves matching of SMILES and SMARTS strings. In fact, both SMILES and SMARTS strings are first converted to internal graph representations which are searched for
2423:
Byers JA, Birgersson G, Löfqvist J, Appelgren M, Bergström G (March 1990). "Isolation of pheromone synergists of bark beetle, Pityogenes chalcographus, from complex insect-plant odors by fractionation and subtractive-combination bioassay".
1285:
While the order in which branches are specified in SMILES is normally unimportant, in this case it matters; swapping any two groups requires reversing the chirality indicator. If the branches are reversed so alanine is written as
237:. Algorithms have been developed to generate the same SMILES string for a given molecule; of the many possible strings, these algorithms choose only one of them. This SMILES is unique for each structure, although dependent on the
273:, 1,2-dicyclopropylethane) and cannot be considered a correct method for representing a graph canonically. There is currently no systematic comparison across commercial software to test if such flaws exist in those packages.
204:
as a standard for formula representation. SMILES is generally considered to have the advantage of being more human-readable than InChI; it also has a wide base of software support with extensive theoretical backing (such as
329:
From the view point of a formal language theory, SMILES is a word. A SMILES is parsable with a context-free parser. The use of this representation has been in the prediction of biochemical properties (incl. toxicity and
1157:. When alternating single-double bonds are present, the groups are larger than two, with the middle directional symbols being adjacent to two double bonds. For example, the common form of (2,4)-hexadiene is written
2195:
The
History and Heritage of Scientific and Technological Information Systems: Proceedings of the 2002 Conference of the American Society of Information Science and Technology and the Chemical Heritage Foundation
977:. The first atom within the parentheses, and the first atom after the parenthesized group, are both bonded to the same branch point atom. The bond symbol must appear inside the parentheses; outside (E.g.:
1019:, or the like. Generally, a SMILES form is easiest to read if the simpler branch comes first, with the final, unparenthesized portion being the most complex. The only caveats to such rearrangements are:
218:
from the context. The terms "canonical" and "isomeric" can lead to some confusion when applied to SMILES. The terms describe different attributes of SMILES strings and are not mutually exclusive.
170:
269:
The original paper that described the CANGEN algorithm claimed to generate unique SMILES strings for graphs representing molecules, but the algorithm fails for a number of simple cases (e.g.
2002:
SMIRKS, a superset of "reaction SMILES" and a subset of "reaction SMARTS", is a line notation for specifying reaction transforms. The general syntax for the reaction extensions is
486:). If there is more than one charge, it is normally written as digit; however, it is also possible to repeat the sign as many times as the ion has charges: one may write either
162:) for supporting the work, and Arthur Weininger (Pomona; Daylight CIS) and Jeremy Scofield (Cedar River Software, Renton, WA) for assistance in programming the system." The
1023:
If ring numbers are reused, they are paired according to their order of appearance in the SMILES string. Some adjustments may be required to preserve the correct pairing.
17:
419:
have the number of hydrogens attached implied by the SMILES valence model (typically their normal valence, but for N and P it is 3 or 5, and for S it is 2, 4 or 6), and
3351:
1809:
307:. Where cycles have been broken, numeric suffix labels are included to indicate the connected nodes. Parentheses are used to indicate points of branching on the tree.
1983:
is a line notation for specification of substructural patterns in molecules. While it uses many of the same symbols as SMILES, it also allows specification of
934:
is required. (In fact, most SMILES software can correctly infer that the bond between the two rings cannot be aromatic and so will accept the nonstandard form
2378:
Sidorova J, Garcia J (November 2015). "Bridging from syntactic to statistical methods: Classification with automatically segmented features from sequences".
1775:
681:
Ring structures are written by breaking each ring at an arbitrary point (although some choices will lead to a more legible SMILES than others) to make an
2637:
1317:
Normally, the first of the four bonds appears to the left of the carbon atom, but if the SMILES is written beginning with the chiral carbon, such as
561:
atoms are assumed to be single unless specified otherwise and are implied by adjacency in the SMILES string. Although single bonds may be written as
197:
1299:
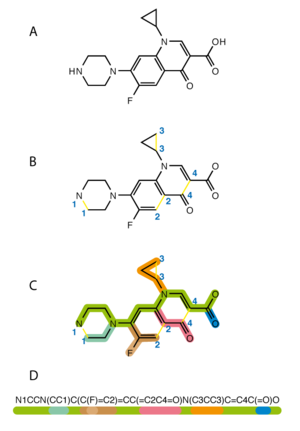
1255:
1279:
997:
989:
1895:
1520:
459:
is added if the atom in brackets is bonded to one or more hydrogen, followed by the number of hydrogen atoms if greater than 1, then by the sign
2844:
2273:
2255:
735:, where the final carbon participates in both ring-closing bonds 1 and 2. If two-digit ring numbers are required, the label is preceded by
2791:
1797:
2186:
2101:
Weininger D (February 1988). "SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules".
1000:) which encode the 3 and 4-cyanoanisole isomers. Writing SMILES for substituted rings in this way can make them more human-readable.
731:
Multiple digits after a single atom indicate multiple ring-closing bonds. For example, an alternative SMILES notation for decalin is
255:
186:
1951:
CC(C)(O1)C(O)1(O2)(C)3CC=C43(C2)C(=O)C54CC(C6)5(C)Cc(n7)c6nc(C89(C))c7C8CC%109C(O)%11(C)C%10=C(O%12)%11(O)(C)%12(O%13)(O)C%13(C)CO
1137:
1119:
3173:
1340:
247:
154:
in the 1980s. Acknowledged for their parts in the early development were "Gilman Veith and Rose Russo (USEPA) and Albert Leo and
437:
All other elements must be enclosed in brackets, and have charges and hydrogens shown explicitly. For instance, the SMILES for
800:
Choosing a ring-break point adjacent to attached groups can lead to a simpler SMILES form by avoiding branches. For example,
3543:
3316:
2630:
2604:
2320:
2207:
2035:
1980:
292:
In terms of a graph-based computational procedure, SMILES is a string obtained by printing the symbol nodes encountered in a
2006:(without spaces), where any of the fields can either be left blank or filled with multiple molecules delineated with a dot (
2269:
3648:
1816:
746:
Either or both of the digits may be preceded by a bond type to indicate the type of the ring-closing bond. For example,
877:
In the latter case, bonds between two aromatic atoms are assumed (if not explicitly shown) to be aromatic bonds. Thus,
2998:
1643:
1607:
1054:. Choosing ring-closing bonds adjacent to branch points can reduce the number of parentheses required. For example,
2758:
2047:
201:
2131:
Weininger D, Weininger A, Weininger JL (May 1989). "SMILES. 2. Algorithm for generation of unique SMILES notation".
3603:
2784:
2623:
2010:), and other descriptions dependent on the base language. Atoms can additionally be identified with a number (e.g.
130:
The original SMILES specification was initiated in the 1980s. It has since been modified and extended. In 2007, an
1939:
3608:
3232:
3133:
163:
1751:
3843:
3833:
3370:
2343:
Sidorova J, Anisimova M (August 2014). "NLP-inspired structural pattern recognition in chemical application".
2224:
1715:
1539:
941:
The
Daylight and OpenEye algorithms for generating canonical SMILES differ in their treatment of aromaticity.
3793:
3461:
3301:
3078:
1993:
808:; choosing a different ring-break location produces a branched structure that requires parentheses to write.
2516:
3633:
3513:
3143:
1679:
2777:
1149:
Bond direction symbols always come in groups of at least two, of which the first is arbitrary. That is,
1132:, in which the fluorine atoms are on opposite sides of the double bond (as shown in the figure), whereas
304:
243:
3558:
2246:
1844:
1004:
303:. The chemical graph is first trimmed to remove hydrogen atoms and cycles are broken to turn it into a
1503:
3783:
3487:
3153:
2839:
2301:"Assigning Unique Keys to Chemical Compounds for Data Integration: Some Interesting Counter Examples"
2067:
259:
112:
685:
structure and adding numerical ring closure labels to show connectivity between non-adjacent atoms.
2992:
2057:
1930:
1575:
1463:
984:
Substituted rings can be written with the branching point in the ring as illustrated by the SMILES
251:
3113:
2882:
2800:
1880:
277:
262:. A common application of canonical SMILES is indexing and ensuring uniqueness of molecules in a
221:
Typically, a number of equally valid SMILES strings can be written for a molecule. For example,
182:
31:
2193:
3658:
3548:
3492:
1354:
are specified with a number equal to the integer isotopic mass preceding the atomic symbol.
2158:
Weininger D (August 1990). "SMILES. 3. DEPICT. Graphical depiction of chemical structures".
3293:
3214:
2931:
2874:
2722:
2387:
2352:
1129:
801:
72:
2494:
8:
3737:
3573:
3167:
3108:
3017:
2986:
2727:
2307:. Lecture Notes in Computer Science. Vol. 3615. Berlin: Springer. pp. 145–157.
2041:
1142:
682:
190:
2391:
2356:
849:
Most commonly, by writing the constituent B, C, N, O, P and S atoms in lower-case forms
3838:
3672:
3553:
3446:
3321:
3246:
2952:
2926:
2682:
2564:
2539:
2538:
Lin TS, Coley CW, Mochigase H, Beech HK, Wang W, Wang Z, et al. (September 2019).
2449:
1984:
1431:
1146:-1,2-difluoroethylene, in which the fluorines are on the same side of the double bond.
558:
293:
45:
1179:
has a very long backbone of alternating single and double bonds, which may be written
3578:
3345:
3088:
3012:
2967:
2906:
2695:
2672:
2667:
2600:
2569:
2441:
2316:
2203:
2199:
1988:
1903:
1411:
1367:
263:
151:
770:
is illegal, as it explicitly specifies conflicting types for the ring-closing bond.
3799:
3476:
3406:
3401:
2892:
2769:
2700:
2652:
2592:
2587:
Helson HE (1999). "Structure
Diagram Generation". In Lipkowitz KB, Boyd DB (eds.).
2559:
2551:
2467:
2433:
2403:
2395:
2360:
2308:
2167:
2140:
2110:
1083:
629:
621:
438:
351:
331:
238:
105:
2453:
828:
754:, but if the double bond is chosen as the ring-closing bond, it may be written as
3788:
3222:
2921:
2809:
2748:
2714:
2690:
2399:
2364:
640:
147:
120:
116:
1181:
CC1CCC/C(C)=C1/C=C/C(C)=C/C=C/C(C)=C/C=C/C=C(C)/C=C/C=C(C)/C=C/C2=C(C)/CCCC2(C)C
448:. Hydrogen may also be written as a separate atom; water may also be written as
242:
have been developed and include those by
Daylight Chemical Information Systems,
3714:
3613:
3083:
2555:
2277:
2061:
1339:
symbol to indicate stereochemistry around more complex chiral centers, such as
966:
609:
300:
296:
159:
2596:
3827:
3482:
3255:
3194:
3181:
2818:
2540:"BigSMILES: A Structurally-Based Line Notation for Describing Macromolecules"
1588:
1176:
1168:
1088:
589:
413:
174:
155:
131:
101:
85:
2248:
SMILES: A line notation and computerized interpreter for chemical structures
2064:
allowing also numerical properties, e.g. physicochemical values or distances
1164:
639:, to indicate that two parts are not bonded together. For example, aqueous
3742:
3148:
3118:
2834:
2732:
2573:
2445:
2038:(SMARTS), an extension of SMILES for specification of substructural queries
1926:
1923:
1769:
1187:
1124:
1100:
773:
Ring-closing bonds may not be used to denote multiple bonds. For example,
747:
717:
430:
206:
178:
2615:
1324:
bond in this case) is used as the reference to order the following three:
324:
3568:
3563:
2887:
1996:
1114:
to show directional single bonds adjacent to a double bond. For example,
816:
689:
135:
2312:
2300:
2258:, Environmental Research Laboratory-Duluth. Report No. EPA/600/M-87/021.
2171:
2144:
2114:
944:
3732:
3688:
3497:
3123:
3098:
2859:
2437:
2408:
1987:
atoms and bonds, which can be used to define substructural queries for
1805:
1396:
1247:
1225:
1213:
974:
386:
181:
open-source chemistry community. Other 'linear' notations include the
57:
3809:
3719:
3698:
3693:
3643:
3623:
2829:
2073:
1801:
1765:
1656:
1624:
1480:
1359:
1240:
912:
704:
respectively. For a second ring, the label will be 2. For example,
514:
507:
497:
124:
1106:
Configuration around double bonds is specified using the characters
3772:
3727:
3638:
3628:
3598:
3466:
3391:
3386:
3189:
3128:
3093:
3037:
2947:
1899:
1857:
1728:
1556:
1457:
1351:
923:
882:
782:
493:
471:
398:
394:
378:
169:
It has since been modified and extended by others, most notably by
1959:
appears in front of the index of ring closure labels above 9; see
3426:
3411:
3268:
3199:
3057:
3047:
3032:
2972:
2962:
2957:
2472:
1829:
1692:
1355:
1228:
1055:
905:
878:
820:
790:
705:
693:
566:
423:
402:
270:
234:
2422:
1038:
require parentheses are ring-closing bonds: the SMILES fragment
922:
When aromatic atoms are singly bonded to each other, such as in
3804:
3653:
3618:
3593:
3588:
3583:
3538:
3533:
3528:
3456:
3451:
3441:
3421:
3381:
3376:
3365:
3326:
3311:
3283:
3278:
3273:
3227:
3138:
3062:
3027:
2849:
2753:
2262:
2077:
1321:, then all four are to the right, but the first to appear (the
1026:
If stereochemistry is specified, adjustments must be made; see
661:
Single bonds adjacent to double bonds may be represented using
533:
529:
406:
390:
382:
374:
281:
3767:
3762:
3523:
3518:
3471:
3431:
3416:
3306:
3204:
3103:
3052:
2864:
2854:
2081:
2051:
1732:
930:. This is one of the few cases where the single bond symbol
886:
500:
370:
361:. Brackets may be omitted in the common case of atoms which:
109:
317:
of the starting atom used for the depth-first traversal, and
3436:
3396:
3331:
3263:
3042:
3022:
2130:
635:
An additional type of bond is a "non-bond", indicated with
366:
358:
347:
320:
of the order in which branches are listed when encountered.
3352:
List of quantum chemistry and solid-state physics software
2187:"The Entrance of Informatics into Combinatorial Chemistry"
1894:
To illustrate a molecule with more than 9 rings, consider
150:
at the USEPA Mid-Continent
Ecology Division Laboratory in
88:: break cycles, then write as branches off a main backbone
1258:). Looking from the nitrogen–carbon bond, the hydrogen (
80:
1946:
Starting with the left-most methyl group in the figure:
650:
An aromatic "one and a half" bond may be indicated with
565:, this is usually omitted. For example, the SMILES for
2591:. Vol. 13. New York: Wiley-VCH. pp. 313–398.
2225:"Acknowledgements on Daylight Tutorial smiles-etc page"
1099:
SMILES permits, but does not require, specification of
325:
SMILES definition as strings of a context-free language
2537:
2244:
1650:
CC1=C(C(=O)C1OC(=O)2(C2(C)C)/C=C(\C)/C(=O)OC)C/C=C\C=C
1335:
The SMILES specification includes elaborations on the
2103:
Journal of
Chemical Information and Computer Sciences
2799:
1003:
Branches may be written in any order. For example,
831:
form with alternating single and double bonds, e.g.
350:
are represented by the standard abbreviation of the
146:The original SMILES specification was initiated by
1066:, avoiding the parentheses required if written as
904:Aromatic nitrogen bonded to hydrogen, as found in
310:The resultant SMILES form depends on the choices:
2342:
2298:
789:is a peculiar but legal alternative way to write
3825:
2184:
2126:
2124:
961:Branches are described with parentheses, as in
550:A bond is represented using one of the symbols
2460:
2377:
1210:symbol itself is a counter-clockwise spiral).
889:can be represented respectively by the SMILES
669:to indicate stereochemical configuration; see
166:funded the initial project to develop SMILES.
2785:
2631:
2216:
2151:
2121:
2094:
785:. However, they may be used with non-bonds;
30:"SMILES" redirects here. For other uses, see
2519:. Daylight Chemical Information Systems, Inc
2497:. Daylight Chemical Information Systems, Inc
2238:
2160:Journal of Chemical Information and Modeling
2133:Journal of Chemical Information and Modeling
276:SMILES notation allows the specification of
94:Simplified Molecular Input Line Entry System
18:Simplified molecular-input line-entry system
2645:
2580:
2178:
1966:
1686:O1C=C(1O2)c3c2cc(OC)c4c3OC(=O)C5=C4CCC(=O)5
1171:, with the eleven double bonds highlighted.
2792:
2778:
2638:
2624:
2245:Anderson E, Veith GD, Weininger D (1987).
926:, a single bond must be shown explicitly:
743:is a single ring-closing bond of ring 12.
604:respectively as illustrated by the SMILES
2563:
2407:
2222:
2157:
2100:
708:(decahydronaphthalene) may be written as
287:
177:called "OpenSMILES" was developed by the
115:. SMILES strings can be imported by most
1290:, then the configuration also reverses;
1212:
1163:
1087:
943:
79:
3174:List of protein-ligand docking software
1758:OC(O1)(O)(O)21c3c(O)c(OC)c(O)cc3C(=O)O2
1341:trigonal bipyramidal molecular geometry
134:called OpenSMILES was developed in the
14:
3826:
2586:
823:may be written in one of three forms:
467:for a negative charge. For example,
100:) is a specification in the form of a
2773:
2619:
2305:Data Integration in the Life Sciences
2299:Neglur G, Grossman RL, Liu B (2005).
2036:SMILES arbitrary target specification
1302:). Other ways of writing it include
171:Daylight Chemical Information Systems
1140:) is one possible representation of
1027:
670:
314:of the bonds chosen to break cycles,
278:configuration at tetrahedral centers
2192:. In Rayward WB, Bowden ME (eds.).
2070:, 2D layout and conversion software
1046:, both denoting a bond between the
948:Visualization of 3-cyanoanisole as
766:. (The first form is preferred.)
455:When brackets are used, the symbol
27:Chemical species structure notation
24:
2999:List of molecular graphics systems
2589:Reviews in Computational Chemistry
2270:"SMILES Tutorial: What is SMILES?"
1077:
1034:The one form of branch which does
25:
3855:
2048:International Chemical Identifier
915:is written in SMILES notation as
2013:) for mapping, for example in .
1938:
1879:
1843:
1815:
1774:
1750:
1714:
1678:
1642:
1606:
1574:
1538:
1502:
1462:
724:, but it may also be written as
655:
104:for describing the structure of
84:SMILES generation algorithm for
3233:Molecular Operating Environment
3134:Molecular Operating Environment
2531:
2509:
2487:
2416:
2371:
1782:CC(=O)OCCC(/C)=C\C(C(C)=C)CCC=C
838:Using the aromatic bond symbol
592:are represented by the symbols
164:Environmental Protection Agency
2826:Avalon Cheminformatics Toolkit
2336:
2292:
1618:CCC(O)CC/C=C/C=C/C#CC#C/C=C/CO
1614:CCC(O)CC\C=C\C=C\C#CC#C\C=C\CO
1231:. One of its SMILES forms is
811:
777:is not a valid alternative to
588:Double, triple, and quadruple
354:, in square brackets, such as
337:
212:
13:
1:
2088:
2020:
1975:
1328:-alanine may also be written
233:all specify the structure of
2517:"Reaction SMILES and SMIRKS"
2400:10.1016/j.patcog.2015.05.001
2365:10.1016/j.patrec.2014.02.012
2004:REACTANT>AGENT>PRODUCT
1270:) groups appear clockwise.
1206:, respectively (because the
956:
463:for a positive charge or by
7:
2426:Journal of Chemical Ecology
2345:Pattern Recognition Letters
2029:
1887:OCCc1c(C)(cs1)Cc2cnc(C)nc2N
1546:CCc(c1)ccc21ccc3c2c4c3cccc4
1510:CC(=O)NCCC1=CNc2c1cc(OC)cc2
1377:
1346:
1274:-Alanine can be written as
1175:As a more complex example,
1122:) is one representation of
244:OpenEye Scientific Software
10:
3860:
2556:10.1021/acscentsci.9b00476
1960:
1898:-1, a steroidic 13-ringed
1224:For example, consider the
1081:
1005:bromochlorodifluoromethane
804:is most simply written as
647:to show the dissociation.
141:
63:chemical/x-daylight-smiles
29:
3755:
3707:
3681:
3671:
3506:
3358:
3344:
3292:
3254:
3245:
3213:
3180:
3166:
3071:
3005:
2985:
2940:
2914:
2905:
2873:
2840:Chemistry Development Kit
2817:
2808:
2741:
2713:
2681:
2660:
2651:
2597:10.1002/9780470125908.ch6
2068:Chemistry Development Kit
1550:CCc1c2ccc3c4ccccc4c3c2cc1
1514:CC(=O)NCCc1cc2ccc(OC)cc12
581:, but is usually written
441:may be written as either
260:Chemistry Development Kit
127:models of the molecules.
119:for conversion back into
68:
56:
44:
2303:. In Ludäscher B (ed.).
2058:Molecular Query Language
2054:'s alternative to SMILES
1967:Other examples of SMILES
1931:Cephalodiscus gilchristi
1810:Pityogenes chalcographus
1235:, more fully written as
793:, more commonly written
676:
545:
342:
252:Chemical Computing Group
58:Internet media type
2801:Computational chemistry
2661:Non-structural formulas
2646:Molecular visualization
2044:, another line notation
1294:-alanine is written as
1058:is normally written as
908:must be represented as
183:Wiswesser Line Notation
32:Smiles (disambiguation)
2026:conversion utilities.
1221:
1172:
1096:
1028:§ Stereochemistry
953:
720:is usually written as
671:§ Stereochemistry
288:Graph-based definition
89:
3844:Chemical file formats
3834:Chemical nomenclature
1358:in which one atom is
1216:
1167:
1095:-1,2-difluoroethylene
1091:
947:
552:. - = # $ : / \
138:chemistry community.
83:
2723:Ball-and-stick model
2223:Weininger D (1998).
1266:), and carboxylate (
1130:1,2-difluoroethylene
802:cyclohexane-1,2-diol
510:) is represented by
73:chemical file format
3738:JME Molecule Editor
2987:Molecular modelling
2728:Space-filling model
2683:Structural formulas
2544:ACS Central Science
2392:2015PatRe..48.3749S
2380:Pattern Recognition
2357:2014PaReL..45...11S
2313:10.1007/11530084_13
2185:Swanson RP (2004).
2172:10.1021/ci00067a005
2145:10.1021/ci00062a008
2115:10.1021/ci00057a005
2042:SYBYL Line Notation
1851:CC(C)12C1(C)C(=O)C2
1768:of the Californian
1722:OC(O1)(O)(O)(O)(O)1
750:is usually written
496:(IV) Ti. Thus, the
41:
3673:Skeletal structure
3247:Molecular dynamics
2953:Chemical WorkBench
2438:10.1007/BF01016496
1922:isolated from the
1699:-glucopyranose) (C
1470:O=Cc1ccc(O)c(OC)c1
1432:Copper(II) sulfate
1246:, the more common
1222:
1188:tetrahedral carbon
1173:
1097:
1007:may be written as
954:
696:may be written as
656:§ Aromaticity
643:may be written as
569:may be written as
196:In July 2006, the
90:
46:Filename extension
39:
3821:
3820:
3751:
3750:
3667:
3666:
3346:Quantum chemistry
3340:
3339:
3241:
3240:
3168:Molecular docking
3162:
3161:
3089:Atomistix ToolKit
3013:Ascalaph Designer
2981:
2980:
2907:Chemical kinetics
2901:
2900:
2767:
2766:
2709:
2708:
2696:Condensed formula
2673:Molecular formula
2668:Empirical formula
2653:Chemical formulas
2606:978-0-470-12590-8
2495:"SMIRKS Tutorial"
2386:(11): 3749–3756.
2322:978-3-540-27967-9
2280:on March 28, 2008
2209:978-1-57387-229-4
2200:Information Today
1989:chemical database
1904:empirical formula
1892:
1891:
1698:
1412:Methyl isocyanate
1368:deuterochloroform
1327:
1293:
1273:
1243:
1219:
1186:Configuration at
1042:is equivalent to
928:c1ccccc1-c2ccccc2
352:chemical elements
125:three-dimensional
78:
77:
16:(Redirected from
3851:
3800:Materials Studio
3679:
3678:
3477:Quantum ESPRESSO
3356:
3355:
3252:
3251:
3178:
3177:
3003:
3002:
2912:
2911:
2893:Discovery Studio
2815:
2814:
2794:
2787:
2780:
2771:
2770:
2715:Molecular models
2701:Skeletal formula
2658:
2657:
2640:
2633:
2626:
2617:
2616:
2611:
2610:
2584:
2578:
2577:
2567:
2550:(9): 1523–1531.
2535:
2529:
2528:
2526:
2524:
2513:
2507:
2506:
2504:
2502:
2491:
2485:
2484:
2482:
2480:
2464:
2458:
2457:
2420:
2414:
2413:
2411:
2375:
2369:
2368:
2340:
2334:
2333:
2331:
2329:
2296:
2290:
2289:
2287:
2285:
2276:. Archived from
2266:
2260:
2259:
2253:
2242:
2236:
2235:
2233:
2231:
2220:
2214:
2213:
2191:
2182:
2176:
2175:
2155:
2149:
2148:
2128:
2119:
2118:
2098:
2012:
2009:
2005:
1958:
1952:
1942:
1888:
1883:
1852:
1847:
1824:
1819:
1783:
1778:
1759:
1754:
1723:
1718:
1696:
1687:
1682:
1651:
1646:
1619:
1615:
1610:
1583:
1578:
1551:
1547:
1542:
1515:
1511:
1506:
1475:
1474:COc1cc(C=O)ccc1O
1471:
1466:
1452:
1447:
1446:
1445:
1426:
1406:
1382:
1381:
1373:
1365:
1338:
1331:
1325:
1323:
1320:
1313:
1309:
1305:
1297:
1291:
1289:
1277:
1271:
1269:
1265:
1261:
1253:
1250:, is written as
1241:
1238:
1234:
1217:
1209:
1205:
1201:
1197:
1193:
1190:is specified by
1182:
1160:
1156:
1152:
1135:
1117:
1113:
1109:
1084:Skeletal formula
1073:
1069:
1065:
1061:
1053:
1049:
1045:
1041:
1018:
1014:
1010:
995:
987:
980:
972:
964:
951:
937:
936:c1ccccc1c2ccccc2
933:
929:
918:
910:
900:
896:
892:
872:
868:
864:
860:
856:
852:
845:
841:
834:
807:
796:
788:
780:
776:
769:
765:
761:
757:
753:
742:
738:
734:
727:
726:C0CCCCC0C0CCCCC0
723:
722:C1CCCCC1C2CCCCC2
711:
703:
699:
668:
664:
653:
646:
638:
630:gallium arsenide
627:
622:hydrogen cyanide
619:
607:
603:
599:
595:
584:
580:
576:
572:
564:
553:
541:
538:
527:
524:
512:
506:
491:
488:
485:
484:
483:
469:
466:
462:
458:
451:
447:
444:
356:
332:biodegradability
239:canonicalization
232:
228:
224:
117:molecule editors
106:chemical species
42:
38:
21:
3859:
3858:
3854:
3853:
3852:
3850:
3849:
3848:
3824:
3823:
3822:
3817:
3789:CrystalExplorer
3747:
3703:
3663:
3502:
3354:
3336:
3288:
3237:
3209:
3176:
3158:
3067:
3001:
2991:
2989:
2977:
2936:
2897:
2869:
2810:Cheminformatics
2804:
2798:
2768:
2763:
2749:Molecular graph
2737:
2705:
2691:Lewis structure
2677:
2647:
2644:
2614:
2607:
2585:
2581:
2536:
2532:
2522:
2520:
2515:
2514:
2510:
2500:
2498:
2493:
2492:
2488:
2478:
2476:
2466:
2465:
2461:
2421:
2417:
2376:
2372:
2341:
2337:
2327:
2325:
2323:
2297:
2293:
2283:
2281:
2268:
2267:
2263:
2251:
2243:
2239:
2229:
2227:
2221:
2217:
2210:
2202:. p. 205.
2198:. Medford, NJ:
2189:
2183:
2179:
2156:
2152:
2129:
2122:
2099:
2095:
2091:
2032:
2023:
2011:
2007:
2003:
1978:
1969:
1956:
1950:
1921:
1917:
1913:
1909:
1886:
1875:
1871:
1867:
1863:
1850:
1839:
1835:
1823:CC(O1)CC12CCCO2
1822:
1781:
1757:
1746:
1742:
1738:
1721:
1710:
1706:
1702:
1685:
1674:
1670:
1666:
1662:
1649:
1638:
1634:
1630:
1617:
1616:
1613:
1602:
1598:
1594:
1582:CN1CCC1c2cccnc2
1581:
1570:
1566:
1562:
1549:
1548:
1545:
1534:
1530:
1526:
1513:
1512:
1509:
1498:
1494:
1490:
1486:
1473:
1472:
1469:
1450:
1444:
1441:
1440:
1439:
1437:
1424:
1420:
1404:
1391:SMILES formula
1380:
1371:
1363:
1349:
1336:
1329:
1322:
1318:
1311:
1307:
1303:
1295:
1287:
1275:
1267:
1263:
1259:
1251:
1236:
1232:
1207:
1203:
1199:
1195:
1191:
1180:
1158:
1154:
1153:is the same as
1150:
1133:
1115:
1111:
1107:
1086:
1080:
1078:Stereochemistry
1071:
1067:
1063:
1059:
1051:
1047:
1043:
1039:
1016:
1012:
1008:
994:COc(cc1)ccc1C#N
993:
986:COc(c1)cccc1C#N
985:
978:
970:
962:
959:
950:COc(c1)cccc1C#N
949:
935:
931:
927:
916:
909:
898:
894:
890:
873:, respectively.
870:
866:
862:
858:
854:
850:
843:
839:
832:
814:
805:
794:
786:
778:
774:
767:
763:
759:
755:
751:
740:
736:
732:
725:
721:
709:
701:
697:
679:
666:
662:
651:
644:
641:sodium chloride
636:
625:
617:
615:
605:
601:
597:
593:
582:
578:
574:
570:
562:
551:
548:
540:
537:
536:(Co) is either
526:
522:
518:
511:
504:
490:
487:
482:
479:
478:
477:
475:
468:
464:
460:
456:
449:
446:
442:
422:are the normal
355:
345:
340:
327:
290:
284:are specified.
230:
226:
222:
215:
200:introduced the
148:David Weininger
144:
121:two-dimensional
64:
52:
35:
28:
23:
22:
15:
12:
11:
5:
3857:
3847:
3846:
3841:
3836:
3819:
3818:
3816:
3815:
3812:
3807:
3802:
3797:
3791:
3786:
3781:
3778:
3775:
3770:
3765:
3759:
3757:
3753:
3752:
3749:
3748:
3746:
3745:
3740:
3735:
3730:
3725:
3722:
3717:
3715:ACD/ChemSketch
3711:
3709:
3705:
3704:
3702:
3701:
3696:
3691:
3685:
3683:
3676:
3669:
3668:
3665:
3664:
3662:
3661:
3656:
3651:
3646:
3641:
3636:
3631:
3626:
3621:
3616:
3611:
3606:
3601:
3596:
3591:
3586:
3581:
3576:
3571:
3566:
3561:
3556:
3551:
3546:
3541:
3536:
3531:
3526:
3521:
3516:
3510:
3508:
3504:
3503:
3501:
3500:
3495:
3490:
3485:
3480:
3474:
3469:
3464:
3459:
3454:
3449:
3444:
3439:
3434:
3429:
3424:
3419:
3414:
3409:
3404:
3399:
3394:
3389:
3384:
3379:
3374:
3368:
3362:
3360:
3350:
3348:
3342:
3341:
3338:
3337:
3335:
3334:
3329:
3324:
3319:
3314:
3309:
3304:
3298:
3296:
3290:
3289:
3287:
3286:
3281:
3276:
3271:
3266:
3260:
3258:
3249:
3243:
3242:
3239:
3238:
3236:
3235:
3230:
3225:
3219:
3217:
3211:
3210:
3208:
3207:
3202:
3197:
3192:
3186:
3184:
3172:
3170:
3164:
3163:
3160:
3159:
3157:
3156:
3151:
3146:
3141:
3136:
3131:
3126:
3121:
3116:
3111:
3106:
3101:
3096:
3091:
3086:
3084:ACD/ChemSketch
3081:
3075:
3073:
3069:
3068:
3066:
3065:
3060:
3055:
3050:
3045:
3040:
3035:
3030:
3025:
3020:
3015:
3009:
3007:
2997:
2995:
2983:
2982:
2979:
2978:
2976:
2975:
2970:
2965:
2960:
2955:
2950:
2944:
2942:
2938:
2937:
2935:
2934:
2929:
2924:
2918:
2916:
2909:
2903:
2902:
2899:
2898:
2896:
2895:
2890:
2885:
2879:
2877:
2871:
2870:
2868:
2867:
2862:
2857:
2852:
2847:
2842:
2837:
2832:
2827:
2823:
2821:
2812:
2806:
2805:
2797:
2796:
2789:
2782:
2774:
2765:
2764:
2762:
2761:
2756:
2751:
2745:
2743:
2739:
2738:
2736:
2735:
2730:
2725:
2719:
2717:
2711:
2710:
2707:
2706:
2704:
2703:
2698:
2693:
2687:
2685:
2679:
2678:
2676:
2675:
2670:
2664:
2662:
2655:
2649:
2648:
2643:
2642:
2635:
2628:
2620:
2613:
2612:
2605:
2579:
2530:
2508:
2486:
2459:
2432:(3): 861–876.
2415:
2370:
2335:
2321:
2291:
2261:
2254:. Duluth, MN:
2237:
2215:
2208:
2177:
2150:
2120:
2092:
2090:
2087:
2086:
2085:
2071:
2065:
2062:query language
2055:
2045:
2039:
2031:
2028:
2022:
2019:
1977:
1974:
1968:
1965:
1954:
1953:
1944:
1943:
1919:
1915:
1911:
1907:
1890:
1889:
1884:
1877:
1873:
1869:
1865:
1861:
1854:
1853:
1848:
1841:
1837:
1833:
1826:
1825:
1820:
1813:
1785:
1784:
1779:
1772:
1761:
1760:
1755:
1748:
1744:
1740:
1736:
1725:
1724:
1719:
1712:
1708:
1704:
1700:
1689:
1688:
1683:
1676:
1672:
1668:
1664:
1660:
1653:
1652:
1647:
1640:
1636:
1632:
1628:
1621:
1620:
1611:
1604:
1600:
1596:
1592:
1585:
1584:
1579:
1572:
1568:
1564:
1560:
1553:
1552:
1543:
1536:
1532:
1528:
1524:
1517:
1516:
1507:
1500:
1496:
1492:
1488:
1484:
1477:
1476:
1467:
1460:
1454:
1453:
1448:
1442:
1434:
1428:
1427:
1422:
1418:
1415:
1408:
1407:
1402:
1399:
1393:
1392:
1389:
1386:
1379:
1376:
1362:is written as
1348:
1345:
1079:
1076:
1032:
1031:
1024:
981:) is invalid.
967:propionic acid
958:
955:
875:
874:
847:
844:C:1:C:C:C:C:C1
836:
819:rings such as
813:
810:
733:C1CCCC2CCCCC12
710:C1CCCC2C1CCCC2
678:
675:
613:
610:carbon dioxide
557:Bonds between
547:
544:
520:
480:
435:
434:
431:chiral centers
427:
420:
417:
410:
367:organic subset
344:
341:
339:
336:
326:
323:
322:
321:
318:
315:
301:chemical graph
297:tree traversal
289:
286:
214:
211:
193:(Tripos Inc).
173:. In 2007, an
160:Pomona College
143:
140:
76:
75:
70:
69:Type of format
66:
65:
62:
60:
54:
53:
50:
48:
26:
9:
6:
4:
3:
2:
3856:
3845:
3842:
3840:
3837:
3835:
3832:
3831:
3829:
3813:
3811:
3808:
3806:
3803:
3801:
3798:
3796:(ICM-Browser)
3795:
3792:
3790:
3787:
3785:
3782:
3779:
3776:
3774:
3771:
3769:
3766:
3764:
3761:
3760:
3758:
3754:
3744:
3741:
3739:
3736:
3734:
3731:
3729:
3726:
3723:
3721:
3718:
3716:
3713:
3712:
3710:
3706:
3700:
3697:
3695:
3692:
3690:
3687:
3686:
3684:
3682:Free software
3680:
3677:
3674:
3670:
3660:
3657:
3655:
3652:
3650:
3647:
3645:
3642:
3640:
3637:
3635:
3632:
3630:
3627:
3625:
3622:
3620:
3617:
3615:
3612:
3610:
3607:
3605:
3602:
3600:
3597:
3595:
3592:
3590:
3587:
3585:
3582:
3580:
3577:
3575:
3572:
3570:
3567:
3565:
3562:
3560:
3557:
3555:
3552:
3550:
3547:
3545:
3542:
3540:
3537:
3535:
3532:
3530:
3527:
3525:
3522:
3520:
3517:
3515:
3512:
3511:
3509:
3505:
3499:
3496:
3494:
3491:
3489:
3486:
3484:
3481:
3478:
3475:
3473:
3470:
3468:
3465:
3463:
3460:
3458:
3455:
3453:
3450:
3448:
3445:
3443:
3440:
3438:
3435:
3433:
3430:
3428:
3425:
3423:
3420:
3418:
3415:
3413:
3410:
3408:
3405:
3403:
3400:
3398:
3395:
3393:
3390:
3388:
3385:
3383:
3380:
3378:
3375:
3372:
3369:
3367:
3364:
3363:
3361:
3359:Free software
3357:
3353:
3349:
3347:
3343:
3333:
3330:
3328:
3325:
3323:
3320:
3318:
3315:
3313:
3310:
3308:
3305:
3303:
3300:
3299:
3297:
3295:
3291:
3285:
3282:
3280:
3277:
3275:
3272:
3270:
3267:
3265:
3262:
3261:
3259:
3257:
3256:Free software
3253:
3250:
3248:
3244:
3234:
3231:
3229:
3226:
3224:
3221:
3220:
3218:
3216:
3212:
3206:
3203:
3201:
3198:
3196:
3195:AutoDock Vina
3193:
3191:
3188:
3187:
3185:
3183:
3182:Free software
3179:
3175:
3171:
3169:
3165:
3155:
3152:
3150:
3147:
3145:
3142:
3140:
3137:
3135:
3132:
3130:
3127:
3125:
3122:
3120:
3117:
3115:
3112:
3110:
3107:
3105:
3102:
3100:
3097:
3095:
3092:
3090:
3087:
3085:
3082:
3080:
3077:
3076:
3074:
3070:
3064:
3061:
3059:
3056:
3054:
3051:
3049:
3046:
3044:
3041:
3039:
3036:
3034:
3031:
3029:
3026:
3024:
3021:
3019:
3016:
3014:
3011:
3010:
3008:
3006:Free software
3004:
3000:
2996:
2994:
2993:visualization
2988:
2984:
2974:
2971:
2969:
2966:
2964:
2961:
2959:
2956:
2954:
2951:
2949:
2946:
2945:
2943:
2939:
2933:
2930:
2928:
2925:
2923:
2920:
2919:
2917:
2915:Free software
2913:
2910:
2908:
2904:
2894:
2891:
2889:
2886:
2884:
2881:
2880:
2878:
2876:
2872:
2866:
2863:
2861:
2858:
2856:
2853:
2851:
2848:
2846:
2843:
2841:
2838:
2836:
2833:
2831:
2828:
2825:
2824:
2822:
2820:
2819:Free software
2816:
2813:
2811:
2807:
2802:
2795:
2790:
2788:
2783:
2781:
2776:
2775:
2772:
2760:
2757:
2755:
2752:
2750:
2747:
2746:
2744:
2740:
2734:
2731:
2729:
2726:
2724:
2721:
2720:
2718:
2716:
2712:
2702:
2699:
2697:
2694:
2692:
2689:
2688:
2686:
2684:
2680:
2674:
2671:
2669:
2666:
2665:
2663:
2659:
2656:
2654:
2650:
2641:
2636:
2634:
2629:
2627:
2622:
2621:
2618:
2608:
2602:
2598:
2594:
2590:
2583:
2575:
2571:
2566:
2561:
2557:
2553:
2549:
2545:
2541:
2534:
2518:
2512:
2496:
2490:
2475:
2474:
2469:
2463:
2455:
2451:
2447:
2443:
2439:
2435:
2431:
2427:
2419:
2410:
2405:
2401:
2397:
2393:
2389:
2385:
2381:
2374:
2366:
2362:
2358:
2354:
2350:
2346:
2339:
2324:
2318:
2314:
2310:
2306:
2302:
2295:
2284:September 23,
2279:
2275:
2271:
2265:
2257:
2250:
2249:
2241:
2226:
2219:
2211:
2205:
2201:
2197:
2196:
2188:
2181:
2173:
2169:
2166:(3): 237–43.
2165:
2161:
2154:
2146:
2142:
2139:(2): 97–101.
2138:
2134:
2127:
2125:
2116:
2112:
2108:
2104:
2097:
2093:
2083:
2079:
2075:
2072:
2069:
2066:
2063:
2059:
2056:
2053:
2050:(InChI), the
2049:
2046:
2043:
2040:
2037:
2034:
2033:
2027:
2018:
2014:
2000:
1998:
1995:
1990:
1986:
1982:
1973:
1964:
1962:
1949:
1948:
1947:
1941:
1937:
1936:
1935:
1933:
1932:
1928:
1925:
1905:
1901:
1897:
1896:cephalostatin
1885:
1882:
1878:
1859:
1856:
1855:
1849:
1846:
1842:
1831:
1828:
1827:
1821:
1818:
1814:
1812:
1811:
1807:
1803:
1799:
1795:
1791:
1787:
1786:
1780:
1777:
1773:
1771:
1767:
1763:
1762:
1756:
1753:
1749:
1734:
1731:(cuscutin, a
1730:
1727:
1726:
1720:
1717:
1713:
1694:
1691:
1690:
1684:
1681:
1677:
1658:
1655:
1654:
1648:
1645:
1641:
1626:
1623:
1622:
1612:
1609:
1605:
1590:
1589:Oenanthotoxin
1587:
1586:
1580:
1577:
1573:
1558:
1555:
1554:
1544:
1541:
1537:
1522:
1521:Flavopereirin
1519:
1518:
1508:
1505:
1501:
1482:
1479:
1478:
1468:
1465:
1461:
1459:
1456:
1455:
1449:
1435:
1433:
1430:
1429:
1423:
1416:
1413:
1410:
1409:
1403:
1400:
1398:
1395:
1394:
1390:
1387:
1384:
1383:
1375:
1369:
1361:
1357:
1353:
1344:
1342:
1333:
1319:C(C)(N)C(=O)O
1315:
1301:
1300:see depiction
1283:
1281:
1280:see depiction
1257:
1256:see depiction
1249:
1245:
1230:
1227:
1215:
1211:
1189:
1184:
1178:
1177:beta-carotene
1170:
1169:Beta-carotene
1166:
1162:
1147:
1145:
1144:
1139:
1138:see depiction
1131:
1127:
1126:
1121:
1120:see depiction
1104:
1102:
1101:stereoisomers
1094:
1090:
1085:
1075:
1057:
1037:
1029:
1025:
1022:
1021:
1020:
1017:C(F)(Cl)(F)Br
1006:
1001:
999:
998:see depiction
991:
990:see depiction
982:
976:
968:
946:
942:
939:
925:
920:
914:
907:
902:
888:
884:
880:
848:
837:
830:
826:
825:
824:
822:
818:
809:
803:
798:
792:
784:
771:
749:
744:
729:
719:
713:
707:
695:
691:
688:For example,
686:
684:
674:
672:
659:
657:
648:
642:
633:
631:
623:
611:
591:
586:
568:
560:
555:
543:
535:
531:
516:
509:
502:
499:
495:
473:
453:
440:
432:
428:
425:
421:
418:
415:
414:formal charge
411:
408:
404:
400:
396:
392:
388:
384:
380:
376:
372:
368:
364:
363:
362:
360:
353:
349:
335:
333:
319:
316:
313:
312:
311:
308:
306:
305:spanning tree
302:
298:
295:
285:
283:
279:
274:
272:
267:
265:
261:
258:LLC, and the
257:
253:
249:
245:
240:
236:
219:
210:
208:
203:
199:
194:
192:
188:
184:
180:
176:
175:open standard
172:
167:
165:
161:
157:
156:Corwin Hansch
153:
149:
139:
137:
133:
132:open standard
128:
126:
122:
118:
114:
111:
107:
103:
102:line notation
99:
95:
87:
86:ciprofloxacin
82:
74:
71:
67:
61:
59:
55:
49:
47:
43:
37:
33:
19:
3743:MarvinSketch
3149:UCSF Chimera
3119:MarvinSketch
2835:Blue Obelisk
2733:CPK coloring
2588:
2582:
2547:
2543:
2533:
2521:. Retrieved
2511:
2499:. Retrieved
2489:
2477:. Retrieved
2471:
2468:"CID 183413"
2462:
2429:
2425:
2418:
2383:
2379:
2373:
2348:
2344:
2338:
2328:February 12,
2326:. Retrieved
2304:
2294:
2282:. Retrieved
2278:the original
2264:
2247:
2240:
2228:. Retrieved
2218:
2194:
2180:
2163:
2159:
2153:
2136:
2132:
2106:
2102:
2096:
2084:(conversion)
2024:
2015:
2001:
1979:
1970:
1961:§ Rings
1955:
1945:
1929:
1927:hemichordate
1924:Indian Ocean
1893:
1808:
1793:
1789:
1770:scale insect
1350:
1334:
1330:(C)(N)C(=O)O
1316:
1284:
1223:
1185:
1174:
1148:
1141:
1123:
1105:
1098:
1092:
1035:
1033:
1002:
983:
960:
940:
921:
903:
876:
815:
799:
772:
748:cyclopropene
745:
730:
718:bicyclohexyl
714:
687:
680:
660:
649:
634:
587:
556:
549:
454:
436:
365:are in the "
346:
328:
309:
291:
275:
268:
220:
216:
207:graph theory
195:
179:Blue Obelisk
168:
145:
129:
123:drawings or
108:using short
97:
93:
91:
36:
3720:BIOVIA Draw
3708:Proprietary
3569:GAMESS (US)
3564:GAMESS (UK)
3507:Proprietary
3294:Proprietary
3215:Proprietary
3072:Proprietary
2941:Proprietary
2888:Chemicalize
2875:Proprietary
2523:October 29,
2501:October 29,
2409:10016/33552
2109:(1): 31–6.
1997:isomorphism
1806:bark beetle
1372:C(Cl)(Cl)Cl
1288:NC(C(=O)O)C
1262:), methyl (
1233:NC(C)C(=O)O
1159:C/C=C/C=C/C
1072:c1cc(ccc1)C
1068:c1cc(C)ccc1
1013:BrC(F)(F)Cl
1009:FC(Br)(Cl)F
833:C1=CC=CC=C1
812:Aromaticity
690:cyclohexane
338:Description
294:depth-first
213:Terminology
136:open source
3828:Categories
3733:ChemWindow
3724:ChemDoodle
3689:JChemPaint
3498:YAMBO code
3452:OpenMolcas
3124:MarvinView
3099:ChemWindow
2860:Open Babel
2742:Other ways
2089:References
2021:Conversion
1976:Extensions
1860:(vitamin B
1798:Chalcogran
1451:.S(=O)(=O)
1397:Dinitrogen
1312:OC(=O)(C)N
1308:OC(=O)(N)C
1304:C(N)C(=O)O
1296:N(C(=O)O)C
1276:N(C)C(=O)O
1252:N(C)C(=O)O
1248:enantiomer
1237:N(C)C(=O)O
1226:amino acid
1082:See also:
975:fluoroform
806:OC1CCCCC1O
3839:Encodings
3810:OpenChrom
3699:XDrawChem
3694:Molsketch
3644:TURBOMOLE
3624:Quantemol
2830:Bioclipse
2351:: 11–16.
2074:OpenBabel
1902:with the
1830:α-Thujone
1802:pheromone
1766:pheromone
1657:Aflatoxin
1625:Pyrethrin
1481:Melatonin
1388:Structure
1360:carbon-14
1064:c1ccccc1C
1060:Cc1ccccc1
957:Branching
913:imidazole
787:C1.C2.C12
624:HCN) and
559:aliphatic
515:hydronium
498:hydroxide
3773:EXC code
3728:ChemDraw
3639:TeraChem
3629:Scigress
3599:OpenAtom
3574:Gaussian
3467:PyQuante
3392:CONQUEST
3387:COLUMBUS
3190:AutoDock
3129:MODELLER
3109:Gaussian
3094:ChemDraw
3038:Ghemical
3018:Avogadro
2948:Autochem
2803:software
2574:31572779
2446:24263601
2274:U.S. EPA
2256:U.S. EPA
2230:June 24,
2030:See also
1994:subgraph
1985:wildcard
1900:pyrazine
1858:Thiamine
1729:Bergenin
1557:Nicotine
1458:Vanillin
1385:Molecule
1378:Examples
1352:Isotopes
1347:Isotopes
1244:-Alanine
1220:-Alanine
1050:and the
979:CCC=(O)O
963:CCC(=O)O
924:biphenyl
895:n1ccccc1
891:c1ccccc1
883:pyridine
817:Aromatic
783:ethylene
702:O1CCOCC1
698:C1CCCCC1
528:and the
517:cation (
494:titanium
472:ammonium
429:are not
424:isotopes
412:have no
264:database
3814:SASHIMI
3784:Mercury
3675:drawing
3634:Spartan
3559:Firefly
3554:CRYSTAL
3479:(PWscf)
3447:Octopus
3427:MADNESS
3412:DP code
3373:(CFOUR)
3322:Desmond
3302:Abalone
3269:GROMACS
3200:FlexAID
3144:Spartan
3114:Maestro
3079:Abalone
3058:QuteMol
3048:Molekel
3033:Gabedit
2973:Khimera
2963:COSILAB
2958:CHEMKIN
2927:Cantera
2565:6764162
2479:May 12,
2473:PubChem
2388:Bibcode
2353:Bibcode
1963:above.
1804:of the
1693:Glucose
1421:−N=C=O
1364:1ccccc1
1356:Benzene
1229:alanine
1155:F/C=C/F
1151:F\C=C\F
1134:F/C=C\F
1116:F/C=C/F
1056:toluene
911:; thus
906:pyrrole
899:o1cccc1
879:benzene
842:, e.g.
821:benzene
791:propane
768:C=1CC-1
764:C=1CC=1
706:decalin
694:dioxane
683:acyclic
673:below.
658:below.
567:ethanol
505:
282:isomers
271:cuneane
256:MolSoft
235:ethanol
185:(WLN),
142:History
113:strings
3805:Molden
3756:Others
3654:WIEN2k
3619:Q-Chem
3594:ONETEP
3589:MOLPRO
3584:MOLCAS
3579:Jaguar
3549:CRUNCH
3539:CASTEP
3534:CASINO
3529:CADPAC
3493:VB2000
3488:SIESTA
3457:PARSEC
3442:NWChem
3422:FreeON
3402:Dalton
3382:BigDFT
3377:AIMAll
3366:ABINIT
3327:GROMOS
3312:CHARMM
3284:PLUMED
3279:OpenMM
3274:LAMMPS
3228:LeDock
3139:SAMSON
3063:RasMol
3028:Biskit
2968:DelPhi
2883:Canvas
2850:JOELib
2754:SMILES
2603:
2572:
2562:
2454:226090
2452:
2444:
2319:
2206:
2078:JOELib
1981:SMARTS
1972:tool.
1425:CN=C=O
1414:(MIC)
1268:C(=O)O
1030:below.
992:) and
971:FC(F)F
917:n1ccc1
829:Kekulé
760:C1CC=1
756:C=1CC1
752:C1=CC1
654:; see
600:, and
534:cation
532:(III)
530:cobalt
513:, the
187:ROSDAL
152:Duluth
98:SMILES
40:SMILES
3768:Eulim
3763:Aqion
3609:PLATO
3524:DMol3
3519:AMPAC
3472:PySCF
3432:MOPAC
3417:FLEUR
3407:DIRAC
3307:AMBER
3223:Glide
3205:rDock
3104:EzMol
3053:PyMOL
2865:RDKit
2855:OELib
2759:InChl
2450:S2CID
2252:(PDF)
2190:(PDF)
2082:OELib
2052:IUPAC
1733:resin
1627:II (C
1125:trans
1093:trans
1044:C(1)N
887:furan
762:, or
739:, so
677:Rings
606:O=C=O
590:bonds
571:C-C-O
546:Bonds
525:) is
501:anion
439:water
426:, and
416:, and
409:, and
405:, or
369:" of
348:Atoms
343:Atoms
299:of a
248:MEDIT
231:C(O)C
202:InChI
198:IUPAC
110:ASCII
3780:GSim
3777:GenX
3659:XMVB
3649:VASP
3604:ORCA
3544:CPMD
3437:MPQC
3397:CP2K
3371:ACES
3332:NAMD
3317:CPMD
3264:CP2K
3043:Jmol
3023:BALL
2990:and
2922:APBS
2845:ECCE
2601:ISBN
2570:PMID
2525:2018
2503:2018
2481:2012
2442:PMID
2330:2013
2317:ISBN
2286:2012
2232:2013
2204:ISBN
2060:, a
1876:OS)
1800:: a
1735:) (C
1401:N≡N
1366:and
1310:and
1202:and
1110:and
973:for
969:and
965:for
897:and
885:and
869:and
846:, or
781:for
775:C1C1
741:C%12
700:and
692:and
579:C-CO
575:CC-O
492:for
470:for
359:gold
357:for
229:and
189:and
92:The
51:.smi
3794:ICM
3614:PQS
3514:ADF
3483:RMG
3462:PSI
3154:VMD
2932:KPP
2593:doi
2560:PMC
2552:doi
2434:doi
2404:hdl
2396:doi
2361:doi
2309:doi
2168:doi
2141:doi
2111:doi
1864:, C
1840:O)
1695:(β-
1405:N#N
1370:is
1282:).
1194:or
1143:cis
1070:or
1062:or
1040:C1N
1036:not
938:.)
827:In
795:CCC
779:C=C
665:or
632:).
618:C#N
616:),
583:CCO
577:or
539:or
489:or
445:or
227:OCC
223:CCO
209:).
191:SLN
3830::
2599:.
2568:.
2558:.
2546:.
2542:.
2470:.
2448:.
2440:.
2430:16
2428:.
2402:.
2394:.
2384:48
2382:.
2359:.
2349:45
2347:.
2315:.
2272:.
2164:30
2162:.
2137:29
2135:.
2123:^
2107:28
2105:.
2080:,
2076:,
1999:.
1934::
1920:10
1912:74
1908:54
1870:17
1866:12
1838:16
1834:10
1832:(C
1796:)-
1792:,5
1788:(2
1764:A
1747:)
1741:16
1737:14
1711:)
1705:12
1675:)
1669:12
1665:17
1663:(C
1639:)
1633:28
1629:22
1603:)
1597:22
1593:17
1591:(C
1571:)
1565:14
1561:10
1559:(C
1535:)
1529:15
1525:17
1523:(C
1499:)
1489:16
1485:13
1483:(C
1438:SO
1436:Cu
1417:CH
1374:.
1343:.
1332:.
1314:.
1306:,
1239:.
1200:@@
1196:@@
1183:.
1161:.
1103:.
1074:.
1015:,
1011:,
919:.
901:.
893:,
881:,
865:,
861:,
857:,
853:,
797:.
758:,
728:.
712:.
626:$
612:CO
602:$
596:,
585:.
573:,
554:.
542:.
508:OH
476:NH
452:.
403:Br
401:,
399:Cl
397:,
393:,
389:,
385:,
381:,
377:,
373:,
266:.
254:,
250:,
246:,
225:,
2793:e
2786:t
2779:v
2639:e
2632:t
2625:v
2609:.
2595::
2576:.
2554::
2548:5
2527:.
2505:.
2483:.
2456:.
2436::
2412:.
2406::
2398::
2390::
2367:.
2363::
2355::
2332:.
2311::
2288:.
2234:.
2212:.
2174:.
2170::
2147:.
2143::
2117:.
2113::
2008:.
1957:%
1918:O
1916:2
1914:N
1910:H
1906:C
1874:4
1872:N
1868:H
1862:1
1836:H
1794:R
1790:S
1745:9
1743:O
1739:H
1709:6
1707:O
1703:H
1701:6
1697:D
1673:6
1671:O
1667:H
1661:1
1659:B
1637:5
1635:O
1631:H
1601:2
1599:O
1595:H
1569:2
1567:N
1563:H
1533:2
1531:N
1527:H
1497:2
1495:O
1493:2
1491:N
1487:H
1443:4
1419:3
1337:@
1326:L
1298:(
1292:L
1278:(
1272:D
1264:C
1260:H
1254:(
1242:L
1218:L
1208:@
1204:@
1192:@
1136:(
1128:-
1118:(
1112:\
1108:/
1052:N
1048:C
996:(
988:(
952:.
932:-
871:s
867:p
863:o
859:n
855:c
851:b
840::
835:,
737:%
667:\
663:/
652::
645:.
637:.
628:(
620:(
614:2
608:(
598:#
594:=
563:-
523:O
521:3
519:H
503:(
481:4
474:(
465:-
461:+
457:H
450:O
443:O
433:.
407:I
395:F
391:S
387:P
383:O
379:N
375:C
371:B
158:(
96:(
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.