619:, where the nucleus remains intact, changes in the permeability barrier of the nuclear envelope (NE) are attributed to alterations within the NPC. These changes facilitate the entry of mitotic regulators into the nucleus. Studies in Aspergillys nidulans suggest that the NPC composition appears to be effeveted by the mitotiv kinase NIMA. NIMA potentially phosphorylates nucleoporins Nup98 and Gle2/Rae1, leading to NPC remodeling. This remodeling allows the nuclear entry of the protein complex cdc2/cyclinB and various other proteins, including soluble tubulin. The NPC scaffold remains intact throughout the whole closed mitosis. This seems to preserver the integrity of NE.
351:
31:
533:
578:(NE) formation using electron microscopy. During the interphase of the cell cycle the formation of the prepore would happen within the nucleus, each component being transported in through existing NPCs. These Nups would bind to an importin, once formed, preventing the assembly of a prepore in the cytoplasm. Once transported into the nucleus Ran GTP would bind to the importin and cause it to release the cargo. This Nup would be free to form a prepore. The binding of
601:
unknown in vivo. In metazoans (which undergo open mitosis) the NE degrades quickly after the loss of the peripheral Nups. The reason for this may be due to the change in the NPC's architecture. This change may make the NPC more permeable to enzymes involved in the degradation of the NE such as cytoplasmic tubulin, as well as allowing the entry of key mitotic regulator proteins. In organisms that undergo a semi-open mitosis such as the filamentous fungus
566:. It is then inserted into the double membrane close to the chromatin. This, in turn, leads to the fusing of that membrane. Around this protein complex others eventually bind forming the NPC. This method is possible during every phase of mitosis as the double membrane is present around the chromatin before the membrane fusion proteins complex can insert. Post mitotic cells could form a membrane first with pores being inserted into after formation.
43:
482:, where GTP hydrolysis occurs, releasing the NES-containing protein. The resulting CRM1-RanGDP complex returns to the nucleus, where RanGEFs catalyze the exchange of GDP for GTP on Ran, replenishing the system's energy source. This entire process is energy-dependent and consumes one GTP molecule. Notably, the export activity mediated by CRM1 can be inhibited by compounds like
359:
374:
Nuclear pore complex (NPC) serves highly regulated gateway for the transport of molecules between the nucleus and the cytoplasm. This intricate system enables the selective passage for molecules including proteins, RNA, and signaling molecules, ensuring proper cellular function and homeostasis. Small
454:
While translocation through the NPC is not energy-dependent, the overall import cycle needs the hydrolysis of two GTPs molecules, making it an active transport process. The import cycle is powered by the nucleo-cytoplasmic RanGTP gradient. This gradient arises from the exclusive nuclear localization
600:
remain stable, as cylindrical ring complexes within the nuclear envelope. This disassembly of the NPC peripheral groups is largely thought to be phosphate driven, as several of these nucleoporins are phosphorylated during the stages of mitosis. However, the enzyme involved in the phosphorylation is
557:
There are several theories as to how NPCs are assembled. As the immunodepletion of certain protein complexes, such as the Nup 107–160 complex, leads to the formation of poreless nuclei, it seems likely that the Nup complexes are involved in fusing the outer membrane of the nuclear envelope with the
523:
In highest eukaryotes, mRNA export is believed to be spicling-dependent. Splicing recruits the TREX protein complex to spliced messages, serving as an adapter for TAP, a low-affinity RNA-binding protein
However, there are alternative mRNA export pathways that do not rely on splicing for specialized
434:
Nuclear proteins are synthesized in the cytoplasm and need to be imported through the NPCs into the nucleus. Import can be directed by various signals, of which nuclear localization signal (NLS) are best characterized. Several NLS sequences are known, generally containing a conserved sequence with
569:
Another model for the formation of the NPC is the production of a prepore as a start as opposed to a single protein complex. This prepore would form when several Nup complexes come together and bind to the chromatin. This would have the double membrane form around it in during mitotic reassembly.
308:
encircling the actual pore, forming the outer ring. Additionally, these subunits project a spoke-shaped protein over the pore channel. The central region of the pore may exhibit a plug-like structure; however, its precise nature remains unknown, and it is yet undetermined whether it represents an
544:
stages characterized by high transcription rates is crucial. For example, cycling mammalian and yeast cells double the amount of NPC in the nucleus between the G1 and G2 phase. Similarly, oocytes accumulate abundant NPCs in anticipation of the rapid mitotic activity during early development.
299:
The count of nuclear pore complexes varies across cell types and different stages of the cell's life cycle, with approximately 1,000 NPCs typically found in vertebrate cells. The human nuclear pore complex (hNPC) is a substantial structure, with a molecular weight of 120
607:, 14 out of the 30 nucleoporins disassemble from the core scaffold structure, driven by the activation of the NIMA and Cdk1 kinases that phosphorylate nucleoporins and open nuclear pores thereby widening the nuclear pore and allowing the entry of mitotic regulators.
447:(CAS), an exportin which in the nucleus is bound to RanGTP, displaces Importin-α from the cargo. The NLS-protein is thus free in the nucleoplasm. The Importinβ-RanGTP and Importinα-CAS-RanGTP complex diffuses back to the cytoplasm where
394:. These are a superfamily of nuclear transport receptors that facilitate the translocation of proteins, RNAs, and ribonuclear particles across the NPC in a Ran GTP hydrolase-dependent process. This family is further subdivided to the
582:
has at least been shown to bring Nup 107 and the Nup 153 nucleoporins into the nucleus. NPC assembly is a very rapid process yet defined intermediate states occur which leads to the idea that this assembly occurs in a stepwise
332:(MDa), comprising approximately 30 distinct protein components, each in multiple copies. The mammalian NPCs contain about 800 nucleoporins each that are organized into distinct NPC subcomplexes. Conversely, the yeast
317:
The nuclear pore complex (NPC) is a crucial cellular structure with a diameter of approximately 120 nanometers in vertebrates. Its channel varies from 5.2 nanometers in humans to 10.7 nm in the frog
441:
As the complex reaches the NPC, it diffuses through the pore without the need for additional energy. Upon entry into nucleus, RanGTP binds to
Importin-β and displaces it from the complex. Then the
524:
messages such as histones. Recent work also suggest an interplay between splicing-dependent export and one of these alternative mRNA export pathways for secretory and mitochondrial transcripts.
292:
regulate molecular transport through the nuclear pore. Nucleoporin-mediated transport does not entail direct energy expenditure but instead relies on concentration gradients associated with the
226:
found in the NPC. The other two are the transmembrane Nups and the scaffold Nups. The transmembrane Nups are made up of transmembrane α-helices and play a vital part in anchoring the NPC to the
549:
must maintain NPC generation to sustain consistent NPC levels, as some may incur damage. Furthermore, certain cells can even increase the NPC numbers due to increased transcriptional demand.
635:
Lin, D. H., Stuwe, T., Schilbach, S., Rundlet, E. J., Perriches, T., Mobbs, G., ... Hoelz, A. (2016). Architecture of the nuclear pore complex symmetric core. Science, 352(6283), aaf1015.
504:
export is signal-mediated, with nuclear export signals (NES) present in RNA-binding proteins, except for tRNA which lacks an adapter. It is notable that all viral RNAs and cellular RNAs (
520:) except mRNA are dependent on RanGTP. Conserved mRNA export factors are necessary for mRNA nuclear export. Export factors are Mex67/Tap (large subunit) and Mtr2/p15 (small subunit).
438:
Importation begins with
Importin-α binding to the NLS sequence of cargo proteins, forming a complex. Importin-β then attaches to Importin-α, facilitating transport towards the NPC.
241:
The principal function of nuclear pore complexes is to facilitate selective membrane transport of various molecules across the nuclear envelope. This includes the transportation of
1284:
1314:
2364:
273:
moving into the nucleus. Notably, the nuclear pore complex (NPC) can actively mediate up to 1000 translocations per complex per second. While smaller molecules can
592:
During mitosis the NPC appears to disassemble in stages, except in lower eukaryotes like yeast, where NPC disassembly does not happen during mitosis. Peripheral
104:
558:
inner and not that the fusing of the membrane begins the formation of the pore. There are several ways that this could lead to the formation of the full NPC.
455:
of RanGEFs, proteins that exchange GDP to GTP on Ran molecules. Thus, there is an elevated RanGTP concentration in the nucleus compared to the cytoplasm.
1798:"The Three Fungal Transmembrane Nuclear Pore Complex Proteins of Aspergillus nidulans Are Dispensable in the Presence of an Intact An-Nup84-120 Complex"
324:, with a depth of roughly 45 nm. Additionally, mRNA, being single-stranded, has a thickness ranging from 0.5 to 1 nm. The mammalian NPC has a
1619:
1197:
47:
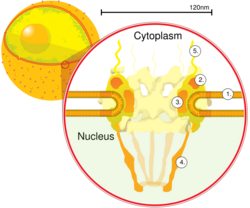
Schematic diagram of a nuclear pore complex within the nuclear envelope (1) with the outer ring (2), spokes (3), basket (4), and filaments (5).
478:
complex with an exportin and RanGTP within the nucleus. Example of such an exportin is CRM1. This complex subsequently translocate to the
1276:
443:
471:, require export from the nucleus to the cytoplasm. This export process mirrors the import mechanism in complexity and importance.
451:
are hydrolyzed to GDP leading to the release of
Importinβ and Importinα which become available for a new NLS-protein import round.
1643:"Nup2 requires a highly divergent partner, NupA, to fulfill functions at nuclear pore complexes and the mitotic chromatin region"
1532:
685:
1887:
1875:
1306:
830:"Evolutionarily Conserved Sequence Features Regulate the Formation of the FG Network at the Center of the Nuclear Pore Complex"
1901:
1395:"Genome analysis reveals interplay between 5'UTR introns and nuclear mRNA export for secretory and mitochondrial genes"
887:"Physical motif clustering within intrinsically disordered nucleoporin sequences reveals universal functional features"
1928:
200:
1447:
Rabut G, Lénárt P, Ellenberg J (June 2004). "Dynamics of nuclear pore complex organization through the cell cycle".
179:
proteins, in 2022 90% of the structure was known, then in 2024 the nuclear basket was decoded. About half of the
648:
Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turoňová B, Zimmerli CE, et al. (June 10, 2022).
354:
The Ran-GTP cycle, which drives the import and export of RNA and proteins through the nuclear protein complex.
2412:
277:
through the pores, larger molecules are often identified by specific signal sequences and are facilitated by
99:
1583:
429:
2298:
1560:
435:
basic residues such as PKKKRKV. Any material with an NLS will be taken up by importins to the nucleus.
1337:
1151:
596:
such as the Nup 153 Nup 98 and Nup 214 disassociate from the NPC. The rest, which can be considered a
92:
2407:
2382:
2136:
1533:"Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos"
2119:
2114:
2109:
2104:
2099:
2094:
1906:
1866:
333:
87:
75:
1897:
3D electron microscopy structures of the NPC and constituent proteins from the EM Data Bank(EMDB)
708:
707:
Singh D, Soni N, Hutchings J, Echeverria I, Shaikh F, Duquette M, et al. (August 9, 2024).
1484:"Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs"
650:"AI-based structure prediction empowers integrative structural analysis of human nuclear pores"
448:
111:
2402:
1921:
175:(Nups). Each human NPC comprises at least 456 individual protein molecules, with 34 distinct
1856:
1703:
898:
841:
603:
1599:
1362:
222:
repeats (FG repeats) in their amino acid sequences. FG-Nups is one of three main types of
8:
1221:"Saccharomyces cerevisiae exhibits a yAP-1-mediated adaptive response to malondialdehyde"
358:
266:
1862:
1707:
1692:"Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans"
902:
845:
375:
molecules such as proteins water and ions can diffuse through NPCs, but cargoes (>40
2059:
1830:
1797:
1773:
1748:
1729:
1667:
1642:
1508:
1483:
1421:
1394:
1127:
1094:
1070:
1045:
1021:
996:
921:
886:
862:
829:
764:
737:
463:
In addition to nuclear import, certain molecules and macromolecular complexes, such as
350:
285:
204:
165:
1353:
1253:
1220:
972:
945:
649:
1884:
1835:
1817:
1778:
1721:
1672:
1611:
1603:
1552:
1513:
1464:
1426:
1375:
1367:
1258:
1240:
1189:
1181:
1132:
1114:
1075:
1026:
977:
926:
867:
810:
769:
677:
669:
540:
Since the NPC regulates genome access, its presence in significant quantities during
474:
In a classical export scenario, proteins with a nuclear export sequence (NES) form a
403:
383:
345:
274:
246:
192:
1872:
1733:
1393:
Cenik C, Chua HN, Zhang H, Tarnawsky SP, Akef A, Derti A, et al. (April 2011).
1236:
962:
379:) such as RNA and protein require the participation of soluble transport receptors.
1950:
1914:
1825:
1809:
1768:
1760:
1711:
1662:
1654:
1595:
1544:
1503:
1495:
1456:
1416:
1406:
1357:
1349:
1248:
1232:
1171:
1163:
1122:
1106:
1065:
1057:
1016:
1008:
967:
957:
916:
906:
857:
849:
800:
759:
754:
749:
716:
661:
597:
575:
329:
227:
153:
149:
145:
2153:
1942:
1891:
1879:
1411:
911:
805:
788:
305:
141:
80:
1046:"Characterisation of the passive permeability barrier of nuclear pore complexes"
2329:
1999:
1955:
1176:
738:"Pore timing: the evolutionary origins of the nucleus and nuclear pore complex"
721:
475:
363:
325:
320:
254:
235:
231:
188:
184:
1896:
1716:
1691:
1460:
1167:
2396:
1821:
1607:
1371:
1244:
1185:
1118:
673:
468:
262:
215:
30:
1813:
1658:
1548:
665:
2183:
1938:
1839:
1782:
1725:
1676:
1615:
1556:
1531:
Kiseleva E, Rutherford S, Cotter LM, Allen TD, Goldberg MW (October 2001).
1468:
1430:
1193:
1136:
1110:
1079:
1061:
1030:
1012:
981:
930:
871:
814:
773:
681:
532:
289:
278:
211:
196:
180:
172:
157:
1517:
1379:
1262:
647:
636:
2335:
2198:
2188:
2071:
1969:
593:
495:
409:
Three models have been suggested to explain the translocation mechanism:
399:
395:
223:
176:
1764:
1499:
2276:
2089:
546:
541:
483:
301:
853:
787:
Nag N, Sasidharan S, Uversky VN, Saudagar P, Tripathi T (April 2022).
2229:
2178:
2081:
1482:
Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ (January 1988).
571:
563:
479:
367:
293:
250:
171:
The nuclear pore complex predominantly consists of proteins known as
1936:
117:
2158:
579:
517:
500:
Different export pathways through the NPC for various RNA classes.
464:
391:
387:
42:
1044:
Mohr D, Frey S, Fischer T, Güttler T, Görlich D (September 2009).
2207:
616:
219:
1581:
2259:
2249:
2239:
2234:
2146:
2129:
2064:
2054:
2049:
2044:
2039:
2034:
2029:
2024:
885:
Ando D, Colvin M, Rexach M, Gopinathan A (September 16, 2013).
789:"Phase separation of FG-nucleoporins in nuclear pore complexes"
270:
1796:
Liu HL, De Souza CP, Osmani AH, Osmani SA (January 15, 2009).
1690:
De Souza CP, Osmani AH, Hashmi SB, Osmani SA (November 2004).
1641:
Markossian S, Suresh S, Osmani AH, Osmani SA (February 2015).
1582:
Hampoelz B, Andres-Pons A, Kastritis P, Beck M (May 6, 2019).
1152:"Mitotic disassembly and reassembly of nuclear pore complexes"
2349:
2254:
2244:
2217:
2212:
2141:
2124:
2019:
2014:
2009:
2004:
1994:
1989:
1984:
1979:
1974:
1530:
793:
Biochimica et
Biophysica Acta (BBA) - Molecular Cell Research
562:
One possibility is that as a protein complex it binds to the
513:
339:
312:
258:
63:
1640:
786:
706:
2314:
2309:
2286:
2281:
2269:
2264:
2222:
1277:"Nuclear Pore Complex - an overview | ScienceDirect Topics"
997:"Nuclear pore proteins and the control of genome functions"
828:
Peyro M, Soheilypour M, Lee BL, Mofrad MR (November 2015).
509:
505:
309:
actual plug or merely cargo transiently caught in transit.
1689:
1442:
1440:
884:
203:, characterized by high flexibility and a lack of ordered
1481:
827:
501:
376:
242:
161:
1795:
1437:
1392:
362:
Scanning and illumination microscopy of nuclear pores,
1859: – Histology Learning System at Boston University
1043:
35:
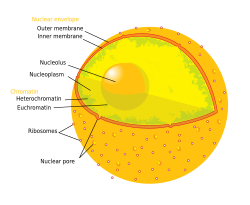
Diagram of the human cell nucleus with nuclear pores.
1584:"Structure and Assembly of the Nuclear Pore Complex"
1446:
336:
possesses a smaller mass, estimated at only 66 MDa.
238:
folds, and create the structural framework of NPCs.
1902:
NCDIR - National Center for the
Dynamic Interactome
1307:"Karyopherin - an overview | ScienceDirect Topics"
1149:
1095:"Perspective on the metazoan nuclear pore complex"
709:"The molecular architecture of the nuclear basket"
570:Possible prepore structures have been observed on
199:exhibit characteristics of "natively unfolded" or
2365:transcription factors and intracellular receptors
1150:Kutay U, Jühlen R, Antonin W (December 1, 2021).
2394:
1218:
16:Openings in nuclear envelope of eukaryotic cells
1746:
1922:
1092:
386:are karyopherin's, these are also knowing as
994:
610:
207:. These disordered proteins, referred to as
183:encompass solenoid protein domains, such as
1929:
1915:
406:include NTF2 and some NTF2-like proteins.
340:Transport through the nuclear pore complex
313:Nuclear Pore Complex: Size and Composition
41:
29:
1865:at the U.S. National Library of Medicine
1829:
1772:
1747:De Souza CP, Osmani SA (September 2007).
1715:
1666:
1507:
1420:
1410:
1361:
1252:
1175:
1126:
1069:
1020:
971:
961:
920:
910:
861:
804:
763:
753:
735:
720:
444:cellular apoptosis susceptibility protein
413:Affinity gradients along the central plug
191:folds, and occasionally both as separate
1037:
531:
357:
349:
1335:
552:
2395:
1317:from the original on February 15, 2024
1287:from the original on February 15, 2024
1219:Turton HE, Dawes IW, Grant CM (1997).
637:http://doi.org/10.1126/science.aaf1015
536:Cell nucleus containing nuclear pores.
527:
1910:
1600:10.1146/annurev-biophys-052118-115308
1200:from the original on February 8, 2024
995:Ibarra A, Hetzer MW (February 2015).
458:
423:
296:(Ras-related nuclear protein cycle).
943:
1622:from the original on April 26, 2021
736:Field MC, Rout MP (April 3, 2019).
230:. The scaffold Nups are made up of
13:
1885:Nuclear Pore Complex illustrations
1749:"Mitosis, not just open or closed"
281:to traverse the nuclear envelope.
14:
2424:
1850:
688:from the original on May 16, 2024
201:intrinsically disordered proteins
489:
304:(MDa). Each NPC comprises eight
1873:Nuclear Pore Complex animations
1789:
1740:
1683:
1634:
1575:
1524:
1475:
1449:Current Opinion in Cell Biology
1386:
1342:Current Opinion in Cell Biology
1329:
1299:
1269:
1237:10.1128/jb.179.4.1096-1101.1997
1212:
1143:
1086:
963:10.1186/gb-2001-2-9-reviews0007
253:, as well as proteins (such as
1363:11858/00-001M-0000-002D-1CC5-E
988:
937:
878:
821:
780:
755:10.12688/f1000research.16402.1
729:
700:
641:
629:
587:
164:and facilitates the selective
1:
1802:Molecular Biology of the Cell
1647:Molecular Biology of the Cell
1354:10.1016/S0955-0674(97)80015-4
622:
1412:10.1371/journal.pgen.1001366
1093:Maimon T, Medalia O (2010).
912:10.1371/journal.pone.0073831
806:10.1016/j.bbamcr.2021.119205
214:(FG-Nups), contain multiple
195:. Conversely, the remaining
132:is a channel as part of the
7:
1588:Annual Review of Biophysics
1488:The Journal of Cell Biology
430:Nuclear localization signal
404:nuclear transport receptors
384:nuclear transport receptors
288:in sequences that code for
10:
2429:
2299:Transition nuclear protein
1336:Görlich D (June 1, 1997).
946:"The nuclear pore complex"
722:10.1016/j.cell.2024.07.020
493:
427:
343:
2378:
2358:
2345:
2325:
2297:
2197:
2174:
2167:
2137:Perinucleolar compartment
2080:
1949:
1890:February 7, 2009, at the
1878:February 7, 2009, at the
1857:Histology image: 20104loa
1717:10.1016/j.cub.2004.10.050
1461:10.1016/j.ceb.2004.04.001
1168:10.1016/j.tcb.2021.06.011
611:Preservation of integrity
110:
98:
86:
74:
62:
57:
52:
40:
28:
23:
1867:Medical Subject Headings
1338:"Nuclear protein import"
416:Brownian affinity gating
334:Saccharomyces cerevisiae
249:from the nucleus to the
1814:10.1091/mbc.E08-06-0628
1659:10.1091/mbc.E14-09-1359
1549:10.1242/jcs.114.20.3607
1537:Journal of Cell Science
1225:Journal of Bacteriology
1001:Genes & Development
666:10.1126/science.abm9506
1156:Trends in Cell Biology
1111:10.4161/nucl.1.5.12332
1062:10.1038/emboj.2009.200
1013:10.1101/gad.256495.114
537:
402:-β subfamilies. Other
382:The largest family of
371:
355:
168:of various molecules.
112:Anatomical terminology
1563:on September 13, 2019
1311:www.sciencedirect.com
1281:www.sciencedirect.com
535:
361:
353:
344:Further information:
2413:Nuclear pore complex
1543:(Pt 20): 3607–3618.
615:In fungi undergoing
604:Aspergillus nidulans
553:Theories of assembly
134:nuclear pore complex
2090:Cajal (coiled) body
1951:Envelope (membrane)
1765:10.1128/EC.00178-07
1708:2004CBio...14.1973D
1500:10.1083/jcb.106.1.1
1177:20.500.11850/518955
903:2013PLoSO...873831A
846:2015NatSR...515795P
528:Assembly of the NPC
267:signaling molecules
156:(NE) surrounds the
1937:Structures of the
956:(9): REVIEWS0007.
834:Scientific Reports
660:(6598): eabm9506.
538:
459:Export of proteins
424:Import of proteins
372:
356:
286:conserved features
247:ribosomal proteins
205:tertiary structure
193:structural domains
166:membrane transport
93:H1.00.01.2.01005
2390:
2389:
2374:
2373:
1702:(22): 1973–1984.
1162:(12): 1019–1033.
1056:(17): 2541–2553.
854:10.1038/srep15795
598:scaffold proteins
346:Nuclear transport
275:passively diffuse
126:
125:
121:
2420:
2408:Membrane biology
2383:nucleus diseases
2332:(Nucleoskeleton)
2172:
2171:
1931:
1924:
1917:
1908:
1907:
1844:
1843:
1833:
1793:
1787:
1786:
1776:
1759:(9): 1521–1527.
1744:
1738:
1737:
1719:
1687:
1681:
1680:
1670:
1638:
1632:
1631:
1629:
1627:
1579:
1573:
1572:
1570:
1568:
1559:. Archived from
1528:
1522:
1521:
1511:
1479:
1473:
1472:
1444:
1435:
1434:
1424:
1414:
1390:
1384:
1383:
1365:
1333:
1327:
1326:
1324:
1322:
1303:
1297:
1296:
1294:
1292:
1273:
1267:
1266:
1256:
1231:(4): 1096–1101.
1216:
1210:
1209:
1207:
1205:
1179:
1147:
1141:
1140:
1130:
1090:
1084:
1083:
1073:
1050:The EMBO Journal
1041:
1035:
1034:
1024:
992:
986:
985:
975:
965:
944:Adam SA (2001).
941:
935:
934:
924:
914:
882:
876:
875:
865:
825:
819:
818:
808:
784:
778:
777:
767:
757:
733:
727:
726:
724:
704:
698:
697:
695:
693:
645:
639:
633:
576:nuclear envelope
547:interphase cells
306:protein subunits
228:nuclear envelope
154:nuclear envelope
150:eukaryotic cells
146:nuclear envelope
118:edit on Wikidata
115:
45:
33:
21:
20:
2428:
2427:
2423:
2422:
2421:
2419:
2418:
2417:
2393:
2392:
2391:
2386:
2370:
2354:
2341:
2321:
2293:
2193:
2163:
2076:
1954:
1945:
1943:nuclear protein
1935:
1892:Wayback Machine
1880:Wayback Machine
1853:
1848:
1847:
1794:
1790:
1753:Eukaryotic Cell
1745:
1741:
1696:Current Biology
1688:
1684:
1639:
1635:
1625:
1623:
1580:
1576:
1566:
1564:
1529:
1525:
1480:
1476:
1445:
1438:
1405:(4): e1001366.
1391:
1387:
1334:
1330:
1320:
1318:
1305:
1304:
1300:
1290:
1288:
1275:
1274:
1270:
1217:
1213:
1203:
1201:
1148:
1144:
1091:
1087:
1042:
1038:
993:
989:
942:
938:
883:
879:
826:
822:
785:
781:
734:
730:
705:
701:
691:
689:
646:
642:
634:
630:
625:
613:
590:
555:
530:
498:
492:
461:
432:
426:
419:Selective phase
348:
342:
315:
185:alpha solenoids
142:protein complex
122:
69:porus nuclearis
48:
36:
17:
12:
11:
5:
2426:
2416:
2415:
2410:
2405:
2388:
2387:
2379:
2376:
2375:
2372:
2371:
2369:
2368:
2359:
2356:
2355:
2353:
2352:
2346:
2343:
2342:
2340:
2339:
2333:
2330:Nuclear matrix
2326:
2323:
2322:
2320:
2319:
2318:
2317:
2312:
2304:
2302:
2295:
2294:
2292:
2291:
2290:
2289:
2284:
2274:
2273:
2272:
2267:
2262:
2257:
2252:
2247:
2242:
2237:
2227:
2226:
2225:
2220:
2215:
2204:
2202:
2195:
2194:
2192:
2191:
2186:
2184:Dot (PML body)
2181:
2175:
2169:
2165:
2164:
2162:
2161:
2156:
2151:
2150:
2149:
2144:
2134:
2133:
2132:
2127:
2122:
2117:
2112:
2107:
2102:
2097:
2086:
2084:
2078:
2077:
2075:
2074:
2069:
2068:
2067:
2062:
2057:
2052:
2047:
2042:
2037:
2032:
2027:
2022:
2017:
2012:
2007:
2002:
1997:
1992:
1987:
1982:
1977:
1967:
1960:
1958:
1956:nuclear lamina
1947:
1946:
1934:
1933:
1926:
1919:
1911:
1905:
1904:
1899:
1894:
1882:
1870:
1860:
1852:
1851:External links
1849:
1846:
1845:
1808:(2): 616–630.
1788:
1739:
1682:
1653:(4): 605–621.
1633:
1594:(1): 515–536.
1574:
1523:
1474:
1455:(3): 314–321.
1436:
1385:
1348:(3): 412–419.
1328:
1298:
1268:
1211:
1142:
1105:(5): 383–386.
1085:
1036:
1007:(4): 337–349.
987:
950:Genome Biology
936:
877:
820:
779:
728:
699:
640:
627:
626:
624:
621:
617:closed mitosis
612:
609:
589:
586:
585:
584:
567:
554:
551:
529:
526:
491:
488:
476:heterotrimeric
469:messenger RNAs
460:
457:
428:Main article:
425:
422:
421:
420:
417:
414:
341:
338:
326:molecular mass
321:Xenopus laevis
314:
311:
255:DNA polymerase
189:beta-propeller
124:
123:
114:
108:
107:
102:
96:
95:
90:
84:
83:
78:
72:
71:
66:
60:
59:
55:
54:
50:
49:
46:
38:
37:
34:
26:
25:
15:
9:
6:
4:
3:
2:
2425:
2414:
2411:
2409:
2406:
2404:
2401:
2400:
2398:
2385:
2384:
2377:
2367:
2366:
2361:
2360:
2357:
2351:
2348:
2347:
2344:
2337:
2334:
2331:
2328:
2327:
2324:
2316:
2313:
2311:
2308:
2307:
2306:
2305:
2303:
2300:
2296:
2288:
2285:
2283:
2280:
2279:
2278:
2275:
2271:
2268:
2266:
2263:
2261:
2258:
2256:
2253:
2251:
2248:
2246:
2243:
2241:
2238:
2236:
2233:
2232:
2231:
2228:
2224:
2221:
2219:
2216:
2214:
2211:
2210:
2209:
2206:
2205:
2203:
2200:
2196:
2190:
2187:
2185:
2182:
2180:
2177:
2176:
2173:
2170:
2166:
2160:
2157:
2155:
2152:
2148:
2145:
2143:
2140:
2139:
2138:
2135:
2131:
2128:
2126:
2123:
2121:
2118:
2116:
2113:
2111:
2108:
2106:
2103:
2101:
2098:
2096:
2093:
2092:
2091:
2088:
2087:
2085:
2083:
2079:
2073:
2070:
2066:
2063:
2061:
2058:
2056:
2053:
2051:
2048:
2046:
2043:
2041:
2038:
2036:
2033:
2031:
2028:
2026:
2023:
2021:
2018:
2016:
2013:
2011:
2008:
2006:
2003:
2001:
1998:
1996:
1993:
1991:
1988:
1986:
1983:
1981:
1978:
1976:
1973:
1972:
1971:
1968:
1965:
1962:
1961:
1959:
1957:
1952:
1948:
1944:
1940:
1932:
1927:
1925:
1920:
1918:
1913:
1912:
1909:
1903:
1900:
1898:
1895:
1893:
1889:
1886:
1883:
1881:
1877:
1874:
1871:
1868:
1864:
1861:
1858:
1855:
1854:
1841:
1837:
1832:
1827:
1823:
1819:
1815:
1811:
1807:
1803:
1799:
1792:
1784:
1780:
1775:
1770:
1766:
1762:
1758:
1754:
1750:
1743:
1735:
1731:
1727:
1723:
1718:
1713:
1709:
1705:
1701:
1697:
1693:
1686:
1678:
1674:
1669:
1664:
1660:
1656:
1652:
1648:
1644:
1637:
1621:
1617:
1613:
1609:
1605:
1601:
1597:
1593:
1589:
1585:
1578:
1562:
1558:
1554:
1550:
1546:
1542:
1538:
1534:
1527:
1519:
1515:
1510:
1505:
1501:
1497:
1493:
1489:
1485:
1478:
1470:
1466:
1462:
1458:
1454:
1450:
1443:
1441:
1432:
1428:
1423:
1418:
1413:
1408:
1404:
1400:
1399:PLOS Genetics
1396:
1389:
1381:
1377:
1373:
1369:
1364:
1359:
1355:
1351:
1347:
1343:
1339:
1332:
1316:
1312:
1308:
1302:
1286:
1282:
1278:
1272:
1264:
1260:
1255:
1250:
1246:
1242:
1238:
1234:
1230:
1226:
1222:
1215:
1199:
1195:
1191:
1187:
1183:
1178:
1173:
1169:
1165:
1161:
1157:
1153:
1146:
1138:
1134:
1129:
1124:
1120:
1116:
1112:
1108:
1104:
1100:
1096:
1089:
1081:
1077:
1072:
1067:
1063:
1059:
1055:
1051:
1047:
1040:
1032:
1028:
1023:
1018:
1014:
1010:
1006:
1002:
998:
991:
983:
979:
974:
969:
964:
959:
955:
951:
947:
940:
932:
928:
923:
918:
913:
908:
904:
900:
897:(9): e73831.
896:
892:
888:
881:
873:
869:
864:
859:
855:
851:
847:
843:
839:
835:
831:
824:
816:
812:
807:
802:
799:(4): 119205.
798:
794:
790:
783:
775:
771:
766:
761:
756:
751:
747:
743:
742:F1000Research
739:
732:
723:
718:
714:
710:
703:
687:
683:
679:
675:
671:
667:
663:
659:
655:
651:
644:
638:
632:
628:
620:
618:
608:
606:
605:
599:
595:
581:
577:
573:
568:
565:
561:
560:
559:
550:
548:
543:
534:
525:
521:
519:
515:
511:
507:
503:
497:
490:Export of RNA
487:
485:
481:
477:
472:
470:
467:subunits and
466:
456:
452:
450:
446:
445:
439:
436:
431:
418:
415:
412:
411:
410:
407:
405:
401:
397:
393:
389:
385:
380:
378:
369:
365:
360:
352:
347:
337:
335:
331:
328:of about 124
327:
323:
322:
310:
307:
303:
297:
295:
291:
287:
284:Evolutionary
282:
280:
276:
272:
268:
264:
263:carbohydrates
260:
256:
252:
248:
244:
239:
237:
233:
229:
225:
221:
217:
216:phenylalanine
213:
210:
206:
202:
198:
194:
190:
186:
182:
178:
174:
169:
167:
163:
159:
155:
151:
147:
144:found in the
143:
139:
135:
131:
119:
113:
109:
106:
103:
101:
97:
94:
91:
89:
85:
82:
79:
77:
73:
70:
67:
65:
61:
56:
51:
44:
39:
32:
27:
22:
19:
2403:Cell nucleus
2380:
2362:
1964:Pore complex
1963:
1939:cell nucleus
1863:Nuclear+pore
1805:
1801:
1791:
1756:
1752:
1742:
1699:
1695:
1685:
1650:
1646:
1636:
1626:February 14,
1624:. Retrieved
1591:
1587:
1577:
1565:. Retrieved
1561:the original
1540:
1536:
1526:
1491:
1487:
1477:
1452:
1448:
1402:
1398:
1388:
1345:
1341:
1331:
1321:February 15,
1319:. Retrieved
1310:
1301:
1291:February 15,
1289:. Retrieved
1280:
1271:
1228:
1224:
1214:
1204:February 12,
1202:. Retrieved
1159:
1155:
1145:
1102:
1098:
1088:
1053:
1049:
1039:
1004:
1000:
990:
953:
949:
939:
894:
890:
880:
837:
833:
823:
796:
792:
782:
745:
741:
731:
712:
702:
690:. Retrieved
657:
653:
643:
631:
614:
602:
594:nucleoporins
591:
556:
539:
522:
499:
473:
462:
453:
442:
440:
437:
433:
408:
381:
373:
319:
316:
298:
290:nucleoporins
283:
279:nucleoporins
240:
224:nucleoporins
212:nucleoporins
208:
197:nucleoporins
181:nucleoporins
173:nucleoporins
170:
158:cell nucleus
137:
133:
130:nuclear pore
129:
127:
68:
24:Nuclear Pore
18:
2338:(Nucleosol)
2336:Nucleoplasm
2199:SMC protein
2189:Paraspeckle
1970:Nucleoporin
1567:November 4,
1494:(1): 1–12.
588:Disassembly
496:Gene gating
400:karyopherin
398:-α and the
396:karyopherin
330:megadaltons
302:megadaltons
236:β-propeller
177:nucleoporin
160:containing
140:), a large
58:Identifiers
2397:Categories
2277:DNA repair
623:References
545:Moreover,
542:cell cycle
494:See also:
484:Leptomycin
232:α-solenoid
2381:see also
2363:see also
2230:Condensin
2179:Chromatin
2082:Nucleolus
1822:1059-1524
1608:1936-122X
1372:0955-0674
1245:0021-9193
1186:0962-8924
1119:1949-1034
840:: 15795.
674:0036-8075
580:importins
572:chromatin
564:chromatin
480:cytoplasm
392:exportins
388:importins
368:chromatin
294:RAN cycle
251:cytoplasm
1888:Archived
1876:Archived
1840:19019988
1783:17660363
1734:14782686
1726:15556859
1677:25540430
1620:Archived
1616:30943044
1557:11707513
1469:15145357
1431:21533221
1315:Archived
1285:Archived
1198:Archived
1194:34294532
1137:21326819
1080:19680228
1031:25691464
982:11574060
931:24066078
891:PLOS ONE
872:26541386
815:34995711
774:31001417
686:Archived
682:35679397
583:fashion.
518:microRNA
465:ribosome
2208:Cohesin
1831:2626566
1774:2043359
1704:Bibcode
1668:4325833
1518:3339085
1509:2114961
1422:3077370
1380:9159081
1263:9023189
1128:3037531
1099:Nucleus
1071:2728435
1022:4335290
922:3774778
899:Bibcode
863:4635341
842:Bibcode
765:6449795
748:: 369.
692:May 16,
654:Science
574:before
514:U snRNA
220:glycine
81:D022022
53:Details
2260:NCAPH2
2250:NCAPG2
2240:NCAPD3
2235:NCAPD2
2147:CUGBP1
2120:GEMIN7
2115:GEMIN6
2110:GEMIN5
2105:GEMIN4
2100:GEMIN2
2065:NUP214
2060:NUP210
2055:NUP205
2050:NUP188
2045:NUP160
2040:NUP155
2035:NUP153
2030:NUP133
2025:NUP107
1869:(MeSH)
1838:
1828:
1820:
1781:
1771:
1732:
1724:
1675:
1665:
1614:
1606:
1555:
1516:
1506:
1467:
1429:
1419:
1378:
1370:
1261:
1254:178803
1251:
1243:
1192:
1184:
1135:
1125:
1117:
1078:
1068:
1029:
1019:
980:
973:138961
970:
929:
919:
870:
860:
813:
772:
762:
680:
672:
366:, and
364:lamina
271:lipids
269:, and
259:lamins
152:. The
2350:LITAF
2255:NCAPH
2245:NCAPG
2218:SMC1B
2213:SMC1A
2168:Other
2159:ATXN7
2142:PTBP1
2125:DDX20
2020:NUP98
2015:NUP93
2010:NUP88
2005:NUP85
2000:NUP62
1995:NUP54
1990:NUP50
1985:NUP43
1980:NUP37
1975:NUP35
1730:S2CID
116:[
105:63148
64:Latin
2315:TNP2
2310:TNP1
2287:SMC6
2282:SMC5
2270:SMC4
2265:SMC2
2223:SMC3
2154:TCOF
2130:COIL
2072:AAAS
1836:PMID
1818:ISSN
1779:PMID
1722:PMID
1673:PMID
1628:2024
1612:PMID
1604:ISSN
1569:2008
1553:PMID
1514:PMID
1465:PMID
1427:PMID
1376:PMID
1368:ISSN
1323:2024
1293:2024
1259:PMID
1241:ISSN
1206:2024
1190:PMID
1182:ISSN
1133:PMID
1115:ISSN
1076:PMID
1027:PMID
978:PMID
927:PMID
868:PMID
811:PMID
797:1869
770:PMID
713:Cell
694:2024
678:PMID
670:ISSN
510:rRNA
506:tRNA
449:GTPs
257:and
245:and
234:and
76:MeSH
2095:SMN
1826:PMC
1810:doi
1769:PMC
1761:doi
1712:doi
1663:PMC
1655:doi
1596:doi
1545:doi
1541:114
1504:PMC
1496:doi
1492:106
1457:doi
1417:PMC
1407:doi
1358:hdl
1350:doi
1249:PMC
1233:doi
1229:179
1172:hdl
1164:doi
1123:PMC
1107:doi
1066:PMC
1058:doi
1017:PMC
1009:doi
968:PMC
958:doi
917:PMC
907:doi
858:PMC
850:doi
801:doi
760:PMC
750:doi
717:doi
662:doi
658:376
502:RNA
390:or
377:KDa
261:),
243:RNA
187:or
162:DNA
148:of
138:NPC
100:FMA
2399::
1941:/
1834:.
1824:.
1816:.
1806:20
1804:.
1800:.
1777:.
1767:.
1755:.
1751:.
1728:.
1720:.
1710:.
1700:14
1698:.
1694:.
1671:.
1661:.
1651:26
1649:.
1645:.
1618:.
1610:.
1602:.
1592:48
1590:.
1586:.
1551:.
1539:.
1535:.
1512:.
1502:.
1490:.
1486:.
1463:.
1453:16
1451:.
1439:^
1425:.
1415:.
1401:.
1397:.
1374:.
1366:.
1356:.
1344:.
1340:.
1313:.
1309:.
1283:.
1279:.
1257:.
1247:.
1239:.
1227:.
1223:.
1196:.
1188:.
1180:.
1170:.
1160:31
1158:.
1154:.
1131:.
1121:.
1113:.
1101:.
1097:.
1074:.
1064:.
1054:28
1052:.
1048:.
1025:.
1015:.
1005:29
1003:.
999:.
976:.
966:.
952:.
948:.
925:.
915:.
905:.
893:.
889:.
866:.
856:.
848:.
836:.
832:.
809:.
795:.
791:.
768:.
758:.
744:.
740:.
715:.
711:.
684:.
676:.
668:.
656:.
652:.
516:,
512:,
508:,
486:B
265:,
209:FG
128:A
88:TH
2301::
2201::
1966::
1953:/
1930:e
1923:t
1916:v
1842:.
1812::
1785:.
1763::
1757:6
1736:.
1714::
1706::
1679:.
1657::
1630:.
1598::
1571:.
1547::
1520:.
1498::
1471:.
1459::
1433:.
1409::
1403:7
1382:.
1360::
1352::
1346:9
1325:.
1295:.
1265:.
1235::
1208:.
1174::
1166::
1139:.
1109::
1103:1
1082:.
1060::
1033:.
1011::
984:.
960::
954:2
933:.
909::
901::
895:8
874:.
852::
844::
838:5
817:.
803::
776:.
752::
746:8
725:.
719::
696:.
664::
370:.
218:–
136:(
120:]
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.