415:
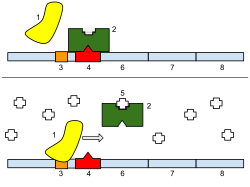
426:. In that study, two main forms of interference were measured. One is when an RNAP is on the downstream promoter, blocking the movement of RNAPs elongating from the upstream promoter. The other is when the two promoters are so close that when an RNAP sits on one of the promoters, it blocks any other RNAP from reaching the other promoter. These events are possible because the RNAP occupies several nucleotides when bound to the DNA, including in transcription start sites. Similar events occur when the promoters are in divergent and convergent formations. The possible events also depend on the distance between them.
644:
411:
divergent, tandem, and convergent directions. They can also be regulated by transcription factors and differ in various features, such as the nucleotide distance between them, the two promoter strengths, etc. The most important aspect of two closely spaced promoters is that they will, most likely, interfere with each other. Several studies have explored this using both analytical and stochastic models. There are also studies that measured gene expression in synthetic genes or from one to a few genes controlled by bidirectional promoters.
751:), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration). Several cell function specific transcription factors (there are about 1,600 transcription factors in a human cell) generally bind to specific motifs on an enhancer and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern the level of transcription of the target gene.
878:, and ACTACAnnTCCC are represented in bidirectional promoters at significantly higher rates than in unidirectional promoters. The absence of TATA boxes in bidirectional promoters suggests that TATA boxes play a role in determining the directionality of promoters, but counterexamples of bidirectional promoters do possess TATA boxes and unidirectional promoters without them indicates that they cannot be the only factor.
20:
588:
759:
may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration). An activated enhancer begins transcription of its RNA before activating a promoter to initiate transcription of messenger RNA from its target gene.
114:: The gene is turned on. Lactose is inhibiting the repressor, allowing the RNA polymerase to bind with the promoter and express the genes, which synthesize lactase. Eventually, the lactase will digest all of the lactose, until there is none to bind to the repressor. The repressor will then bind to the operator, stopping the manufacture of lactase.
995:. Importantly, intervention in the number or structure of promoter-bound proteins is one key to treating a disease without affecting expression of unrelated genes sharing elements with the target gene. Some genes whose change is not desirable are capable of influencing the potential of a cell to become cancerous.
953:
The initiation of the transcription is a multistep sequential process that involves several mechanisms: promoter location, initial reversible binding of RNA polymerase, conformational changes in RNA polymerase, conformational changes in DNA, binding of nucleoside triphosphate (NTP) to the functional
365:
The above consensus sequences, while conserved on average, are not found intact in most promoters. On average, only 3 to 4 of the 6 base pairs in each consensus sequence are found in any given promoter. Few natural promoters have been identified to date that possess intact consensus sequences at both
799:
of bidirectional promoter regions has been shown to downregulate both genes, and demethylation to upregulate both genes. There are exceptions to this, however. In some cases (about 11%), only one gene of a bidirectional pair is expressed. In these cases, the promoter is implicated in suppression of
738:
are regions of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene expression programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes. In a study of brain cortical neurons, 24,937 loops
758:
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two eRNAs as illustrated in the Figure. An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor
434:
Gene promoters are typically located upstream of the gene and can have regulatory elements several kilobases away from the transcriptional start site (enhancers). In eukaryotes, the transcriptional complex can cause the DNA to bend back on itself, which allows for placement of regulatory sequences
775:
in a bidirectional gene pair. A "bidirectional gene pair" refers to two adjacent genes coded on opposite strands, with their 5' ends oriented toward one another. The two genes are often functionally related, and modification of their shared promoter region allows them to be co-regulated and thus
708:
in CpG islands of promoters causes stable silencing of genes. However, the presence or absence of the other elements have relatively small effects on gene expression in experiments. Two sequences, the TATA box and Inr, caused small but significant increases in expression (45% and 28% increases,
410:
Promoters can be very closely located in the DNA. Such "closely spaced promoters" have been observed in the DNAs of all life forms, from humans to prokaryotes and are highly conserved. Therefore, they may provide some (presently unknown) advantages. These pairs of promoters can be positioned in
1081:. The presence of multiple methylated CpG sites in CpG islands of promoters causes stable silencing of genes. Silencing of a gene may be initiated by other mechanisms, but this is often followed by methylation of CpG sites in the promoter CpG island to cause the stable silencing of the gene.
188:
Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All DNAs have "Closely spaced promoters". Divergent, tandem, and convergent orientations are possible. Two closely spaced promoters will likely interfere. Regulatory elements can be several kilobases away from the
212:
Hypermethylation downregulates both genes, while demethylation upregulates them. Non-coding RNAs are linked to mRNA promoter regions. Subgenomic promoters range from 24 to 100 nucleotides (Beet necrotic yellow vein virus). Gene expression depends on promoter binding. Unwanted gene changes can
979:
Most diseases are heterogeneous in cause, meaning that one "disease" is often many different diseases at the molecular level, though symptoms exhibited and response to treatment may be identical. How diseases of different molecular origin respond to treatments is partially addressed in the
216:
MicroRNA promoters often contain CpG islands. DNA methylation forms 5-methylcytosines at the 5' pyrimidine ring of CpG cytosine residues. Some cancer genes are silenced by mutation, but most are silenced by DNA methylation. Others are regulated promoters. Selection may favor less energetic
938:
methods. A promoter region is located before the -35 and -10 Consensus sequences. The closer the promoter region is to the consensus sequences the more often transcription of that gene will take place. There is not a set pattern for promoter regions as there are for consensus sequences.
1157:
However, natural selection may favor less energetic binding as a way of regulating transcriptional output. In this case, we may call the most common sequence in a population the wild-type sequence. It may not even be the most advantageous sequence to have under prevailing conditions.
739:
were found, bringing enhancers to promoters. Multiple enhancers, each often at tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and coordinate with each other to control expression of their common target gene.
970:
shows high affinity to non-specific sites of the DNA, this characteristic does not allow us to clarify the process of promoter location. This process of promoter location has been attributed to the structure of the holoenzyme to DNA and sigma 4 to DNA complexes.
897:
usually begins bidirectionally, but divergent transcription is halted at a checkpoint later during elongation. Possible mechanisms behind this regulation include sequences in the promoter region, chromatin modification, and the spatial orientation of the DNA.
715:
that are localized in DNA regions distant from the promoters of genes can have very large effects on gene expression, with some genes undergoing up to 100-fold increased expression due to such a cis-regulatory module. These cis-regulatory modules include
192:
In eukaryotes, the transcriptional complex can bend DNA, allowing regulatory sequences to be placed far from the transcription site. The distal promoter is upstream of the gene and may contain additional regulatory elements with a weaker influence.
3681:
Copland JA, Sheffield-Moore M, Koldzic-Zivanovic N, Gentry S, Lamprou G, Tzortzatou-Stathopoulou F, et al. (June 2009). "Sex steroid receptors in skeletal differentiation and epithelial neoplasia: is tissue-specific intervention possible?".
704:(present in about 49% of promoters), upstream and downstream TFIIB recognition elements (BREu and BREd) (present in about 22% of promoters), and downstream core promoter element (DPE) (present in about 12% of promoters). The presence of multiple
109:: The transcription of the gene is turned off. There is no lactose to inhibit the repressor, so the repressor binds to the operator, which obstructs the RNA polymerase from binding to the promoter and making the mRNA encoding the lactase gene.
391:← upstream downstream → 5'-XXXXXXXPPPPPPXXXXXXPPPPPPXXXXGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGXXXX-3' -35 -10 Gene to be transcribed
295:) to direct the level of transcription of a given gene. A promoter is induced in response to changes in abundance or conformation of regulatory proteins in a cell, which enable activating transcription factors to recruit RNA polymerase.
683:
shown by a small red star on a transcription factor on the enhancer) the enhancer is activated and can now activate its target promoter. The active enhancer is transcribed on each strand of DNA in opposite directions by bound RNAP IIs.
755:(a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.
811:
Some functional classes of genes are more likely to be bidirectionally paired than others. Genes implicated in DNA repair are five times more likely to be regulated by bidirectional promoters than by unidirectional promoters.
742:
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of
957:
The promoter binding process is crucial in the understanding of the process of gene expression. Tuning synthetic genetic systems relies on precisely engineered synthetic promoters with known levels of transcription rates.
794:
analysis has shown bidirectionally paired genes to be co-expressed to a higher degree than random genes or neighboring unidirectional genes. Although co-expression does not necessarily indicate co-regulation,
670:
of the gene. The loop is stabilized by one architectural protein anchored to the enhancer and one anchored to the promoter and these proteins are joined to form a dimer (red zigzags). Specific regulatory
3217:
Koyanagi KO, Hagiwara M, Itoh T, Gojobori T, Imanishi T (July 2005). "Comparative genomics of bidirectional gene pairs and its implications for the evolution of a transcriptional regulation system".
1851:
Sneppen K, Dodd IB, Shearwin KE, Palmer AC, Schubert RA, Callen BP, Egan JB (February 2005). "A mathematical model for transcriptional interference by RNA polymerase traffic in
Escherichia coli".
1616:
Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, et al. (November 1993). "A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase".
366:
the -10 and -35; artificial promoters with complete conservation of the -10 and -35 elements have been found to transcribe at lower frequencies than those with a few mismatches with the consensus.
3999:
Burchard EG, Silverman EK, Rosenwasser LJ, Borish L, Yandava C, Pillari A, et al. (September 1999). "Association between a sequence variant in the IL-4 gene promoter and FEV(1) in asthma".
1886:
Martins L, Mäkelä J, Häkkinen A, Kandhavelu M, Yli-Harja O, Fonseca JM, Ribeiro AS (May 2012). "Dynamics of transcription of closely spaced promoters in
Escherichia coli, one event at a time".
692:
Up-regulated expression of genes in mammals is initiated when signals are transmitted to the promoters associated with the genes. Promoter DNA sequences may include different elements such as
1154:
In the case of a transcription factor binding site, there may be a single sequence that binds the protein most strongly under specified cellular conditions. This might be called canonical.
320:, where transcription of DNA begins for a particular gene (i.e., positions upstream are negative numbers counting back from -1, for example -100 is a position 100 base pairs upstream).
4075:
Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, et al. (July 1995). "Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP".
1808:
Korbel JO, Jensen LJ, von Mering C, Bork P (July 2004). "Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs".
461:
Eukaryotic promoter regulatory sequences typically bind proteins called transcription factors that are involved in the formation of the transcriptional complex. An example is the
4707:
3964:
Hobbs K, Negri J, Klinnert M, Rosenwasser LJ, Borish L (December 1998). "Interleukin-10 and transforming growth factor-beta promoter polymorphisms in allergies and asthma".
1117:
1090:
688:(a complex consisting of about 26 proteins in an interacting structure) communicates regulatory signals from the enhancer DNA-bound transcription factors to the promoter.
824:
and cellular metabolic genes are regulated by bidirectional promoters. The overrepresentation of bidirectionally paired DNA repair genes associates these promoters with
709:
respectively). The BREu and the BREd elements significantly decreased expression by 35% and 20%, respectively, and the DPE element had no detected effect on expression.
858:, and a symmetry around the midpoint of dominant Cs and As on one side and Gs and Ts on the other. A motif with the consensus sequence of TCTCGCGAGA, also called the
1188:. For example, to overexpress an important gene in a network, to yield higher production of target protein, synthetic biologists design promoters to upregulate its
1136:
1980:
Bordoy AE, Varanasi US, Courtney CM, Chatterjee A (December 2016). "Transcriptional
Interference in Convergent Promoters as a Means for Tunable Gene Expression".
918:(sgRNA) as one of the common infection techniques used by these viruses and generally transcribe late viral genes. Subgenomic promoters range from 24 nucleotide (
1093:. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation (see
934:
A wide variety of algorithms have been developed to facilitate detection of promoters in genomic sequence, and promoter prediction is a common element of many
376:
5'-AAAAAARNR-3' when centered in the -42 region; consensus sequence 5'-AWWWWWTTTTT-3' when centered in the -52 region; W = A or T; R = A or G; N = any base).
220:
Variations in promoters or transcription factors cause some diseases. Misunderstandings can result from using a canonical sequence to describe a promoter.
1661:"Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit"
835:
seem to be regulated by bidirectional promoters – significantly more than non-cancer causing genes. Hypermethylation of the promoters between gene pairs
477:). Some promoters that are targeted by multiple transcription factors might achieve a hyperactive state, leading to increased transcriptional activity.
3254:"Genome-wide analysis of the transcription factor binding preference of human bi-directional promoters and functional annotation of related gene pairs"
2304:
1135:
Silencing of DNA repair genes through methylation of CpG islands in their promoters appears to be especially important in progression to cancer (see
565:– the distal sequence upstream of the gene that may contain additional regulatory elements, often with a weaker influence than the proximal promoter
4036:"Thalassemia intermedia: moderate reduction of beta globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element"
2369:"Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters"
1151:
to refer to a promoter is often problematic, and can lead to misunderstandings about promoter sequences. Canonical implies perfect, in some sense.
859:
893:
are frequently associated with the promoter regions of mRNA-encoding genes. It has been hypothesized that the recruitment and initiation of
1520:"Promoter organization of the interferon-A genes differentially affects virus-induced expression and responsiveness to TBK1 and IKKepsilon"
4655:
4162:– a research project with an aim to generate 160 fully characterized, human DNA promoters of less than 4 kb (MiniPromoters) to drive
4127:
2155:"New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB"
608:
Text about mammals highly duplicated among uses of the same picture -- can we make a "canonical" version and redirect people there?
4169:
1261:
316:
As promoters are typically immediately adjacent to the gene in question, positions in the promoter are designated relative to the
4200:
458:. The TATA element and BRE typically are located close to the transcriptional start site (typically within 30 to 40 base pairs).
568:
Anything further upstream (but not an enhancer or other regulatory region whose influence is positional/orientation independent)
543:
Many other elements/motifs may be present. There is no such thing as a set of "universal elements" found in every core promoter.
3725:
Vlahopoulos SA, Logotheti S, Mikas D, Giarika A, Gorgoulis V, Zoumpourlis V (April 2008). "The role of ATF-2 in oncogenesis".
159:
4823:
4346:
1004:
1268:
system (Ptac). Notice how tac is written as a tac promoter, while in fact tac is actually both a promoter and an operator.
612:
414:
808:
to upregulate transcription of one gene, or remove bound transcription factors to downregulate transcription of one gene.
298:
Given the short sequences of most promoter elements, promoters can rapidly evolve from random sequences. For instance, in
4578:
4687:
1041:
4795:
1288:
1009:
In humans, about 70% of promoters located near the transcription start site of a gene (proximal promoters) contain a
630:
555:
3111:"Comprehensive annotation of bidirectional promoters identifies co-regulation among breast and ovarian cancer genes"
1053:, where the CpG island-containing promoter is located about 5,400 nucleotides upstream of the coding region of the
4949:
4893:
4550:
3770:"A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters"
3646:
Borukhov S, Nudler E (April 2003). "RNA polymerase holoenzyme: structure, function and biological implications".
3004:"Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems"
2906:"The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription"
987:
Not listed here are the many kinds of cancers involving aberrant transcriptional regulation owing to creation of
923:
548:
Proximal promoter – the proximal sequence upstream of the gene that tends to contain primary regulatory elements
308:
with only one mutation, and that ~10% of random sequences can serve as active promoters even without evolution.
4888:
4603:
3918:
2617:
Spitz F, Furlong EE (September 2012). "Transcription factors: from enhancer binding to developmental control".
4545:
1931:"Transcription closed and open complex formation coordinate expression of genes with a shared promoter region"
889:
assays have shown that over half of human genes do not have a strong directional bias. Research suggests that
197:(RNAP II) bound to the transcription start site promoter can start mRNA synthesis. It also typically contains
181:. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an
4976:
4907:
4648:
4598:
3401:"A methyl-sensitive element induces bidirectional transcription in TATA-less CpG island-associated promoters"
1298:
1233:
1232:
Some promoters are called constitutive as they are active in all circumstances in the cell, while others are
1196:
can be used to design neutral DNA or insulators that do not trigger gene expression of downstream sequences.
185:
binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters.
155:
4566:
4540:
2663:
Schoenfelder S, Fraser P (August 2019). "Long-range enhancer-promoter contacts in gene expression control".
1073:
In humans, DNA methylation occurs at the 5' position of the pyrimidine ring of the cytosine residues within
4712:
676:
444:
170:
long, the sequence of which is highly dependent on the gene and product of transcription, type or class of
1371:"Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria"
1204:
Some cases of many genetic diseases are associated with variations in promoters or transcription factors.
435:
far from the actual site of transcription. Eukaryotic RNA-polymerase-II-dependent promoters can contain a
275:
The process is more complicated, and at least seven different factors are necessary for the binding of an
4193:
388:. RNA polymerase holoenzymes containing other sigma factors recognize different core promoter sequences.
790:
database shared at least one database-assigned functional category with their partners 47% of the time.
4927:
4226:
2955:"MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300"
2519:"Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression"
248:
sequences of corresponding nucleotides that attach to specific promoters and regulate gene expression.
4159:
3002:
Carullo NV, Phillips Iii RA, Simon RC, Soto SA, Hinds JE, Salisbury AJ, et al. (September 2020).
1058:
4423:
4359:
4329:
4307:
2253:
Liefke R, Windhof-Jaidhauser IM, Gaedcke J, Salinas-Riester G, Wu F, Ghadimi M, Dango S (June 2015).
1221:
1094:
992:
603:
206:
2904:
Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, Furlong EE (January 2018).
850:
Certain sequence characteristics have been observed in bidirectional promoters, including a lack of
800:
the non-expressed gene. The mechanism behind this could be competition for the same polymerases, or
418:
Depiction the phenomenon of interference between tandem promoters. Figure created with BioRender.com
5030:
4971:
4759:
4672:
4641:
4217:
4209:
348:
317:
262:, which in turn are often brought to the promoter DNA by an activator protein's binding to its own
135:
2465:
Weingarten-Gabbay S, Nir R, Lubliner S, Sharon E, Kalma Y, Weinberger A, Segal E (February 2019).
1047:
Distal promoters also frequently contain CpG islands, such as the promoter of the DNA repair gene
4932:
4749:
4734:
4524:
4213:
1189:
667:
2015:
Chauhan V, Bahrudeen MN, Palma CS, Baptista IS, Almeida BL, Dash S, et al. (January 2022).
954:
RNA polymerase-promoter complex, and nonproductive and productive initiation of RNA synthesis.
481:
Core promoter – the minimal portion of the promoter required to properly initiate transcription
4937:
4862:
4754:
4433:
4186:
752:
712:
685:
466:
3917:
Hossain A, Lopez E, Halper SM, Cetnar DP, Reis AC, Strickland D, et al. (December 2020).
679:
bind to the promoter. When a transcription factor is activated by a signal (here indicated as
283:
Promoters represent critical elements that can work in concert with other regulatory regions (
4852:
4837:
4717:
4617:
4612:
4480:
4382:
3919:"Automated design of thousands of nonrepetitive parts for engineering stable genetic systems"
2706:
Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. (December 2017).
2320:"Regulation of gene expression via the core promoter and the basal transcriptional machinery"
2298:
2090:
729:
728:
and tethering elements. Among this constellation of elements, enhancers and their associated
537:
451:
237:
142:
transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (
1426:
Vaishnav ED, de Boer CG, Molinet J, Yassour M, Fan L, Adiconis X, et al. (March 2022).
1335:
4959:
4857:
4775:
4514:
4428:
4394:
4288:
4084:
3781:
3412:
3122:
2809:
2211:
2028:
1895:
1762:
1625:
1572:
1439:
1382:
1308:
1278:
725:
672:
447:
292:
241:
3878:"Role of ERCC1 promoter hypermethylation in drug resistance to cisplatin in human gliomas"
2114:"Synergy of human Pol II core promoter elements revealed by statistical sequence analysis"
8:
4497:
4351:
4325:
2517:
Beagan JA, Pastuzyn ED, Fernandez LR, Guo MH, Feng K, Titus KR, et al. (June 2020).
2255:"The oxidative demethylase ALKBH3 marks hyperactive gene promoters in human cancer cells"
1283:
735:
721:
717:
651:
643:
284:
4178:
4088:
3785:
3416:
3126:
3060:
Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM (January 2004).
2813:
2755:
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. (February 2018).
2215:
2032:
1899:
1766:
1629:
1576:
1443:
1386:
1336:"Analysis of Biological Networks: Transcriptional Networks – Promoter Sequence Analysis"
4920:
4803:
4266:
4108:
3946:
3853:
3828:
3804:
3769:
3750:
3707:
3484:
3459:
3435:
3400:
3376:
3351:
3280:
3253:
3199:
3145:
3110:
3028:
3003:
2930:
2905:
2881:
2856:
2832:
2797:
2732:
2707:
2688:
2642:
2594:
2567:
2543:
2518:
2491:
2466:
2393:
2368:
2344:
2319:
2281:
2254:
2235:
2051:
2016:
1957:
1930:
1833:
1593:
1560:
1460:
1427:
1403:
1370:
1256:; i.e., the lac promoter is IPTG inducible, meaning that besides the lac promoter, the
1253:
1244:
A tissue-specific promoter is a promoter that has activity in only certain cell types.
1148:
862:, was recently shown to drive PolII-driven bidirectional transcription in CpG islands.
817:
813:
516:
440:
373:
356:
232:, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as
3659:
3623:
3606:
3582:
3557:
3533:
3508:
3186:
3169:
3086:
3061:
2979:
2954:
2179:
2154:
1785:
1750:
1726:
1709:
1685:
1660:
881:
Although the term "bidirectional promoter" refers specifically to promoter regions of
236:
that provide a secure initial binding site for RNA polymerase and for proteins called
4585:
4100:
4057:
4016:
3981:
3950:
3938:
3899:
3858:
3809:
3742:
3711:
3699:
3663:
3628:
3587:
3573:
3538:
3524:
3489:
3460:"Transcription factor binding and modified histones in human bidirectional promoters"
3440:
3381:
3329:
3285:
3234:
3191:
3170:"Bidirectional gene organization: a common architectural feature of the human genome"
3150:
3091:
3033:
2984:
2935:
2886:
2837:
2778:
2737:
2692:
2680:
2646:
2634:
2599:
2548:
2496:
2444:
2398:
2349:
2286:
2227:
2184:
2135:
2094:
2056:
1997:
1962:
1911:
1868:
1825:
1790:
1731:
1710:"Bidirectional gene organization: a common architectural feature of the human genome"
1690:
1641:
1598:
1541:
1500:
1465:
1408:
1185:
1181:
948:
894:
829:
821:
701:
663:
598:
507:
499:
276:
233:
194:
5004:
2130:
2113:
1496:
372:
Some promoters contain one or more upstream promoter element (UP element) subsites (
304:, ~60% of random sequences can evolve expression levels comparable to the wild-type
4507:
4490:
4173:
4112:
4092:
4047:
4008:
3973:
3930:
3889:
3848:
3840:
3799:
3789:
3734:
3691:
3655:
3618:
3577:
3569:
3528:
3520:
3479:
3471:
3430:
3420:
3371:
3363:
3319:
3275:
3265:
3226:
3203:
3181:
3140:
3130:
3081:
3073:
3023:
3015:
2974:
2966:
2925:
2917:
2876:
2868:
2827:
2817:
2798:"Positional specificity of different transcription factor classes within enhancers"
2768:
2727:
2719:
2672:
2626:
2589:
2579:
2538:
2530:
2486:
2478:
2434:
2388:
2380:
2339:
2331:
2276:
2266:
2239:
2219:
2174:
2166:
2125:
2086:
2046:
2036:
1989:
1952:
1942:
1903:
1860:
1837:
1817:
1780:
1770:
1721:
1680:
1672:
1633:
1588:
1580:
1531:
1492:
1455:
1447:
1398:
1390:
1313:
1216:
1078:
981:
524:
490:
299:
263:
4052:
4035:
3754:
3324:
3307:
2953:
Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M (January 2003).
910:
gene, resulting in the formation of mRNA for that gene alone. Many positive-sense
4981:
4818:
4664:
4371:
4163:
4012:
3977:
3425:
3135:
2041:
935:
915:
890:
866:
are common, as they are in many promoters that lack TATA boxes. In addition, the
705:
680:
562:
3558:"RNA polymerase-promoter interactions: the comings and goings of RNA polymerase"
3509:"A positive-strand RNA virus with three very different subgenomic RNA promoters"
2335:
2202:
Levine M, Tjian R (July 2003). "Transcription regulation and animal diversity".
1659:
Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL (August 1999).
1483:
Yaniv M (September 2014). "Chromatin remodeling: from transcription to cancer".
654:
regulatory region is enabled to interact with the promoter region of its target
4808:
4780:
4581:
4569:
4519:
3774:
Proceedings of the
National Academy of Sciences of the United States of America
3607:"Promoter search by Escherichia coli RNA polymerase on a circular DNA template"
3230:
2802:
Proceedings of the
National Academy of Sciences of the United States of America
2773:
2756:
2723:
2384:
1907:
1755:
Proceedings of the
National Academy of Sciences of the United States of America
1584:
1451:
1394:
1162:
1010:
867:
422:
More recently, one study measured most genes controlled by tandem promoters in
288:
229:
171:
32:
4153:
3934:
3367:
3270:
2796:
Grossman SR, Engreitz J, Ray JP, Nguyen TH, Hacohen N, Lander ES (July 2018).
2676:
2584:
2534:
2271:
2252:
1993:
1864:
1125:
1102:
5024:
5009:
4696:
4283:
3680:
1097:). DNA methylation causing silencing in cancer typically occurs at multiple
988:
919:
787:
659:
503:
494:
3794:
3458:
Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, Weng Z (June 2007).
2822:
1637:
4883:
4813:
4682:
4020:
3942:
3903:
3862:
3813:
3746:
3703:
3695:
3667:
3542:
3493:
3444:
3385:
3333:
3306:
Shu J, Jelinek J, Chang H, Shen L, Qin T, Chung W, et al. (May 2006).
3289:
3238:
3195:
3154:
3095:
3037:
2988:
2970:
2939:
2921:
2890:
2841:
2782:
2741:
2684:
2638:
2603:
2552:
2500:
2448:
2402:
2353:
2290:
2231:
2139:
2098:
2060:
2001:
1966:
1947:
1915:
1872:
1829:
1775:
1751:"Overlapping promoters and their control in Escherichia coli: the gal case"
1735:
1694:
1676:
1602:
1545:
1536:
1519:
1504:
1469:
1412:
1265:
1169:
1033:
907:
520:
259:
228:
For transcription to take place, the enzyme that synthesizes RNA, known as
163:
4104:
4061:
3985:
3632:
3591:
3308:"Silencing of bidirectional promoters by DNA methylation in tumorigenesis"
3019:
2482:
2188:
1794:
1645:
1113:
484:
Includes the transcription start site (TSS) and elements directly upstream
402:
for -35 sequence T T G A C A 69% 79% 61% 56% 54% 54%
4986:
4915:
4470:
4438:
4341:
2153:
Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH (January 1998).
805:
796:
470:
399:
for -10 sequence T A T A A T 77% 76% 60% 61% 56% 82%
340:
3844:
2568:"The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1"
2223:
2170:
362:
The sequence at -35 (the -35 element) has the consensus sequence TTGACA.
4944:
4622:
4316:
4271:
4261:
4256:
4251:
4246:
3738:
3475:
3077:
2439:
2422:
1257:
1025:
967:
886:
791:
381:
305:
270:
240:
that recruit RNA polymerase. These transcription factors have specific
198:
167:
3894:
3877:
1929:
Häkkinen A, Oliveira SM, Neeli-Venkata R, Ribeiro AS (December 2019).
1013:. CpG islands are generally 200 to 2000 base pairs long, have a C:G
339:, the promoter contains two short sequence elements approximately 10 (
4278:
4096:
3399:
Mahpour A, Scruggs BS, Smiraglia D, Ouchi T, Gelman IH (2018-10-17).
2077:
Smale ST, Kadonaga JT (2003). "The RNA polymerase II core promoter".
1303:
1193:
1037:
1014:
911:
863:
832:
801:
245:
178:
154:. Promoters are located near the transcription start sites of genes,
45:
4034:
Kulozik AE, Bellan-Koch A, Bail S, Kohne E, Kleihauer E (May 1991).
2903:
2872:
2630:
1928:
1428:"The evolution, evolvability and engineering of gene regulatory DNA"
1236:, becoming active in the cell only in response to specific stimuli.
906:
A subgenomic promoter is a promoter added to a virus for a specific
767:
Bidirectional promoters are short (<1 kbp) intergenic regions of
4964:
4954:
4878:
4485:
4475:
2565:
1821:
1252:
When referring to a promoter some authors actually mean promoter +
1129:
1121:
1109:
1098:
1074:
1068:
1062:
1022:
855:
851:
697:
693:
533:
511:
436:
385:
380:
The above promoter sequences are recognized only by RNA polymerase
336:
253:
202:
119:
4633:
3998:
2464:
1199:
1112:
also silence or activate many genes in progression to cancer (see
4700:
4445:
4312:
1161:
Recent evidence also indicates that several genes (including the
1029:
131:
84:
3724:
2367:
Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E (March 2007).
2017:"Analytical kinetic model of native tandem promoters in E. coli"
1979:
1180:
Promoters are important gene regulatory elements used in tuning
1084:
4241:
3059:
1885:
1293:
1211:
844:
840:
825:
780:
777:
454:(BRE), which is recognized by the general transcription factor
394:
369:
The optimal spacing between the -35 and -10 sequences is 17 bp.
258:
The promoter is recognized by RNA polymerase and an associated
182:
71:
776:
co-expressed. Bidirectional promoters are a common feature of
4722:
4502:
4354:
3963:
3349:
3062:"An abundance of bidirectional promoters in the human genome"
2857:"The Mediator complex: a central integrator of transcription"
2152:
2014:
1518:
Civas A, Génin P, Morin P, Lin R, Hiscott J (February 2006).
1425:
1165:
1049:
974:
926:) and are usually found upstream of the transcription start.
871:
836:
772:
462:
455:
19:
3398:
3216:
3001:
2566:
Verheul TC, van Hijfte L, Perenthaler E, Barakat TS (2020).
1175:
465:(sequence CACGTG), which binds transcription factors in the
4573:
4403:
4334:
4033:
3350:
Wei W, Pelechano V, Järvelin AI, Steinmetz LM (July 2011).
1264:
would not have an inducible effect. Another example is the
1105:
that are present in the promoters of protein coding genes.
1089:
Generally, in progression to cancer, hundreds of genes are
882:
744:
655:
474:
151:
147:
143:
4172:
RNA and chromatin modification patterns around promoters.
4148:
4001:
American
Journal of Respiratory and Critical Care Medicine
3966:
American
Journal of Respiratory and Critical Care Medicine
3876:
Chen HY, Shao CJ, Chen FR, Kwan AL, Chen ZP (April 2010).
2708:"YY1 Is a Structural Regulator of Enhancer-Promoter Loops"
2516:
1807:
1260:
is also present. If the lac operator were not present the
1057:
gene. CpG islands also occur frequently in promoters for
732:
have a leading role in the regulation of gene expression.
189:
transcriptional start site in gene promoters (enhancers).
4692:
4208:
2795:
2754:
1850:
1615:
1018:
875:
768:
748:
139:
127:
4074:
3916:
2705:
1561:"Random sequences rapidly evolve into de novo promoters"
783:. About 11% of human genes are bidirectionally paired.
551:
Approximately 250 base pairs upstream of the start site
3555:
2366:
658:
by formation of a chromosome loop. This can initiate
311:
3457:
2317:
4154:
Identifying a
Protein Binding Sites on DNA molecule
3352:"Functional consequences of bidirectional promoters"
2460:
2458:
1142:
1032:
nucleotide and this occurs frequently in the linear
146:), or can have a function in and of itself, such as
3305:
2111:
1368:
4166:in defined brain regions of therapeutic interests.
3767:
3108:
2662:
1658:
1517:
1369:LaFleur TL, Hossain A, Salis HM (September 2022).
804:modification. Divergent transcription could shift
177:Promoters control gene expression in bacteria and
3829:"CpG islands and the regulation of transcription"
3556:deHaseth PL, Zupancic ML, Record MT (June 1998).
2952:
2455:
532:General transcription factor binding sites, e.g.
5022:
3875:
2423:"DNA methylation patterns and epigenetic memory"
1069:Methylation of CpG islands stably silences genes
174:recruited to the site, and species of organism.
2658:
2656:
1200:Diseases that may be associated with variations
1116:). Altered microRNA expression occurs through
493:: transcribes genes encoding 18S, 5.8S and 28S
4149:ORegAnno – Open Regulatory Annotation Database
3768:Saxonov S, Berg P, Brutlag DL (January 2006).
3645:
3109:Yang MQ, Koehly LM, Elnitski LL (April 2007).
1748:
1128:in promoters controlling transcription of the
405:
355:The sequence at -10 (the -10 element) has the
4649:
4194:
2467:"Systematic interrogation of human promoters"
2303:: CS1 maint: DOI inactive as of April 2024 (
2076:
1922:
1085:Promoter CpG hyper/hypo-methylation in cancer
675:bind to DNA sequence motifs on the enhancer.
3869:
3820:
3761:
3167:
3055:
3053:
3051:
3049:
3047:
2995:
2946:
2897:
2854:
2848:
2789:
2748:
2699:
2653:
2616:
2610:
2559:
2311:
2072:
2070:
2008:
1879:
1844:
1742:
1707:
1227:
762:
395:Probability of occurrence of each nucleotide
3598:
3251:
2572:Frontiers in Cell and Developmental Biology
2318:Juven-Gershon T, Kadonaga JT (March 2010).
2201:
1973:
1801:
1558:
1511:
1476:
1239:
998:
847:/BF195580 has been associated with tumors.
571:Specific transcription factor binding sites
4804:Precursor mRNA (pre-mRNA / hnRNA)
4656:
4642:
4201:
4187:
3826:
3506:
2246:
2112:Gershenzon NI, Ioshikhes IP (April 2005).
1701:
975:Diseases associated with aberrant function
820:are more than twice as likely. Many basic
4051:
3893:
3852:
3803:
3793:
3622:
3581:
3532:
3483:
3434:
3424:
3375:
3323:
3279:
3269:
3185:
3144:
3134:
3085:
3044:
3027:
2978:
2929:
2880:
2831:
2821:
2772:
2731:
2593:
2583:
2542:
2512:
2510:
2490:
2438:
2392:
2343:
2280:
2270:
2178:
2129:
2067:
2050:
2040:
1956:
1946:
1784:
1774:
1725:
1684:
1592:
1535:
1459:
1402:
1176:Synthetic promoter design and engineering
1137:methylation of DNA repair genes in cancer
631:Learn how and when to remove this message
3604:
2091:10.1146/annurev.biochem.72.121801.161520
1609:
1172:motifs as potential regulatory signals.
642:
413:
18:
2416:
2414:
2412:
1935:Journal of the Royal Society, Interface
696:(present in about 70% of promoters), a
666:(RNAP II) bound to the promoter at the
16:Region of DNA encouraging transcription
5023:
3301:
3299:
2861:Nature Reviews. Molecular Cell Biology
2507:
1559:Yona AH, Alm EJ, Gore J (April 2018).
1333:
648:Regulation of transcription in mammals
577:
4824:Histone acetylation and deacetylation
4637:
4347:Histone acetylation and deacetylation
4182:
3345:
3343:
1749:Herbert M, Kolb A, Buc H (May 1986).
1482:
1017:content >50%, and have regions of
1005:Regulation of transcription in cancer
700:(present in about 24% of promoters),
4889:Ribosome-nascent chain complex (RNC)
2420:
2409:
2360:
1364:
1362:
1360:
1358:
1356:
786:Bidirectionally paired genes in the
581:
443:TATAAA), which is recognized by the
4663:
3611:The Journal of Biological Chemistry
3296:
2855:Allen BL, Taatjes DJ (March 2015).
1524:The Journal of Biological Chemistry
312:Identification of relative location
166:). Promoters can be about 100–1000
13:
3340:
3252:Liu B, Chen J, Shen B (May 2011).
1247:
556:transcription factor binding sites
487:A binding site for RNA polymerase
14:
5042:
4142:
4125:
3168:Adachi N, Lieber MR (June 2002).
2757:"The Human Transcription Factors"
1708:Adachi N, Lieber MR (June 2002).
1353:
1289:Glossary of gene expression terms
1143:Canonical sequences and wild-type
816:are three times more likely, and
3605:Singer P, Wu CW (October 1987).
3574:10.1128/jb.180.12.3019-3025.1998
3525:10.1128/jvi.74.13.5988-5996.2000
586:
4894:Post-translational modification
4551:Archaeal transcription factor B
4119:
4068:
4027:
3992:
3957:
3910:
3882:International Journal of Cancer
3718:
3674:
3648:Current Opinion in Microbiology
3639:
3549:
3507:Koev G, Miller WA (July 2000).
3500:
3451:
3392:
3245:
3210:
3161:
3102:
2195:
2146:
2105:
1497:10.1016/j.cancergen.2014.03.006
924:Beet necrotic yellow vein virus
213:increase a cell's cancer risk.
3827:Deaton AM, Bird A (May 2011).
1888:Journal of Theoretical Biology
1652:
1552:
1419:
1327:
828:. Forty-five percent of human
1:
4053:10.1182/blood.V77.9.2054.2054
3660:10.1016/s1369-5274(03)00036-5
3624:10.1016/S0021-9258(18)47921-5
3325:10.1158/0008-5472.CAN-05-2629
3187:10.1016/S0092-8674(02)00758-4
2131:10.1093/bioinformatics/bti172
2079:Annual Review of Biochemistry
1727:10.1016/S0092-8674(02)00758-4
1320:
1299:Regulation of gene expression
901:
677:General transcription factors
519:: transcribes genes encoding
502:: transcribes genes encoding
429:
4013:10.1164/ajrccm.160.3.9812024
3978:10.1164/ajrccm.158.6.9804011
3426:10.1371/journal.pone.0205608
3136:10.1371/journal.pcbi.0030072
2042:10.1371/journal.pcbi.1009824
1853:Journal of Molecular Biology
929:
445:general transcription factor
330:
7:
2336:10.1016/j.ydbio.2009.08.009
1334:Sharan R (4 January 2007).
1271:
961:
922:) to over 100 nucleotides (
771:between the 5' ends of the
606:. The specific problem is:
406:Bidirectional (prokaryotic)
323:
223:
10:
5047:
4227:Transcriptional regulation
4132:San Diego State University
3231:10.1016/j.gene.2005.04.027
3115:PLOS Computational Biology
2774:10.1016/j.cell.2018.01.029
2724:10.1016/j.cell.2017.11.008
2385:10.1016/j.gene.2006.09.029
2021:PLOS Computational Biology
1908:10.1016/j.jtbi.2012.02.015
1585:10.1038/s41467-018-04026-w
1452:10.1038/s41586-022-04506-6
1395:10.1038/s41467-022-32829-5
1002:
946:
942:
318:transcriptional start site
207:TFIIB recognition elements
4997:
4906:
4871:
4845:
4836:
4794:
4768:
4742:
4733:
4671:
4594:
4559:
4533:
4458:
4424:Transcription coregulator
4416:
4393:
4370:
4360:Histone acetyltransferase
4330:Histone methyltransferase
4308:Histone-modifying enzymes
4306:
4299:
4234:
4225:
4160:Pleiades Promoter Project
3935:10.1038/s41587-020-0584-2
3368:10.1016/j.tig.2011.04.002
3271:10.1186/1752-0509-5-S1-S2
2677:10.1038/s41576-019-0128-0
2585:10.3389/fcell.2020.592164
2535:10.1038/s41593-020-0634-6
2272:10.1186/s13073-015-0180-0
1994:10.1021/acssynbio.5b00223
1865:10.1016/j.jmb.2004.11.075
1228:Constitutive vs regulated
1222:Rubinstein-Taybi syndrome
1095:DNA methylation in cancer
1059:functional noncoding RNAs
993:chromosomal translocation
763:Bidirectional (mammalian)
217:transcriptional binding.
4955:sequestration (P-bodies)
2665:Nature Reviews. Genetics
2619:Nature Reviews. Genetics
1240:Tissue-specific promoter
999:CpG islands in promoters
966:Although RNA polymerase
668:transcription start site
349:transcription start site
158:on the DNA (towards the
4933:Gene regulatory network
4525:Internal control region
4170:ENCODE threads Explorer
3833:Genes & Development
3795:10.1073/pnas.0510310103
3562:Journal of Bacteriology
2910:Genes & Development
2823:10.1073/pnas.1804663115
2427:Genes & Development
2421:Bird A (January 2002).
2275:(inactive 2024-04-27).
2159:Genes & Development
1665:Genes & Development
1638:10.1126/science.8248780
1108:Altered expressions of
4938:cis-regulatory element
4156:YouTube tutorial video
3696:10.1002/bies.200800138
3008:Nucleic Acids Research
2922:10.1101/gad.308619.117
1948:10.1098/rsif.2019.0507
1776:10.1073/pnas.83.9.2807
1677:10.1101/gad.13.16.2134
1537:10.1074/jbc.M506812200
1182:synthetically designed
1147:The usage of the term
1118:hyper/hypo-methylation
753:Mediator (coactivator)
713:Cis-regulatory modules
689:
686:Mediator (coactivator)
467:basic helix-loop-helix
419:
115:
4618:Intrinsic termination
4383:DNA methyltransferase
2483:10.1101/gr.236075.118
2324:Developmental Biology
1982:ACS Synthetic Biology
1565:Nature Communications
1375:Nature Communications
1341:. Tel Aviv University
1184:genetic circuits and
1091:silenced or activated
991:through pathological
730:transcription factors
673:transcription factors
646:
538:B recognition element
452:B recognition element
417:
343:) and 35 nucleotides
238:transcription factors
22:
4960:alternative splicing
4950:Post-transcriptional
4776:Transcription factor
4395:Chromatin remodeling
4128:"Expression vectors"
3923:Nature Biotechnology
2971:10.1093/emboj/cdg028
2718:(7): 1573–1588.e28.
1810:Nature Biotechnology
1309:Transcription factor
1279:Activator (genetics)
706:methylated CpG sites
662:(mRNA) synthesis by
613:improve this section
602:to meet Knowledge's
527:and other small RNAs
469:(bHLH) family (e.g.
448:TATA-binding protein
291:, boundary elements/
4884:Transfer RNA (tRNA)
4352:Histone deacetylase
4342:Histone demethylase
4326:Histone methylation
4089:1995Natur.376..348P
3845:10.1101/gad.2037511
3786:2006PNAS..103.1412S
3617:(29): 14178–14189.
3513:Journal of Virology
3417:2018PLoSO..1305608M
3258:BMC Systems Biology
3127:2007PLSCB...3...72Y
3020:10.1093/nar/gkaa671
2814:2018PNAS..115E7222G
2808:(30): E7222–E7230.
2523:Nature Neuroscience
2224:10.1038/nature01763
2216:2003Natur.424..147L
2171:10.1101/gad.12.1.34
2033:2022PLSCB..18E9824C
1900:2012JThBi.301...83M
1767:1986PNAS...83.2807H
1630:1993Sci...262.1407R
1624:(5138): 1407–1413.
1577:2018NatCo...9.1530Y
1444:2022Natur.603..455V
1387:2022NatCo..13.5159L
1284:Enhancer (genetics)
1114:microRNAs in cancer
818:mitochondrial genes
578:Mammalian promoters
4998:Influential people
4977:Post-translational
4796:Post-transcription
3739:10.1002/bies.20734
3476:10.1101/gr.5623407
3356:Trends in Genetics
3078:10.1101/gr.1982804
2440:10.1101/gad.947102
1207:Examples include:
1186:metabolic networks
1149:canonical sequence
854:, an abundance of
814:Chaperone proteins
690:
517:RNA polymerase III
508:small nuclear RNAs
441:consensus sequence
420:
374:consensus sequence
357:consensus sequence
116:
5018:
5017:
4902:
4901:
4832:
4831:
4708:Special transfers
4631:
4630:
4586:RNA polymerase II
4454:
4453:
4412:
4411:
4083:(6538): 348–351.
3929:(12): 1466–1475.
3895:10.1002/ijc.24772
3839:(10): 1010–1022.
3568:(12): 3019–3025.
3519:(13): 5988–5996.
3318:(10): 5077–5084.
3014:(17): 9550–9570.
2210:(6945): 147–151.
1988:(12): 1331–1341.
1941:(161): 20190507.
1671:(16): 2134–2147.
1438:(7901): 455–463.
1079:5-methylcytosines
1042:5' → 3' direction
1028:is followed by a
949:Promoter activity
895:RNA polymerase II
885:-encoding genes,
664:RNA polymerase II
641:
640:
633:
604:quality standards
595:This section may
525:5s ribosomal RNAs
500:RNA polymerase II
277:RNA polymerase II
234:response elements
195:RNA polymerase II
134:bind to initiate
126:is a sequence of
5038:
4843:
4842:
4740:
4739:
4658:
4651:
4644:
4635:
4634:
4508:Response element
4491:Response element
4304:
4303:
4232:
4231:
4203:
4196:
4189:
4180:
4179:
4174:Nature (journal)
4136:
4135:
4123:
4117:
4116:
4097:10.1038/376348a0
4072:
4066:
4065:
4055:
4046:(9): 2054–2058.
4031:
4025:
4024:
3996:
3990:
3989:
3972:(6): 1958–1962.
3961:
3955:
3954:
3914:
3908:
3907:
3897:
3888:(8): 1944–1954.
3873:
3867:
3866:
3856:
3824:
3818:
3817:
3807:
3797:
3780:(5): 1412–1417.
3765:
3759:
3758:
3722:
3716:
3715:
3678:
3672:
3671:
3643:
3637:
3636:
3626:
3602:
3596:
3595:
3585:
3553:
3547:
3546:
3536:
3504:
3498:
3497:
3487:
3455:
3449:
3448:
3438:
3428:
3411:(10): e0205608.
3396:
3390:
3389:
3379:
3347:
3338:
3337:
3327:
3303:
3294:
3293:
3283:
3273:
3249:
3243:
3242:
3214:
3208:
3207:
3189:
3165:
3159:
3158:
3148:
3138:
3106:
3100:
3099:
3089:
3057:
3042:
3041:
3031:
2999:
2993:
2992:
2982:
2959:The EMBO Journal
2950:
2944:
2943:
2933:
2901:
2895:
2894:
2884:
2852:
2846:
2845:
2835:
2825:
2793:
2787:
2786:
2776:
2752:
2746:
2745:
2735:
2703:
2697:
2696:
2660:
2651:
2650:
2614:
2608:
2607:
2597:
2587:
2563:
2557:
2556:
2546:
2514:
2505:
2504:
2494:
2462:
2453:
2452:
2442:
2418:
2407:
2406:
2396:
2364:
2358:
2357:
2347:
2315:
2309:
2308:
2302:
2294:
2284:
2274:
2250:
2244:
2243:
2199:
2193:
2192:
2182:
2150:
2144:
2143:
2133:
2124:(8): 1295–1300.
2109:
2103:
2102:
2074:
2065:
2064:
2054:
2044:
2012:
2006:
2005:
1977:
1971:
1970:
1960:
1950:
1926:
1920:
1919:
1883:
1877:
1876:
1848:
1842:
1841:
1805:
1799:
1798:
1788:
1778:
1761:(9): 2807–2811.
1746:
1740:
1739:
1729:
1705:
1699:
1698:
1688:
1656:
1650:
1649:
1613:
1607:
1606:
1596:
1556:
1550:
1549:
1539:
1515:
1509:
1508:
1480:
1474:
1473:
1463:
1423:
1417:
1416:
1406:
1366:
1351:
1350:
1348:
1346:
1340:
1331:
1314:Promoter bashing
1217:Beta thalassemia
982:pharmacogenomics
916:subgenomic mRNAs
636:
629:
625:
622:
616:
590:
589:
582:
491:RNA polymerase I
279:to the promoter.
264:DNA binding site
91:
78:
65:
52:
39:
26:
5046:
5045:
5041:
5040:
5039:
5037:
5036:
5035:
5031:Gene expression
5021:
5020:
5019:
5014:
4993:
4928:Transcriptional
4898:
4867:
4828:
4819:Polyadenylation
4790:
4764:
4729:
4723:Protein→Protein
4674:
4667:
4665:Gene expression
4662:
4632:
4627:
4602:
4596:
4590:
4555:
4529:
4450:
4408:
4389:
4372:DNA methylation
4366:
4310:
4295:
4221:
4207:
4164:gene expression
4145:
4140:
4139:
4124:
4120:
4073:
4069:
4032:
4028:
3997:
3993:
3962:
3958:
3915:
3911:
3874:
3870:
3825:
3821:
3766:
3762:
3723:
3719:
3679:
3675:
3644:
3640:
3603:
3599:
3554:
3550:
3505:
3501:
3464:Genome Research
3456:
3452:
3397:
3393:
3348:
3341:
3312:Cancer Research
3304:
3297:
3264:(Suppl 1): S2.
3250:
3246:
3215:
3211:
3166:
3162:
3107:
3103:
3066:Genome Research
3058:
3045:
3000:
2996:
2951:
2947:
2902:
2898:
2873:10.1038/nrm3951
2853:
2849:
2794:
2790:
2753:
2749:
2704:
2700:
2661:
2654:
2631:10.1038/nrg3207
2615:
2611:
2564:
2560:
2515:
2508:
2471:Genome Research
2463:
2456:
2419:
2410:
2365:
2361:
2316:
2312:
2296:
2295:
2259:Genome Medicine
2251:
2247:
2200:
2196:
2151:
2147:
2110:
2106:
2075:
2068:
2027:(1): e1009824.
2013:
2009:
1978:
1974:
1927:
1923:
1884:
1880:
1849:
1845:
1806:
1802:
1747:
1743:
1706:
1702:
1657:
1653:
1614:
1610:
1557:
1553:
1516:
1512:
1485:Cancer Genetics
1481:
1477:
1424:
1420:
1367:
1354:
1344:
1342:
1338:
1332:
1328:
1323:
1318:
1274:
1250:
1248:Use of the term
1242:
1230:
1202:
1178:
1145:
1087:
1071:
1007:
1001:
977:
964:
951:
945:
936:gene prediction
932:
904:
891:non-coding RNAs
843:/BC040563, and
765:
702:initiator (Inr)
681:phosphorylation
637:
626:
620:
617:
610:
591:
587:
580:
563:Distal promoter
432:
408:
403:
400:
397:
392:
333:
326:
314:
226:
110:
105:
89:
87:
76:
74:
63:
61:
50:
48:
37:
35:
24:
17:
12:
11:
5:
5044:
5034:
5033:
5016:
5015:
5013:
5012:
5007:
5005:François Jacob
5001:
4999:
4995:
4994:
4992:
4991:
4990:
4989:
4984:
4974:
4969:
4968:
4967:
4962:
4957:
4947:
4942:
4941:
4940:
4935:
4925:
4924:
4923:
4912:
4910:
4904:
4903:
4900:
4899:
4897:
4896:
4891:
4886:
4881:
4875:
4873:
4869:
4868:
4866:
4865:
4860:
4855:
4849:
4847:
4840:
4834:
4833:
4830:
4829:
4827:
4826:
4821:
4816:
4811:
4806:
4800:
4798:
4792:
4791:
4789:
4788:
4783:
4781:RNA polymerase
4778:
4772:
4770:
4766:
4765:
4763:
4762:
4757:
4752:
4746:
4744:
4737:
4731:
4730:
4728:
4727:
4726:
4725:
4720:
4715:
4705:
4704:
4703:
4685:
4679:
4677:
4669:
4668:
4661:
4660:
4653:
4646:
4638:
4629:
4628:
4626:
4625:
4620:
4615:
4609:
4607:
4592:
4591:
4589:
4588:
4582:RNA polymerase
4576:
4570:RNA polymerase
4563:
4561:
4557:
4556:
4554:
4553:
4548:
4543:
4537:
4535:
4531:
4530:
4528:
4527:
4522:
4517:
4512:
4511:
4510:
4505:
4495:
4494:
4493:
4488:
4483:
4478:
4473:
4462:
4460:
4456:
4455:
4452:
4451:
4449:
4448:
4443:
4442:
4441:
4436:
4431:
4420:
4418:
4414:
4413:
4410:
4409:
4407:
4406:
4400:
4398:
4391:
4390:
4388:
4387:
4386:
4385:
4377:
4375:
4368:
4367:
4365:
4364:
4363:
4362:
4357:
4344:
4339:
4338:
4337:
4322:
4320:
4301:
4297:
4296:
4294:
4293:
4292:
4291:
4286:
4276:
4275:
4274:
4269:
4264:
4259:
4254:
4249:
4238:
4236:
4229:
4223:
4222:
4206:
4205:
4198:
4191:
4183:
4177:
4176:
4167:
4157:
4151:
4144:
4143:External links
4141:
4138:
4137:
4118:
4067:
4026:
4007:(3): 919–922.
3991:
3956:
3909:
3868:
3819:
3760:
3733:(4): 314–327.
3717:
3690:(6): 629–641.
3673:
3638:
3597:
3548:
3499:
3470:(6): 818–827.
3450:
3391:
3362:(7): 267–276.
3339:
3295:
3244:
3225:(2): 169–176.
3209:
3180:(7): 807–809.
3160:
3101:
3043:
2994:
2965:(2): 281–291.
2945:
2896:
2867:(3): 155–166.
2847:
2788:
2767:(4): 650–665.
2747:
2698:
2671:(8): 437–455.
2652:
2625:(9): 613–626.
2609:
2558:
2529:(6): 707–717.
2506:
2477:(2): 171–183.
2454:
2408:
2359:
2330:(2): 225–229.
2310:
2245:
2194:
2145:
2118:Bioinformatics
2104:
2066:
2007:
1972:
1921:
1878:
1859:(2): 399–409.
1843:
1822:10.1038/nbt988
1816:(7): 911–917.
1800:
1741:
1720:(7): 807–809.
1700:
1651:
1608:
1551:
1530:(8): 4856–66.
1510:
1475:
1418:
1352:
1325:
1324:
1322:
1319:
1317:
1316:
1311:
1306:
1301:
1296:
1291:
1286:
1281:
1275:
1273:
1270:
1249:
1246:
1241:
1238:
1229:
1226:
1225:
1224:
1219:
1214:
1201:
1198:
1177:
1174:
1163:proto-oncogene
1144:
1141:
1086:
1083:
1070:
1067:
1003:Main article:
1000:
997:
989:chimeric genes
980:discipline of
976:
973:
963:
960:
944:
941:
931:
928:
914:produce these
903:
900:
764:
761:
639:
638:
621:September 2021
594:
592:
585:
579:
576:
575:
574:
573:
572:
569:
560:
559:
558:
552:
546:
545:
544:
541:
530:
529:
528:
514:
497:
495:ribosomal RNAs
485:
431:
428:
407:
404:
401:
398:
396:
393:
390:
378:
377:
370:
367:
363:
360:
332:
329:
325:
322:
313:
310:
281:
280:
273:
267:
256:
230:RNA polymerase
225:
222:
172:RNA polymerase
88:
75:
62:
49:
36:
33:RNA Polymerase
23:
15:
9:
6:
4:
3:
2:
5043:
5032:
5029:
5028:
5026:
5011:
5010:Jacques Monod
5008:
5006:
5003:
5002:
5000:
4996:
4988:
4985:
4983:
4980:
4979:
4978:
4975:
4973:
4972:Translational
4970:
4966:
4963:
4961:
4958:
4956:
4953:
4952:
4951:
4948:
4946:
4943:
4939:
4936:
4934:
4931:
4930:
4929:
4926:
4922:
4919:
4918:
4917:
4914:
4913:
4911:
4909:
4905:
4895:
4892:
4890:
4887:
4885:
4882:
4880:
4877:
4876:
4874:
4870:
4864:
4861:
4859:
4856:
4854:
4851:
4850:
4848:
4844:
4841:
4839:
4835:
4825:
4822:
4820:
4817:
4815:
4812:
4810:
4807:
4805:
4802:
4801:
4799:
4797:
4793:
4787:
4784:
4782:
4779:
4777:
4774:
4773:
4771:
4767:
4761:
4758:
4756:
4753:
4751:
4748:
4747:
4745:
4741:
4738:
4736:
4735:Transcription
4732:
4724:
4721:
4719:
4716:
4714:
4711:
4710:
4709:
4706:
4702:
4698:
4694:
4691:
4690:
4689:
4688:Central dogma
4686:
4684:
4681:
4680:
4678:
4676:
4670:
4666:
4659:
4654:
4652:
4647:
4645:
4640:
4639:
4636:
4624:
4621:
4619:
4616:
4614:
4611:
4610:
4608:
4605:
4600:
4593:
4587:
4583:
4580:
4577:
4575:
4571:
4568:
4565:
4564:
4562:
4558:
4552:
4549:
4547:
4544:
4542:
4539:
4538:
4536:
4532:
4526:
4523:
4521:
4518:
4516:
4513:
4509:
4506:
4504:
4501:
4500:
4499:
4496:
4492:
4489:
4487:
4484:
4482:
4479:
4477:
4474:
4472:
4469:
4468:
4467:
4464:
4463:
4461:
4457:
4447:
4444:
4440:
4437:
4435:
4432:
4430:
4427:
4426:
4425:
4422:
4421:
4419:
4415:
4405:
4402:
4401:
4399:
4396:
4392:
4384:
4381:
4380:
4379:
4378:
4376:
4373:
4369:
4361:
4358:
4356:
4353:
4350:
4349:
4348:
4345:
4343:
4340:
4336:
4333:
4332:
4331:
4327:
4324:
4323:
4321:
4318:
4314:
4309:
4305:
4302:
4298:
4290:
4289:trp repressor
4287:
4285:
4284:lac repressor
4282:
4281:
4280:
4277:
4273:
4270:
4268:
4265:
4263:
4260:
4258:
4255:
4253:
4250:
4248:
4245:
4244:
4243:
4240:
4239:
4237:
4233:
4230:
4228:
4224:
4219:
4215:
4211:
4210:Transcription
4204:
4199:
4197:
4192:
4190:
4185:
4184:
4181:
4175:
4171:
4168:
4165:
4161:
4158:
4155:
4152:
4150:
4147:
4146:
4133:
4129:
4122:
4114:
4110:
4106:
4102:
4098:
4094:
4090:
4086:
4082:
4078:
4071:
4063:
4059:
4054:
4049:
4045:
4041:
4037:
4030:
4022:
4018:
4014:
4010:
4006:
4002:
3995:
3987:
3983:
3979:
3975:
3971:
3967:
3960:
3952:
3948:
3944:
3940:
3936:
3932:
3928:
3924:
3920:
3913:
3905:
3901:
3896:
3891:
3887:
3883:
3879:
3872:
3864:
3860:
3855:
3850:
3846:
3842:
3838:
3834:
3830:
3823:
3815:
3811:
3806:
3801:
3796:
3791:
3787:
3783:
3779:
3775:
3771:
3764:
3756:
3752:
3748:
3744:
3740:
3736:
3732:
3728:
3721:
3713:
3709:
3705:
3701:
3697:
3693:
3689:
3685:
3677:
3669:
3665:
3661:
3657:
3654:(2): 93–100.
3653:
3649:
3642:
3634:
3630:
3625:
3620:
3616:
3612:
3608:
3601:
3593:
3589:
3584:
3579:
3575:
3571:
3567:
3563:
3559:
3552:
3544:
3540:
3535:
3530:
3526:
3522:
3518:
3514:
3510:
3503:
3495:
3491:
3486:
3481:
3477:
3473:
3469:
3465:
3461:
3454:
3446:
3442:
3437:
3432:
3427:
3422:
3418:
3414:
3410:
3406:
3402:
3395:
3387:
3383:
3378:
3373:
3369:
3365:
3361:
3357:
3353:
3346:
3344:
3335:
3331:
3326:
3321:
3317:
3313:
3309:
3302:
3300:
3291:
3287:
3282:
3277:
3272:
3267:
3263:
3259:
3255:
3248:
3240:
3236:
3232:
3228:
3224:
3220:
3213:
3205:
3201:
3197:
3193:
3188:
3183:
3179:
3175:
3171:
3164:
3156:
3152:
3147:
3142:
3137:
3132:
3128:
3124:
3120:
3116:
3112:
3105:
3097:
3093:
3088:
3083:
3079:
3075:
3071:
3067:
3063:
3056:
3054:
3052:
3050:
3048:
3039:
3035:
3030:
3025:
3021:
3017:
3013:
3009:
3005:
2998:
2990:
2986:
2981:
2976:
2972:
2968:
2964:
2960:
2956:
2949:
2941:
2937:
2932:
2927:
2923:
2919:
2915:
2911:
2907:
2900:
2892:
2888:
2883:
2878:
2874:
2870:
2866:
2862:
2858:
2851:
2843:
2839:
2834:
2829:
2824:
2819:
2815:
2811:
2807:
2803:
2799:
2792:
2784:
2780:
2775:
2770:
2766:
2762:
2758:
2751:
2743:
2739:
2734:
2729:
2725:
2721:
2717:
2713:
2709:
2702:
2694:
2690:
2686:
2682:
2678:
2674:
2670:
2666:
2659:
2657:
2648:
2644:
2640:
2636:
2632:
2628:
2624:
2620:
2613:
2605:
2601:
2596:
2591:
2586:
2581:
2577:
2573:
2569:
2562:
2554:
2550:
2545:
2540:
2536:
2532:
2528:
2524:
2520:
2513:
2511:
2502:
2498:
2493:
2488:
2484:
2480:
2476:
2472:
2468:
2461:
2459:
2450:
2446:
2441:
2436:
2432:
2428:
2424:
2417:
2415:
2413:
2404:
2400:
2395:
2390:
2386:
2382:
2378:
2374:
2370:
2363:
2355:
2351:
2346:
2341:
2337:
2333:
2329:
2325:
2321:
2314:
2306:
2300:
2292:
2288:
2283:
2278:
2273:
2268:
2264:
2260:
2256:
2249:
2241:
2237:
2233:
2229:
2225:
2221:
2217:
2213:
2209:
2205:
2198:
2190:
2186:
2181:
2176:
2172:
2168:
2164:
2160:
2156:
2149:
2141:
2137:
2132:
2127:
2123:
2119:
2115:
2108:
2100:
2096:
2092:
2088:
2084:
2080:
2073:
2071:
2062:
2058:
2053:
2048:
2043:
2038:
2034:
2030:
2026:
2022:
2018:
2011:
2003:
1999:
1995:
1991:
1987:
1983:
1976:
1968:
1964:
1959:
1954:
1949:
1944:
1940:
1936:
1932:
1925:
1917:
1913:
1909:
1905:
1901:
1897:
1893:
1889:
1882:
1874:
1870:
1866:
1862:
1858:
1854:
1847:
1839:
1835:
1831:
1827:
1823:
1819:
1815:
1811:
1804:
1796:
1792:
1787:
1782:
1777:
1772:
1768:
1764:
1760:
1756:
1752:
1745:
1737:
1733:
1728:
1723:
1719:
1715:
1711:
1704:
1696:
1692:
1687:
1682:
1678:
1674:
1670:
1666:
1662:
1655:
1647:
1643:
1639:
1635:
1631:
1627:
1623:
1619:
1612:
1604:
1600:
1595:
1590:
1586:
1582:
1578:
1574:
1570:
1566:
1562:
1555:
1547:
1543:
1538:
1533:
1529:
1525:
1521:
1514:
1506:
1502:
1498:
1494:
1490:
1486:
1479:
1471:
1467:
1462:
1457:
1453:
1449:
1445:
1441:
1437:
1433:
1429:
1422:
1414:
1410:
1405:
1400:
1396:
1392:
1388:
1384:
1380:
1376:
1372:
1365:
1363:
1361:
1359:
1357:
1337:
1330:
1326:
1315:
1312:
1310:
1307:
1305:
1302:
1300:
1297:
1295:
1292:
1290:
1287:
1285:
1282:
1280:
1277:
1276:
1269:
1267:
1263:
1259:
1255:
1245:
1237:
1235:
1223:
1220:
1218:
1215:
1213:
1210:
1209:
1208:
1205:
1197:
1195:
1191:
1187:
1183:
1173:
1171:
1167:
1164:
1159:
1155:
1152:
1150:
1140:
1138:
1133:
1131:
1127:
1123:
1119:
1115:
1111:
1106:
1104:
1100:
1096:
1092:
1082:
1080:
1076:
1066:
1064:
1060:
1056:
1052:
1051:
1045:
1043:
1039:
1035:
1031:
1027:
1024:
1020:
1016:
1012:
1006:
996:
994:
990:
985:
983:
972:
969:
959:
955:
950:
940:
937:
927:
925:
921:
920:Sindbis virus
917:
913:
909:
899:
896:
892:
888:
884:
879:
877:
873:
869:
865:
861:
857:
853:
848:
846:
842:
838:
834:
831:
827:
823:
819:
815:
809:
807:
803:
798:
793:
789:
788:Gene Ontology
784:
782:
779:
774:
770:
760:
756:
754:
750:
746:
740:
737:
733:
731:
727:
723:
719:
714:
710:
707:
703:
699:
695:
687:
682:
678:
674:
669:
665:
661:
660:messenger RNA
657:
653:
649:
645:
635:
632:
624:
614:
609:
605:
601:
600:
593:
584:
583:
570:
567:
566:
564:
561:
557:
553:
550:
549:
547:
542:
539:
535:
531:
526:
522:
518:
515:
513:
509:
505:
504:messenger RNA
501:
498:
496:
492:
489:
488:
486:
483:
482:
480:
479:
478:
476:
472:
468:
464:
459:
457:
453:
450:(TBP); and a
449:
446:
442:
438:
427:
425:
416:
412:
389:
387:
383:
375:
371:
368:
364:
361:
358:
354:
353:
352:
350:
346:
342:
338:
328:
321:
319:
309:
307:
303:
302:
296:
294:
290:
286:
278:
274:
272:
268:
265:
261:
257:
255:
251:
250:
249:
247:
243:
239:
235:
231:
221:
218:
214:
210:
208:
204:
200:
196:
190:
186:
184:
180:
175:
173:
169:
165:
161:
157:
153:
149:
145:
141:
137:
136:transcription
133:
129:
125:
121:
113:
108:
103:
100:: lacY,
99:
96:: lacZ,
95:
86:
82:
73:
69:
60:
56:
47:
43:
34:
30:
21:
4987:irreversible
4872:Key elements
4785:
4769:Key elements
4683:Genetic code
4673:Introduction
4465:
4131:
4121:
4080:
4076:
4070:
4043:
4039:
4029:
4004:
4000:
3994:
3969:
3965:
3959:
3926:
3922:
3912:
3885:
3881:
3871:
3836:
3832:
3822:
3777:
3773:
3763:
3730:
3726:
3720:
3687:
3683:
3676:
3651:
3647:
3641:
3614:
3610:
3600:
3565:
3561:
3551:
3516:
3512:
3502:
3467:
3463:
3453:
3408:
3404:
3394:
3359:
3355:
3315:
3311:
3261:
3257:
3247:
3222:
3218:
3212:
3177:
3173:
3163:
3118:
3114:
3104:
3072:(1): 62–66.
3069:
3065:
3011:
3007:
2997:
2962:
2958:
2948:
2916:(1): 42–57.
2913:
2909:
2899:
2864:
2860:
2850:
2805:
2801:
2791:
2764:
2760:
2750:
2715:
2711:
2701:
2668:
2664:
2622:
2618:
2612:
2575:
2571:
2561:
2526:
2522:
2474:
2470:
2430:
2426:
2379:(1): 52–65.
2376:
2372:
2362:
2327:
2323:
2313:
2299:cite journal
2262:
2258:
2248:
2207:
2203:
2197:
2165:(1): 34–44.
2162:
2158:
2148:
2121:
2117:
2107:
2082:
2078:
2024:
2020:
2010:
1985:
1981:
1975:
1938:
1934:
1924:
1891:
1887:
1881:
1856:
1852:
1846:
1813:
1809:
1803:
1758:
1754:
1744:
1717:
1713:
1703:
1668:
1664:
1654:
1621:
1617:
1611:
1568:
1564:
1554:
1527:
1523:
1513:
1491:(9): 352–7.
1488:
1484:
1478:
1435:
1431:
1421:
1378:
1374:
1343:. Retrieved
1329:
1266:Tac-Promoter
1251:
1243:
1231:
1206:
1203:
1192:. Automated
1179:
1170:G-quadruplex
1160:
1156:
1153:
1146:
1134:
1107:
1088:
1072:
1054:
1048:
1046:
1008:
986:
978:
965:
956:
952:
933:
908:heterologous
905:
880:
860:CGCG element
849:
822:housekeeping
810:
785:
766:
757:
741:
734:
711:
691:
650:. An active
647:
627:
618:
611:Please help
607:
596:
521:transfer RNA
506:and certain
460:
433:
423:
421:
409:
379:
344:
334:
327:
315:
306:lac promoter
300:
297:
282:
260:sigma factor
227:
219:
215:
211:
191:
187:
176:
164:sense strand
138:of a single
123:
117:
111:
106:
104:: lacA.
101:
97:
93:
80:
67:
58:
54:
41:
28:
4838:Translation
4675:to genetics
4595:Termination
4471:Pribnow box
4439:Corepressor
4434:Coactivator
4235:prokaryotic
2433:(1): 6–21.
2085:: 449–479.
1571:(1): 1530.
1381:(1): 5159.
1345:30 December
1126:CpG islands
1103:CpG islands
912:RNA viruses
864:CCAAT boxes
856:CpG islands
839:/CD558500,
806:nucleosomes
797:methylation
694:CpG islands
615:if you can.
471:BMAL1-Clock
384:containing
341:Pribnow Box
199:CpG islands
4982:reversible
4945:lac operon
4921:imprinting
4916:Epigenetic
4908:Regulation
4863:Eukaryotic
4809:5' capping
4760:Eukaryotic
4623:Rho factor
4613:Terminator
4604:eukaryotic
4579:eukaryotic
4560:Elongation
4546:Eukaryotic
4534:Initiation
4317:nucleosome
4300:eukaryotic
4272:gal operon
4267:ara operon
4262:Gua Operon
4257:gab operon
4252:trp operon
4247:lac operon
4218:Eukaryotic
3121:(4): e72.
2578:: 592164.
1321:References
1258:lac operon
1194:algorithms
1190:expression
1040:along its
1026:nucleotide
1011:CpG island
968:holoenzyme
947:See also:
902:Subgenomic
887:luciferase
852:TATA boxes
792:Microarray
726:insulators
430:Eukaryotic
382:holoenzyme
293:insulators
271:eukaryotes
179:eukaryotes
168:base pairs
4853:Bacterial
4750:Bacterial
4599:bacterial
4567:bacterial
4541:Bacterial
4515:Insulator
4459:Promotion
4429:Activator
4279:Repressor
4214:Bacterial
4126:Maloy S.
3951:220506228
3727:BioEssays
3712:205469320
3684:BioEssays
2693:152283312
2647:205485256
2265:(1): 66.
1894:: 83–94.
1304:Repressor
1234:regulated
1130:microRNAs
1122:CpG sites
1110:microRNAs
1099:CpG sites
1075:CpG sites
1063:microRNAs
1015:base pair
930:Detection
833:oncogenes
802:chromatin
778:mammalian
736:Enhancers
722:silencers
718:enhancers
554:Specific
347:from the
331:Bacterial
289:silencers
285:enhancers
246:repressor
242:activator
160:5' region
130:to which
46:Repressor
5025:Category
4965:microRNA
4879:Ribosome
4858:Archaeal
4814:Splicing
4786:Promoter
4755:Archaeal
4699: →
4695: →
4520:Silencer
4498:Enhancer
4486:CAAT box
4476:TATA box
4466:Promoter
4021:10471619
3943:32661437
3904:19626585
3863:21576262
3814:16432200
3747:18348191
3704:19382224
3668:12732296
3543:10846080
3494:17568000
3445:30332484
3405:PLOS ONE
3386:21601935
3334:16707430
3290:21689477
3239:15944140
3196:12110178
3155:17447839
3096:14707170
3038:32810208
2989:12514134
2940:29378788
2891:25693131
2842:29987030
2783:29425488
2742:29224777
2685:31086298
2639:22868264
2604:33102493
2553:32451484
2501:30622120
2449:11782440
2403:17123746
2354:19682982
2291:26221185
2232:12853946
2140:15572469
2099:12651739
2061:35100257
2002:27346626
1967:31822223
1916:22370562
1873:15670592
1830:15229555
1736:12110178
1695:10465790
1603:29670097
1546:16380379
1505:24825771
1470:35264797
1413:36056029
1272:See also
1254:operator
1077:to form
1061:such as
1034:sequence
1023:cytosine
1021:where a
962:Location
698:TATA box
652:enhancer
597:require
534:TATA box
512:microRNA
437:TATA box
386:sigma-70
345:upstream
337:bacteria
324:Elements
254:bacteria
224:Overview
203:TATA box
156:upstream
132:proteins
124:promoter
120:genetics
72:Operator
59:Promoter
4718:RNA→DNA
4713:RNA→RNA
4701:Protein
4446:Inducer
4313:histone
4113:4254507
4105:7630403
4085:Bibcode
4062:2018842
3986:9847292
3854:3093116
3805:1345710
3782:Bibcode
3633:3308887
3592:9620948
3485:1891341
3436:6192621
3413:Bibcode
3377:3123404
3281:3121118
3204:8556921
3146:1853124
3123:Bibcode
3029:7515708
2931:5828394
2882:4963239
2833:6065035
2810:Bibcode
2733:5785279
2595:7554316
2544:7558717
2492:6360817
2394:1955227
2345:2830304
2282:4517488
2240:4373712
2212:Bibcode
2189:9420329
2052:8830795
2029:Bibcode
1958:6936044
1896:Bibcode
1838:3546895
1795:3010319
1763:Bibcode
1646:8248780
1626:Bibcode
1618:Science
1594:5906472
1573:Bibcode
1461:8934302
1440:Bibcode
1404:9440211
1383:Bibcode
1168:) have
1101:in the
1030:guanine
943:Binding
870:NRF-1,
830:somatic
781:genomes
599:cleanup
424:E. coli
359:TATAAT.
301:E. coli
266:nearby.
162:of the
85:Lactose
83::
70::
57::
44::
31::
4242:Operon
4111:
4103:
4077:Nature
4060:
4019:
3984:
3949:
3941:
3902:
3861:
3851:
3812:
3802:
3755:678541
3753:
3745:
3710:
3702:
3666:
3631:
3590:
3583:107799
3580:
3541:
3534:112095
3531:
3492:
3482:
3443:
3433:
3384:
3374:
3332:
3288:
3278:
3237:
3202:
3194:
3153:
3143:
3094:
3087:314279
3084:
3036:
3026:
2987:
2980:140103
2977:
2938:
2928:
2889:
2879:
2840:
2830:
2781:
2740:
2730:
2691:
2683:
2645:
2637:
2602:
2592:
2551:
2541:
2499:
2489:
2447:
2401:
2391:
2352:
2342:
2289:
2279:
2238:
2230:
2204:Nature
2187:
2180:316406
2177:
2138:
2097:
2059:
2049:
2000:
1965:
1955:
1914:
1871:
1836:
1828:
1793:
1786:323395
1783:
1734:
1693:
1686:316962
1683:
1644:
1601:
1591:
1544:
1503:
1468:
1458:
1432:Nature
1411:
1401:
1294:Operon
1212:Asthma
868:motifs
845:KCNK15
841:CTDSPL
826:cancer
205:, and
183:enzyme
112:Bottom
92:
90:
79:
77:
66:
64:
53:
51:
40:
38:
27:
25:
4846:Types
4743:Types
4503:E-box
4355:HDAC1
4109:S2CID
4040:Blood
3947:S2CID
3751:S2CID
3708:S2CID
3200:S2CID
2689:S2CID
2643:S2CID
2236:S2CID
1834:S2CID
1339:(PDF)
1166:c-myc
1055:ERCC1
1050:ERCC1
1038:bases
872:GABPA
837:WNT9A
773:genes
463:E-box
456:TFIIB
4574:rpoB
4417:both
4404:CHD7
4335:EZH2
4101:PMID
4058:PMID
4017:PMID
3982:PMID
3939:PMID
3900:PMID
3859:PMID
3810:PMID
3743:PMID
3700:PMID
3664:PMID
3629:PMID
3588:PMID
3539:PMID
3490:PMID
3441:PMID
3382:PMID
3330:PMID
3286:PMID
3235:PMID
3219:Gene
3192:PMID
3174:Cell
3151:PMID
3092:PMID
3034:PMID
2985:PMID
2936:PMID
2887:PMID
2838:PMID
2779:PMID
2761:Cell
2738:PMID
2712:Cell
2681:PMID
2635:PMID
2600:PMID
2549:PMID
2497:PMID
2445:PMID
2399:PMID
2373:Gene
2350:PMID
2305:link
2287:PMID
2228:PMID
2185:PMID
2136:PMID
2095:PMID
2057:PMID
1998:PMID
1963:PMID
1912:PMID
1869:PMID
1826:PMID
1791:PMID
1732:PMID
1714:Cell
1691:PMID
1642:PMID
1599:PMID
1542:PMID
1501:PMID
1466:PMID
1409:PMID
1347:2012
1262:IPTG
883:mRNA
745:CTCF
656:gene
510:and
475:cMyc
201:, a
152:rRNA
148:tRNA
144:mRNA
122:, a
4697:RNA
4693:DNA
4481:BRE
4093:doi
4081:376
4048:doi
4009:doi
4005:160
3974:doi
3970:158
3931:doi
3890:doi
3886:126
3849:PMC
3841:doi
3800:PMC
3790:doi
3778:103
3735:doi
3692:doi
3656:doi
3619:doi
3615:262
3578:PMC
3570:doi
3566:180
3529:PMC
3521:doi
3480:PMC
3472:doi
3431:PMC
3421:doi
3372:PMC
3364:doi
3320:doi
3276:PMC
3266:doi
3227:doi
3223:353
3182:doi
3178:109
3141:PMC
3131:doi
3082:PMC
3074:doi
3024:PMC
3016:doi
2975:PMC
2967:doi
2926:PMC
2918:doi
2877:PMC
2869:doi
2828:PMC
2818:doi
2806:115
2769:doi
2765:172
2728:PMC
2720:doi
2716:171
2673:doi
2627:doi
2590:PMC
2580:doi
2539:PMC
2531:doi
2487:PMC
2479:doi
2435:doi
2389:PMC
2381:doi
2377:389
2340:PMC
2332:doi
2328:339
2277:PMC
2267:doi
2220:doi
2208:424
2175:PMC
2167:doi
2126:doi
2087:doi
2047:PMC
2037:doi
1990:doi
1953:PMC
1943:doi
1904:doi
1892:301
1861:doi
1857:346
1818:doi
1781:PMC
1771:doi
1722:doi
1718:109
1681:PMC
1673:doi
1634:doi
1622:262
1589:PMC
1581:doi
1532:doi
1528:281
1493:doi
1489:207
1456:PMC
1448:doi
1436:603
1399:PMC
1391:doi
1139:).
1124:in
1120:of
1036:of
1019:DNA
876:YY1
769:DNA
749:YY1
747:or
335:In
269:In
252:In
244:or
150:or
140:RNA
128:DNA
118:In
107:Top
5027::
4584::
4572::
4319:):
4216:,
4130:.
4107:.
4099:.
4091:.
4079:.
4056:.
4044:77
4042:.
4038:.
4015:.
4003:.
3980:.
3968:.
3945:.
3937:.
3927:38
3925:.
3921:.
3898:.
3884:.
3880:.
3857:.
3847:.
3837:25
3835:.
3831:.
3808:.
3798:.
3788:.
3776:.
3772:.
3749:.
3741:.
3731:30
3729:.
3706:.
3698:.
3688:31
3686:.
3662:.
3650:.
3627:.
3613:.
3609:.
3586:.
3576:.
3564:.
3560:.
3537:.
3527:.
3517:74
3515:.
3511:.
3488:.
3478:.
3468:17
3466:.
3462:.
3439:.
3429:.
3419:.
3409:13
3407:.
3403:.
3380:.
3370:.
3360:27
3358:.
3354:.
3342:^
3328:.
3316:66
3314:.
3310:.
3298:^
3284:.
3274:.
3260:.
3256:.
3233:.
3221:.
3198:.
3190:.
3176:.
3172:.
3149:.
3139:.
3129:.
3117:.
3113:.
3090:.
3080:.
3070:14
3068:.
3064:.
3046:^
3032:.
3022:.
3012:48
3010:.
3006:.
2983:.
2973:.
2963:22
2961:.
2957:.
2934:.
2924:.
2914:32
2912:.
2908:.
2885:.
2875:.
2865:16
2863:.
2859:.
2836:.
2826:.
2816:.
2804:.
2800:.
2777:.
2763:.
2759:.
2736:.
2726:.
2714:.
2710:.
2687:.
2679:.
2669:20
2667:.
2655:^
2641:.
2633:.
2623:13
2621:.
2598:.
2588:.
2574:.
2570:.
2547:.
2537:.
2527:23
2525:.
2521:.
2509:^
2495:.
2485:.
2475:29
2473:.
2469:.
2457:^
2443:.
2431:16
2429:.
2425:.
2411:^
2397:.
2387:.
2375:.
2371:.
2348:.
2338:.
2326:.
2322:.
2301:}}
2297:{{
2285:.
2261:.
2257:.
2234:.
2226:.
2218:.
2206:.
2183:.
2173:.
2163:12
2161:.
2157:.
2134:.
2122:21
2120:.
2116:.
2093:.
2083:72
2081:.
2069:^
2055:.
2045:.
2035:.
2025:18
2023:.
2019:.
1996:.
1984:.
1961:.
1951:.
1939:16
1937:.
1933:.
1910:.
1902:.
1890:.
1867:.
1855:.
1832:.
1824:.
1814:22
1812:.
1789:.
1779:.
1769:.
1759:83
1757:.
1753:.
1730:.
1716:.
1712:.
1689:.
1679:.
1669:13
1667:.
1663:.
1640:.
1632:.
1620:.
1597:.
1587:.
1579:.
1567:.
1563:.
1540:.
1526:.
1522:.
1499:.
1487:.
1464:.
1454:.
1446:.
1434:.
1430:.
1407:.
1397:.
1389:.
1379:13
1377:.
1373:.
1355:^
1132:.
1065:.
1044:.
984:.
874:,
724:,
720:,
536:,
523:,
473:,
351:.
287:,
209:.
4657:e
4650:t
4643:v
4606:)
4601:,
4597:(
4397::
4374::
4328:/
4315:/
4311:(
4220:)
4212:(
4202:e
4195:t
4188:v
4134:.
4115:.
4095::
4087::
4064:.
4050::
4023:.
4011::
3988:.
3976::
3953:.
3933::
3906:.
3892::
3865:.
3843::
3816:.
3792::
3784::
3757:.
3737::
3714:.
3694::
3670:.
3658::
3652:6
3635:.
3621::
3594:.
3572::
3545:.
3523::
3496:.
3474::
3447:.
3423::
3415::
3388:.
3366::
3336:.
3322::
3292:.
3268::
3262:5
3241:.
3229::
3206:.
3184::
3157:.
3133::
3125::
3119:3
3098:.
3076::
3040:.
3018::
2991:.
2969::
2942:.
2920::
2893:.
2871::
2844:.
2820::
2812::
2785:.
2771::
2744:.
2722::
2695:.
2675::
2649:.
2629::
2606:.
2582::
2576:8
2555:.
2533::
2503:.
2481::
2451:.
2437::
2405:.
2383::
2356:.
2334::
2307:)
2293:.
2269::
2263:7
2242:.
2222::
2214::
2191:.
2169::
2142:.
2128::
2101:.
2089::
2063:.
2039::
2031::
2004:.
1992::
1986:5
1969:.
1945::
1918:.
1906::
1898::
1875:.
1863::
1840:.
1820::
1797:.
1773::
1765::
1738:.
1724::
1697:.
1675::
1648:.
1636::
1628::
1605:.
1583::
1575::
1569:9
1548:.
1534::
1507:.
1495::
1472:.
1450::
1442::
1415:.
1393::
1385::
1349:.
634:)
628:(
623:)
619:(
540:.
439:(
102:8
98:7
94:6
81:5
68:4
55:3
42:2
29:1
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.