262:
550:, proviral DNA is incorporated in transcription of the DNA of the cell being infected. Since retroviruses need to change their pre-mRNA into DNA so that this DNA can be integrated within the DNA of the host it is affecting, the formation of that DNA template is a vital step for retrovirus replication. Cell type, the differentiation or changed state of the cell, and the physiological state of the cell, result in a significant change in the availability and activity of certain factors necessary for transcription. These variables create a wide range of viral gene expression. For example, tissue culture cells actively producing infectious virions of avian or murine
368:
of proteins at the same time they are being produced via transcription. Alternatively, pre-mRNA of eukaryotic cells undergo a wide range of modifications prior to their transport from the nucleus to cytoplasm where their mature forms are translated. These modifications are responsible for the different types of encoded messages that lead to translation of various types of products. Furthermore, primary transcript processing provides a control for gene expression as well as a regulatory mechanism for the degradation rates of mRNAs. The processing of pre-mRNA in eukaryotic cells includes
836:
288:
methylation patterns at promoter regions of DNA can regulate RNA polymerase access to a given template. RNA polymerase is often incapable of synthesizing a primary transcript if the targeted gene's promoter region contains specific methylated cytosines— residues that hinder binding of transcription-activating factors and recruit other enzymes to stabilize a tightly bound nucleosome structure, excluding access to RNA polymerase and preventing the production of primary transcripts.
42:
1962:
139:. In particular, alternative splicing directly contributes to the diversity of mRNA found in cells. The modifications of primary transcripts have been further studied in research seeking greater knowledge of the role and significance of these transcripts. Experimental studies based on molecular changes to primary transcripts and the processes before and after transcription have led to greater understanding of diseases involving primary transcripts.
452:
542:
Institute in Genoa, Italy, explains that 1785 nucleotides of the region in the DNA that codes for the estrogen receptor alpha (ER-alpha) are spread over a region that holds more than 300,000 nucleotides in the primary transcript. Splicing of this pre-mRNA frequently leads to variants or different kinds of the mRNA lacking one or more exons or regions necessary for coding proteins. These variants have been associated with
33:
496:(DHFR) mRNA levels, while the level of DHFR pre-mRNA with certain introns remained unaffected. The half-life of DHFR mRNA or pre-mRNA did not change significantly, but the transition rate of DHFR RNA from the nucleus to the cytoplasm decreased, suggesting that FUra may influence mRNA processing and/or nuclear DHFR mRNA stability.
421:
is added. Signals for polyadenylation, which include several RNA sequence elements, are detected by a group of proteins which signal the addition of the poly-A tail (approximately 200 nucleotides in length). The polyadenylation reaction provides a signal for the end of transcription and this reaction
367:
The basic primary transcript modification process is similar for tRNA and rRNA in both eukaryotic and prokaryotic cells. On the other hand, primary transcript processing varies in mRNAs of prokaryotic and eukaryotic cells. For example, some prokaryotic bacterial mRNAs serve as templates for synthesis
153:
The steps contributing to the production of primary transcripts involve a series of molecular interactions that initiate transcription of DNA within a cell's nucleus. Based on the needs of a given cell, certain DNA sequences are transcribed to produce a variety of RNA products to be translated into
554:
viruses (ASLV or MLV) contain such high levels of viral RNA that 5–10% of the mRNA in a cell can be of viral origin. This shows that the primary transcripts produced by these retroviruses do not always follow the normal path to protein production and convert back into DNA in order to multiply and
287:
inhibit transcription. Acetylation of histones induces repulsion between negative components within nucleosomes, allowing for RNA polymerase access. Deacetylation of histones stabilizes tightly coiled nucleosomes, inhibiting RNA polymerase access. In addition to acetylation patterns of histones,
253:
components, causing DNA to be either more or less accessible to RNA polymerase. The unique combinations of either activating or inhibiting transcription factors that bind to enhancer DNA regions determine whether or not the gene that enhancer interacts with is activated for transcription or not.
541:
and differential splicing. The article entitled, "Alternative splicing of the human estrogen receptor alpha primary transcript: mechanisms of exon skipping" by Paola Ferro, Alessandra
Forlani, Marco Muselli and Ulrich Pfeffer from the laboratory of Molecular Oncology at National Cancer Research
358:
Transcription, a highly regulated phase in gene expression, produces primary transcripts. However, transcription is only the first step which should be followed by many modifications that yield functional forms of RNAs. Otherwise stated, the newly synthesized primary transcripts are modified in
36:
Pre-mRNA is the first form of RNA created through transcription in protein synthesis. The pre-mRNA lacks structures that the messenger RNA (mRNA) requires. First all introns have to be removed from the transcribed RNA through a process known as splicing. Before the RNA is ready for export, a
459:
The effect of alternative splicing in gene expression can be seen in complex eukaryotes which have a fixed number of genes in their genome yet produce much larger numbers of different gene products. Most eukaryotic pre-mRNA transcripts contain multiple introns and exons. The various possible
171:
460:
combinations of 5' and 3' splice sites in a pre-mRNA can lead to different excision and combination of exons while the introns are eliminated from the mature mRNA. Thus, various kinds of mature mRNAs are generated. Alternative splicing takes place in a large protein complex called the
273:. Like enhancers, silencers may be located at locations farther up or downstream from the genes they regulate. These DNA sequences bind to factors that contribute to the destabilization of the initiation complex required to activate RNA polymerase, and therefore inhibit transcription.
402:
to the 5' terminal nucleotide of the pre-mRNA in reverse orientation followed by the addition of methyl groups to the G residue. 5' capping is essential for the production of functional mRNAs since the 5' cap is responsible for aligning the mRNA with the ribosome during translation.
127:. Certain factors play key roles in the activation and inhibition of transcription, where they regulate primary transcript production. Transcription produces primary transcripts that are further modified by several processes. These processes include the
158:
connecting compatible nucleic acids of DNA are broken to produce two unconnected single DNA strands. One strand of the DNA template is used for transcription of the single-stranded primary transcript mRNA. This DNA strand is bound by an
526:, with this formation being temperature-dependent and influenced by specific RNA sequences. Pre-mRNA targeting and splicing factor loading in speckles were critical for spliceosome group formation, resulting in a speckled pattern.
447:
In complex eukaryotic cells, one primary transcript is able to prepare large amounts of mature mRNAs due to alternative splicing. Alternative splicing is regulated so that each mature mRNA may encode a multiplicity of proteins.
194:
DNA template in the 5' to 3' direction, and this newly synthesized primary transcript is complementary to the antisense strand of DNA. RNA polymerase II constructs the primary transcript using a set of four specific
475:
cleavage or by endolytic cleavage. Autocatalytic cleavages, in which no proteins are involved, are usually reserved for sections that code for rRNA, whereas endolytic cleavage corresponds to tRNA precursors.
299:
are formed during transcription. An R-loop is a three-stranded nucleic acid structure containing a DNA-RNA hybrid region and an associated non-template single-stranded DNA. Actively transcribed regions of
323:
itself is a source of endogenous DNA damages resulting from the susceptibility of single-stranded DNA to damage. Other sources of DNA damage are conflicts of the primary transcription machinery with the
254:
Activation of transcription depends on whether or not the transcription elongation complex, itself consisting of a variety of transcription factors, can induce RNA polymerase to dissociate from the
190:, only one RNA polymerase exists to create all kinds of RNA molecules. RNA polymerase II of eukaryotes transcribes the primary transcript, a transcript destined to be processed into mRNA, from the
464:. Alternative splicing is crucial for tissue-specific and developmental regulation in gene expression. Alternative splicing can be affected by various factors, including mutations such as
319:
arise in each cell, every day, with the number of damages in each cell reaching tens to hundreds of thousands, and such DNA damages can impede primary transcription. The process of
115:
There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and complete the transcription of
529:
Recruiting pre-mRNA to nuclear speckles significantly increased splicing efficiency and protein levels, indicating that proximity to speckles enhances splicing efficiency.
112:". The term hnRNA is often used as a synonym for pre-mRNA, although, in the strict sense, hnRNA may include nuclear RNA transcripts that do not end up as cytoplasmic mRNA.
1237:
515:
due to its larger genome, despite both species producing mature mRNA of similar size and sequence complexity. This indicates that hnRNA size increases with genome size.
238:
A number of factors contribute to the activation and inhibition of transcription and therefore regulate the production of primary transcripts from a given DNA template.
1161:
Ferro P, Forlani A, Muselli M, Pfeffer U (September 2003). "Alternative splicing of the human estrogen receptor alpha primary transcript: mechanisms of exon skipping".
230:
sequences within primary transcripts explains the size difference between larger primary transcripts and smaller, mature mRNA ready for translation into protein.
336:
enzymes. Even though these processes are tightly regulated and are usually accurate, occasionally they can make mistakes and leave behind DNA breaks that drive
279:
modification by transcription factors is another key regulatory factor for transcription by RNA polymerase. In general, factors that lead to histone
1281:
1230:
973:"5-Fluorouracil inhibits dihydrofolate reductase precursor mRNA processing and/or nuclear mRNA stability in methotrexate-resistant KB cells"
1223:
1444:
1439:
537:
Research has also led to greater knowledge about certain diseases related to changes within primary transcripts. One study involved
359:
several ways to be converted to their mature, functional forms to produce different proteins and RNAs such as mRNA, tRNA, and rRNA.
154:
functional proteins for cellular use. To initiate the transcription process in a cell's nucleus, DNA double helices are unwound and
1449:
1424:
249:, proteins that bind to DNA elements to either activate or repress transcription, bind to enhancers and recruit enzymes that alter
1557:
17:
1522:
1210:
745:
699:
626:
1291:
1643:
1246:
903:
353:
101:
82:
81:. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after
241:
Activation of RNA polymerase activity to produce primary transcripts is often controlled by sequences of DNA called
1930:
316:
305:
1935:
218:
Studies of primary transcripts produced by RNA polymerase II reveal that an average primary transcript is 7,000
1832:
1718:
919:
Wahl MC, Will CL, Lührmann R (February 2009). "The spliceosome: design principles of a dynamic RNP machine".
394:
Shortly after transcription is initiated in eukaryotes, a pre-mRNA's 5' end is modified by the addition of a
261:
1945:
1940:
1925:
1550:
1286:
1012:"hnRNA size and processing as related to different DNA content in two dipterans: Drosophila and Aedes".
100:. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA (hnRNA). Once pre-mRNA has been completely
465:
337:
1072:
148:
97:
58:
1492:
1313:
585:
493:
208:
200:
1965:
1658:
1543:
1067:
761:
Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, et al. (August 2017).
399:
255:
215:(UMP)) that are added continuously to the 3' hydroxyl group on the 3' end of the growing mRNA.
204:
417:
In eukaryotes, polyadenylation further modifies pre-mRNAs during which a structure called the
45:
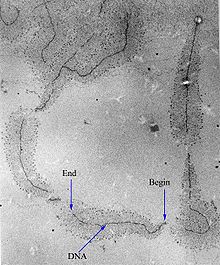
Micrograph of gene transcription of ribosomal RNA illustrating the growing primary transcripts
1842:
1686:
246:
212:
186:, and mRNA—are produced based on the activity of three distinct RNA polymerases, whereas, in
78:
222:
in length, with some growing as long as 20,000 nucleotides in length. The inclusion of both
1877:
1860:
1691:
1376:
431:
377:
333:
136:
105:
1056:"Prespliceosomal assembly on microinjected precursor mRNA takes place in nuclear speckles"
941:
840:
835:
8:
1872:
1855:
1713:
1596:
1369:
164:
1138:
1113:
807:
Milano L, Gautam A, Caldecott KW (January 2024). "DNA damage and transcription stress".
398:, also known as a 5' cap. The 5' capping modification is initiated by the addition of a
1766:
1761:
1037:
954:
422:
ends approximately a few hundred nucleotides downstream from the poly-A tail location.
989:
972:
763:"Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability"
1850:
1812:
1805:
1756:
1708:
1364:
1170:
1143:
1095:
1090:
1055:
1029:
1025:
994:
946:
899:
824:
784:
741:
695:
622:
538:
1215:
1041:
958:
269:
Inhibition of RNA polymerase activity can also be regulated by DNA sequences called
1676:
1133:
1125:
1085:
1077:
1021:
984:
936:
928:
816:
774:
523:
308:. Introns reduce R-loop formation and DNA damage in highly expressed yeast genes.
37:
Poly(A)tail is added to the 3' end of the RNA and a 5' cap is added to the 5' end.
1800:
1771:
1323:
1306:
1301:
1276:
820:
779:
762:
735:
689:
616:
412:
373:
325:
320:
132:
77:. The primary transcripts designated to be mRNAs are modified in preparation for
1892:
1795:
1612:
1487:
1386:
1271:
1266:
1189:
1129:
932:
716:
665:
646:
577:
389:
270:
196:
160:
128:
876:
1980:
1827:
1817:
1698:
1653:
1631:
1114:"Genome organization around nuclear speckles drives mRNA splicing efficiency"
590:
543:
472:
329:
284:
155:
109:
1986:
1910:
1730:
1703:
1648:
1567:
1333:
1254:
1174:
1147:
1099:
950:
828:
788:
564:
489:
485:
120:
93:
998:
1822:
1668:
1535:
1512:
1359:
1318:
1081:
1033:
461:
437:
418:
280:
265:
Role of transcription factors and enhancers in gene expression regulation
187:
41:
1623:
1882:
1591:
1586:
1581:
547:
501:
341:
250:
219:
124:
451:
1746:
519:
395:
369:
242:
191:
174:
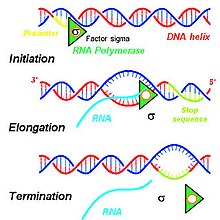
Transcription of DNA by RNA polymerase to produce primary transcript
1900:
1751:
1681:
551:
1905:
1479:
1459:
1454:
276:
170:
1920:
1915:
1865:
1434:
1429:
1409:
1345:
1296:
1196:. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
572:
296:
227:
1723:
1502:
1497:
1419:
1414:
1404:
1399:
1394:
1354:
507:
441:
65:, and processed to yield various mature RNA products such as
760:
1517:
1507:
1349:
653:(3rd ed.). New York: Garland Science – via NCBI.
522:, spliceosome groups on pre-mRNA were found to form within
223:
183:
179:
74:
70:
66:
32:
1160:
883:(2nd ed.). Sunderland (MA): Sinauer Associates; 2000.
283:
activate transcription while factors that lead to histone
258:
complex that connects an enhancer region to the promoter.
1785:
1606:
839: This article incorporates text available under the
492:-resistant KB cells led to a two-fold reduction in total
301:
116:
89:
62:
54:
694:. Sinauer Associates, Incorporated. pp. 38–39, 50.
436:
Eukaryotic pre-mRNAs have their introns spliced out by
328:
machinery, and the activity of certain enzymes such as
1245:
806:
1187:
717:"Assembly of the Transcription Initiation Complex"
610:
608:
606:
1978:
918:
898:. New York, NY: McGraw-Hill. pp. 432–448.
603:
1188:Coffin JM, Hughes SH, Varmus HE, eds. (1997).
455:Alternative splicing of the primary transcript
1551:
1231:
683:
681:
679:
870:
802:
800:
798:
640:
638:
614:
1163:International Journal of Molecular Medicine
868:
866:
864:
862:
860:
858:
856:
854:
852:
850:
754:
1565:
1558:
1544:
1238:
1224:
676:
304:often form R-loops that are vulnerable to
1137:
1089:
1071:
988:
940:
795:
778:
727:
635:
53:is the single-stranded ribonucleic acid (
847:
450:
260:
169:
40:
31:
687:
644:
425:
311:
14:
1979:
893:
874:
733:
511:, hnRNA (pre-mRNA) size was larger in
1539:
1219:
714:
666:"An Introduction to Genetic Analysis"
663:
615:Strachan T, Read AP (January 2004).
471:In prokaryotes, splicing is done by
977:The Journal of Biological Chemistry
740:. W. H. Freeman. pp. 303–306.
723:(2nd ed.). Oxford: Wiley-Liss.
621:. Garland Science. pp. 16–17.
532:
24:
647:"RNA Synthesis and RNA Processing"
546:progression. In the life cycle of
406:
178:In eukaryotes, three kinds of RNA—
25:
1998:
1682:Micro
1247:Post-transcriptional modification
1204:
354:Post-transcriptional modification
347:
1961:
1960:
834:
442:small nuclear ribonucleoproteins
1637:precursor, heterogenous nuclear
1181:
1154:
1106:
1048:
1005:
983:(35): 21413–21. December 1989.
965:
912:
887:
88:Pre-mRNA is synthesized from a
1767:Trans-acting small interfering
1731:Enhancer RNAs
1649:Transfer
942:11858/00-001M-0000-000F-9EAB-8
881:The Cell: A Molecular Approach
708:
657:
13:
1:
1654:Ribosomal
1632:Messenger
1124:(8014): 1165–1173. May 2024.
1066:(2): 393–406. February 2001.
1060:Molecular Biology of the Cell
990:10.1016/S0021-9258(19)30096-1
877:"RNA Processing and Turnover"
651:Molecular Biology of the Cell
596:
383:
362:
233:
142:
27:RNA produced by transcription
1026:10.1016/0092-8674(75)90103-8
821:10.1016/j.molcel.2023.11.014
780:10.1016/j.molcel.2017.07.002
7:
688:Gilbert SF (15 July 2013).
558:
479:
10:
2003:
1833:Multicopy single-stranded
1677:Interferential
1211:Scienceden.com RNA Article
1130:10.1038/s41586-024-07429-6
933:10.1016/j.cell.2009.02.009
618:Human Molecular Genetics 3
429:
410:
387:
351:
338:chromosomal rearrangements
291:
146:
1956:
1891:
1841:
1784:
1747:Guide
1739:
1667:
1622:
1605:
1574:
1478:
1385:
1341:
1332:
1262:
1253:
672:. New York: W.H. Freeman.
466:chromosomal translocation
57:) product synthesized by
1709:Small nuclear
1020:(3): 281–90. July 1975.
199:monophosphate residues (
149:Transcription (genetics)
1823:Genomic
1314:Poly(A)-binding protein
586:Transcription (biology)
494:dihydrofolate reductase
209:guanosine monophosphate
201:adenosine monophosphate
18:Precursor messenger RNA
1926:Artificial chromosomes
1714:Small nucleolar
737:Molecular Cell Biology
456:
266:
205:cytidine monophosphate
175:
46:
38:
1719:Small Cajal Body RNAs
691:Developmental Biology
454:
396:7-methylguanosine cap
264:
247:Transcription factors
213:uridine monophosphate
173:
44:
35:
1772:Subgenomic messenger
1687:Small interfering
1659:Transfer-messenger
1377:Alternative splicing
1082:10.1091/mbc.12.2.393
432:Alternative splicing
426:Alternative splicing
378:alternative splicing
334:base excision repair
312:Transcription stress
137:alternative splicing
106:mature messenger RNA
488:(FUra) exposure in
167:region of the DNA.
1801:Chloroplast
1644:modified Messenger
1607:Ribonucleic acids
1488:5′ cap methylation
894:Weaver RF (2005).
875:Cooper GM (2000).
645:Alberts B (1994).
539:estrogen receptors
457:
374:3' polyadenylation
267:
176:
133:3'-polyadenylation
51:primary transcript
47:
39:
1974:
1973:
1851:Xeno
1813:Complementary
1786:Deoxyribonucleic
1780:
1779:
1757:Small hairpin
1533:
1532:
1474:
1473:
1470:
1469:
1387:pre-mRNA factors
896:Molecular Biology
773:(4): 608–621.e6.
747:978-0-7167-7601-7
734:Lodish H (2008).
715:Brown TA (2002).
701:978-1-60535-173-5
628:978-0-8153-4184-0
16:(Redirected from
1994:
1964:
1963:
1941:Yeast
1762:Small temporal
1692:Piwi-interacting
1620:
1619:
1616:
1597:Deoxynucleotides
1560:
1553:
1546:
1537:
1536:
1339:
1338:
1272:5′ cap formation
1260:
1259:
1240:
1233:
1226:
1217:
1216:
1198:
1197:
1185:
1179:
1178:
1158:
1152:
1151:
1141:
1110:
1104:
1103:
1093:
1075:
1052:
1046:
1045:
1009:
1003:
1002:
992:
969:
963:
962:
944:
916:
910:
909:
891:
885:
884:
872:
845:
838:
832:
804:
793:
792:
782:
758:
752:
751:
731:
725:
724:
712:
706:
705:
685:
674:
673:
661:
655:
654:
642:
633:
632:
612:
533:Related diseases
524:nuclear speckles
104:, it is termed "
92:template in the
21:
2002:
2001:
1997:
1996:
1995:
1993:
1992:
1991:
1977:
1976:
1975:
1970:
1952:
1893:Cloning vectors
1887:
1873:Locked
1837:
1787:
1776:
1735:
1663:
1610:
1609:
1601:
1570:
1564:
1534:
1529:
1466:
1381:
1328:
1324:Polyuridylation
1277:Polyadenylation
1249:
1244:
1207:
1202:
1201:
1190:"Transcription"
1186:
1182:
1159:
1155:
1112:
1111:
1107:
1073:10.1.1.324.8865
1054:
1053:
1049:
1011:
1010:
1006:
971:
970:
966:
917:
913:
906:
892:
888:
873:
848:
805:
796:
759:
755:
748:
732:
728:
713:
709:
702:
686:
677:
662:
658:
643:
636:
629:
613:
604:
599:
561:
535:
482:
434:
428:
415:
413:Polyadenylation
409:
407:Polyadenylation
392:
386:
365:
356:
350:
326:DNA replication
321:gene expression
314:
294:
236:
151:
145:
28:
23:
22:
15:
12:
11:
5:
2000:
1990:
1989:
1972:
1971:
1969:
1968:
1957:
1954:
1953:
1951:
1950:
1949:
1948:
1943:
1938:
1933:
1923:
1918:
1913:
1908:
1903:
1897:
1895:
1889:
1888:
1886:
1885:
1880:
1878:Peptide
1875:
1870:
1869:
1868:
1863:
1858:
1856:Glycol
1847:
1845:
1839:
1838:
1836:
1835:
1830:
1825:
1820:
1815:
1810:
1809:
1808:
1803:
1792:
1790:
1782:
1781:
1778:
1777:
1775:
1774:
1769:
1764:
1759:
1754:
1749:
1743:
1741:
1737:
1736:
1734:
1733:
1728:
1727:
1726:
1721:
1716:
1711:
1701:
1696:
1695:
1694:
1689:
1684:
1673:
1671:
1665:
1664:
1662:
1661:
1656:
1651:
1646:
1641:
1640:
1639:
1628:
1626:
1617:
1603:
1602:
1600:
1599:
1594:
1589:
1584:
1578:
1576:
1572:
1571:
1568:nucleic acids
1563:
1562:
1555:
1548:
1540:
1531:
1530:
1528:
1527:
1526:
1525:
1520:
1515:
1510:
1505:
1500:
1493:mRNA decapping
1490:
1484:
1482:
1476:
1475:
1472:
1471:
1468:
1467:
1465:
1464:
1463:
1462:
1457:
1452:
1447:
1442:
1437:
1432:
1427:
1422:
1417:
1412:
1407:
1402:
1391:
1389:
1383:
1382:
1380:
1379:
1374:
1373:
1372:
1367:
1357:
1352:
1342:
1336:
1330:
1329:
1327:
1326:
1321:
1316:
1311:
1310:
1309:
1304:
1299:
1294:
1289:
1284:
1274:
1269:
1267:Precursor mRNA
1263:
1257:
1251:
1250:
1243:
1242:
1235:
1228:
1220:
1214:
1213:
1206:
1205:External links
1203:
1200:
1199:
1180:
1153:
1105:
1047:
1004:
964:
911:
904:
886:
846:
794:
767:Molecular Cell
753:
746:
726:
707:
700:
675:
664:Griffiths AJ.
656:
634:
627:
601:
600:
598:
595:
594:
593:
588:
583:
575:
570:
560:
557:
534:
531:
481:
478:
430:Main article:
427:
424:
411:Main article:
408:
405:
390:Five-prime cap
388:Main article:
385:
382:
364:
361:
352:Main article:
349:
348:RNA processing
346:
330:topoisomerases
313:
310:
293:
290:
235:
232:
197:ribonucleoside
161:RNA polymerase
156:hydrogen bonds
147:Main article:
144:
141:
108:", or simply "
26:
9:
6:
4:
3:
2:
1999:
1988:
1985:
1984:
1982:
1967:
1959:
1958:
1955:
1947:
1944:
1942:
1939:
1937:
1934:
1932:
1929:
1928:
1927:
1924:
1922:
1919:
1917:
1914:
1912:
1909:
1907:
1904:
1902:
1899:
1898:
1896:
1894:
1890:
1884:
1881:
1879:
1876:
1874:
1871:
1867:
1864:
1862:
1861:Threose
1859:
1857:
1854:
1853:
1852:
1849:
1848:
1846:
1844:
1840:
1834:
1831:
1829:
1826:
1824:
1821:
1819:
1818:Deoxyribozyme
1816:
1814:
1811:
1807:
1806:Mitochondrial
1804:
1802:
1799:
1798:
1797:
1794:
1793:
1791:
1789:
1783:
1773:
1770:
1768:
1765:
1763:
1760:
1758:
1755:
1753:
1750:
1748:
1745:
1744:
1742:
1738:
1732:
1729:
1725:
1722:
1720:
1717:
1715:
1712:
1710:
1707:
1706:
1705:
1702:
1700:
1697:
1693:
1690:
1688:
1685:
1683:
1680:
1679:
1678:
1675:
1674:
1672:
1670:
1666:
1660:
1657:
1655:
1652:
1650:
1647:
1645:
1642:
1638:
1635:
1634:
1633:
1630:
1629:
1627:
1625:
1624:Translational
1621:
1618:
1614:
1608:
1604:
1598:
1595:
1593:
1590:
1588:
1585:
1583:
1580:
1579:
1577:
1573:
1569:
1561:
1556:
1554:
1549:
1547:
1542:
1541:
1538:
1524:
1521:
1519:
1516:
1514:
1511:
1509:
1506:
1504:
1501:
1499:
1496:
1495:
1494:
1491:
1489:
1486:
1485:
1483:
1481:
1477:
1461:
1458:
1456:
1453:
1451:
1448:
1446:
1443:
1441:
1438:
1436:
1433:
1431:
1428:
1426:
1423:
1421:
1418:
1416:
1413:
1411:
1408:
1406:
1403:
1401:
1398:
1397:
1396:
1393:
1392:
1390:
1388:
1384:
1378:
1375:
1371:
1368:
1366:
1363:
1362:
1361:
1358:
1356:
1353:
1351:
1347:
1344:
1343:
1340:
1337:
1335:
1331:
1325:
1322:
1320:
1317:
1315:
1312:
1308:
1305:
1303:
1300:
1298:
1295:
1293:
1290:
1288:
1285:
1283:
1280:
1279:
1278:
1275:
1273:
1270:
1268:
1265:
1264:
1261:
1258:
1256:
1252:
1248:
1241:
1236:
1234:
1229:
1227:
1222:
1221:
1218:
1212:
1209:
1208:
1195:
1191:
1184:
1176:
1172:
1169:(3): 355–63.
1168:
1164:
1157:
1149:
1145:
1140:
1135:
1131:
1127:
1123:
1119:
1115:
1109:
1101:
1097:
1092:
1087:
1083:
1079:
1074:
1069:
1065:
1061:
1057:
1051:
1043:
1039:
1035:
1031:
1027:
1023:
1019:
1015:
1008:
1000:
996:
991:
986:
982:
978:
974:
968:
960:
956:
952:
948:
943:
938:
934:
930:
927:(4): 701–18.
926:
922:
915:
907:
905:0-07-284611-9
901:
897:
890:
882:
878:
871:
869:
867:
865:
863:
861:
859:
857:
855:
853:
851:
844:
842:
837:
830:
826:
822:
818:
814:
810:
803:
801:
799:
790:
786:
781:
776:
772:
768:
764:
757:
749:
743:
739:
738:
730:
722:
718:
711:
703:
697:
693:
692:
684:
682:
680:
671:
667:
660:
652:
648:
641:
639:
630:
624:
620:
619:
611:
609:
607:
602:
592:
591:Transcriptome
589:
587:
584:
582:
580:
576:
574:
571:
569:
567:
563:
562:
556:
553:
549:
545:
544:breast cancer
540:
530:
527:
525:
521:
516:
514:
510:
509:
504:
503:
497:
495:
491:
487:
477:
474:
473:autocatalytic
469:
467:
463:
453:
449:
445:
443:
439:
433:
423:
420:
414:
404:
401:
397:
391:
381:
379:
375:
371:
360:
355:
345:
343:
339:
335:
331:
327:
322:
318:
309:
307:
303:
298:
289:
286:
285:deacetylation
282:
278:
274:
272:
263:
259:
257:
252:
248:
244:
239:
231:
229:
225:
221:
216:
214:
210:
206:
202:
198:
193:
189:
185:
181:
172:
168:
166:
162:
157:
150:
140:
138:
134:
130:
126:
122:
118:
113:
111:
110:messenger RNA
107:
103:
99:
98:transcription
95:
91:
86:
84:
80:
76:
72:
68:
64:
60:
59:transcription
56:
52:
43:
34:
30:
19:
1936:Bacterial
1911:Lambda phage
1636:
1575:Constituents
1348: /
1334:RNA splicing
1194:Retroviruses
1193:
1183:
1166:
1162:
1156:
1121:
1117:
1108:
1063:
1059:
1050:
1017:
1013:
1007:
980:
976:
967:
924:
920:
914:
895:
889:
880:
833:
815:(1): 70–79.
812:
808:
770:
766:
756:
736:
729:
720:
710:
690:
669:
659:
650:
617:
578:
565:
548:retroviruses
536:
528:
517:
512:
506:
500:
498:
490:methotrexate
486:Fluorouracil
483:
470:
458:
446:
438:spliceosomes
435:
416:
393:
366:
357:
315:
295:
275:
268:
240:
237:
217:
177:
152:
114:
94:cell nucleus
87:
50:
48:
29:
1931:P1-derived
1699:Antisense
1592:Nucleotides
1587:Nucleosides
1582:Nucleobases
1360:Spliceosome
1319:RNA editing
462:spliceosome
440:made up of
419:poly-A tail
317:DNA damages
281:acetylation
220:nucleotides
211:(GMP), and
188:prokaryotes
79:translation
1883:Morpholino
1796:Organellar
1704:Processual
1669:Regulatory
1613:non-coding
597:References
520:HeLa cells
502:Drosophila
384:5' capping
370:5' capping
363:Processing
342:cell death
306:DNA damage
251:nucleosome
234:Regulation
143:Production
125:eukaryotes
83:processing
1843:Analogues
1828:Hachimoji
1611:(coding,
1566:Types of
1480:Cytosolic
1068:CiteSeerX
841:CC BY 4.0
581:-splicing
568:-splicing
271:silencers
243:enhancers
192:antisense
102:processed
1981:Category
1966:Category
1901:Phagemid
1752:Ribozyme
1175:12883652
1148:38720076
1139:11164319
1100:11179423
1042:39038640
959:21330280
951:19239890
843:license.
829:38103560
809:Mol Cell
789:28757210
559:See also
555:expand.
552:leukemia
480:Research
256:Mediator
165:promoter
1906:Plasmid
1460:PRPF40B
1455:PRPF40A
1445:PRPF38B
1440:PRPF38A
1255:Nuclear
999:2592384
721:Genomes
297:R-loops
292:R-loops
277:Histone
207:(CMP),
203:(AMP),
163:at the
121:nucleus
119:in the
1921:Fosmid
1916:Cosmid
1866:Hexose
1788:acids
1740:Others
1450:PRPF39
1435:PRPF31
1430:PRPF19
1425:PRPF18
1410:PRPF4B
1346:Intron
1173:
1146:
1136:
1118:Nature
1098:
1088:
1070:
1040:
1034:807333
1032:
997:
957:
949:
902:
827:
787:
744:
698:
625:
573:Outron
376:, and
228:intron
135:, and
129:5' cap
73:, and
1946:Human
1724:Y RNA
1503:DCP1B
1498:DCP1A
1420:PRPF8
1415:PRPF6
1405:PRPF4
1400:PRPF3
1395:PLRG1
1365:minor
1355:snRNP
1091:30951
1038:S2CID
955:S2CID
579:trans
513:Aedes
508:Aedes
75:rRNAs
71:tRNAs
67:mRNAs
1523:EDC4
1518:EDC3
1513:DCPS
1508:DCP2
1350:Exon
1307:CFII
1297:PAB2
1287:CstF
1282:CPSF
1171:PMID
1144:PMID
1096:PMID
1030:PMID
1014:Cell
995:PMID
947:PMID
921:Cell
900:ISBN
825:PMID
785:PMID
742:ISBN
696:ISBN
670:NCBI
623:ISBN
505:and
332:and
226:and
224:exon
184:tRNA
180:rRNA
1987:RNA
1302:CFI
1292:PAP
1134:PMC
1126:doi
1122:629
1086:PMC
1078:doi
1022:doi
985:doi
981:264
937:hdl
929:doi
925:136
817:doi
775:doi
566:cis
518:In
499:In
400:GTP
340:or
302:DNA
123:of
117:DNA
96:by
90:DNA
63:DNA
61:of
55:RNA
1983::
1370:U1
1192:.
1167:12
1165:.
1142:.
1132:.
1120:.
1116:.
1094:.
1084:.
1076:.
1064:12
1062:.
1058:.
1036:.
1028:.
1016:.
993:.
979:.
975:.
953:.
945:.
935:.
923:.
879:.
849:^
823:.
813:84
811:.
797:^
783:.
771:67
769:.
765:.
719:.
678:^
668:.
649:.
637:^
605:^
484:5-
468:.
444:.
380:.
372:,
344:.
245:.
182:,
131:,
85:.
69:,
49:A
1615:)
1559:e
1552:t
1545:v
1239:e
1232:t
1225:v
1177:.
1150:.
1128::
1102:.
1080::
1044:.
1024::
1018:5
1001:.
987::
961:.
939::
931::
908:.
831:.
819::
791:.
777::
750:.
704:.
631:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.