667:
890:
612:
640:
920:
133:
934:
237:
382:, DNA methylation is associated with alternative splicing and gene regulation based on functional genomic research published in 2013. In addition, DNA methylation is associated with expression changes in immune genes when honey bees were under lethal viral infection. Several review papers have been published on the topics of DNA methylation in social insects.
375:. One to two percent of the human genome are CpG clusters, and there is an inverse relationship between CpG methylation and transcriptional activity. Methylation contributing to epigenetic inheritance can occur through either DNA methylation or protein methylation. Improper methylations of human genes can lead to disease development, including cancer.
497:
Methyl metabolism is very ancient and can be found in all organisms on earth, from bacteria to humans, indicating the importance of methyl metabolism for physiology. Indeed, pharmacological inhibition of global methylation in species ranging from human, mouse, fish, fly, roundworm, plant, algae, and
427:
is the most common and abundant methylation modification in RNA molecules (mRNA) present in eukaryotes. 5-methylcytosine (5-mC) also commonly occurs in various RNA molecules. Recent data strongly suggest that m6A and 5-mC RNA methylation affects the regulation of various biological processes such as
2111:
Fustin, Jean-Michel; Ye, Shiqi; Rakers, Christin; Kaneko, Kensuke; Fukumoto, Kazuki; Yamano, Mayu; Versteven, Marijke; Grünewald, Ellen; Cargill, Samantha J.; Tamai, T. Katherine; Xu, Yao; Jabbur, Maria Luísa; Kojima, Rika; Lamberti, Melisa L.; Yoshioka-Kobayashi, Kumiko; Whitmore, David; Tammam,
213:, methylation is a major biochemical process for modifying protein function. The most prevalent protein methylations affect arginine and lysine residue of specific histones. Otherwise histidine, glutamate, asparagine, cysteine are susceptible to methylation. Some of these products include
1508:
Rotondo JC, Borghi A, Selvatici R, Magri E, Bianchini E, Montinari E, Corazza M, Virgili A, Tognon M, Martini F (2016). "Hypermethylation-Induced
Inactivation of the IRF6 Gene as a Possible Early Event in Progression of Vulvar Squamous Cell Carcinoma Associated With Lichen Sclerosus".
1222:
Styblo, M.; Del Razo, L. M.; Vega, L.; Germolec, D. R.; LeCluyse, E. L.; Hamilton, G. A.; Reed, W.; Wang, C.; Cullen, W. R.; Thomas, D. J. (2000). "Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells".
2279:
Tundo, Pietro; Selva, Maurizio; Bomben, Andrea (1999). "Mono-C-methylathion of arylacetonitriles and methyl arylacetates by dimethyl carbonate: a general method for the synthesis of pure 2-arylpropionic acids. 2-Phenylpropionic acid".
269:
In methylcobalamin-dependent forms of the enzyme, the reaction proceeds by two steps in a ping-pong reaction. The enzyme is initially primed into a reactive state by the transfer of a methyl group from N-MeTHF to Co(I) in enzyme-bound
421:, and viral RNA. Different catalytic strategies are employed for RNA methylation by a variety of RNA-methyltransferases. RNA methylation is thought to have existed before DNA methylation in the early forms of life evolving on earth.
1194:
Matthews, R. G.; Smith, A. E.; Zhou, Z. S.; Taurog, R. E.; Bandarian, V.; Evans, J. C.; Ludwig, M. (2003). "Cobalamin-Dependent and
Cobalamin-Independent Methionine Synthases: Are There Two Solutions to the Same Chemical Problem?".
1593:
Li-Byarlay, Hongmei; Li, Yang; Stroud, Hume; Feng, Suhua; Newman, Thomas C.; Kaneda, Megan; Hou, Kirk K.; Worley, Kim C.; Elsik, Christine G.; Wickline, Samuel A.; Jacobsen, Steven E.; Ma, Jian; Robinson, Gene E. (30 July 2013).
825:
358:
in the DNA sequence). In mammals, DNA methylation is common in body cells, and methylation of CpG sites seems to be the default. Human DNA has about 80–90% of CpG sites methylated, but there are certain areas, known as
1821:
Choi, Junhong; Ieong, Ka-Weng; Demirci, Hasan; Chen, Jin; Petrov, Alexey; Prabhakar, Arjun; O'Leary, Seán E; Dominissini, Dan; Rechavi, Gideon; Soltis, S Michael; Ehrenberg, Måns; Puglisi, Joseph D (February 2016).
1309:
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (November 2009).
266:(THF) while transferring a methyl group to Hcy to form Met. Methionine Syntheses can be cobalamin-dependent and cobalamin-independent: Plants have both, animals depend on the methylcobalamin-dependent form.
2350:
Poon, Kevin W. C.; Albiniak, Philip A.; Dudley, Gregory B. (2007). "Protection of alcohols using 2-benzyloxy-1-methylpyridinium trifluoromethanesulfanonate: Methyl (R)-(-)-3-benzyloxy-2-methyl propanoate".
109:
In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals and can regulate gene expression, RNA processing, and protein function. It is a key process underlying
2017:
Nakayama, Jun-ichi; Rice, Judd C.; Strahl, Brian D.; Allis, C. David; Grewal, Shiv I. S. (6 April 2001). "Role of
Histone H3 Lysine 9 Methylation in Epigenetic Control of Heterochromatin Assembly".
278:
to form a reactive thiolate reacts with the Me-Cob. The activated methyl group is transferred from Me-Cob to the Hcy thiolate, which regenerates Co(I) in Cob, and Met is released from the enzyme.
2112:
Stephanie; Howell, P. Lynne; Kageyama, Ryoichiro; Matsuo, Takuya; Stanewsky, Ralf; Golombek, Diego A.; Johnson, Carl
Hirschie; Kakeya, Hideaki; van Ooijen, Gerben; Okamura, Hitoshi (6 May 2020).
458:
274:((Cob), also known as vitamine B12)) , , forming methyl-cobalamin(Me-Cob) that now contains Me-Co(III) and activating the enzyme. Then, a Hcy that has coordinated to an enzyme-bound
462:
3302:
1653:
Li-Byarlay, Hongmei; Boncristiani, Humberto; Howell, Gary; Herman, Jake; Clark, Lindsay; Strand, Micheline K.; Tarpy, David; Rueppell, Olav (24 September 2020).
1410:"Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males"
453:
residues in the protein sequence. Arginine can be methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (
1367:
Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van
Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schübeler D (December 2011).
2708:
2307:
Nenad, Maraš; Polanc, Slovenko; Kočevar, Marijan (2008). "Microwave-assisted methylation of phenols with tetramethylammonium chloride in the presence of K
3364:
2661:
1459:"Methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples of infertile couples correlates with recurrent spontaneous abortion"
431:
In social insects such as honey bees, RNA methylation is studied as a possible epigenetic mechanism underlying aggression via reciprocal crosses.
2780:
696:
666:
611:
498:
cyanobacteria causes the same effects on their biological rhythms, demonstrating conserved physiological roles of methylation during evolution.
3129:
3119:
1872:
1900:"Examining parent-of-origin effects on transcription and RNA methylation in mediating aggressive behavior in honey bees (Apis mellifera)"
129:, involves a series of methylation reactions. These reactions are caused by a set of enzymes harbored by a family of anaerobic microbes.
1898:
Bresnahan, Sean T.; Lee, Ellen; Clark, Lindsay; Ma, Rong; Rangel, Juliana; Grozinger, Christina M.; Li-Byarlay, Hongmei (12 June 2023).
1546:"Association of Retinoic Acid Receptor β Gene With Onset and Progression of Lichen Sclerosus-Associated Vulvar Squamous Cell Carcinoma"
2554:
Negishi, Ei-ichi; Matsushita, Hajime (1984). "Palladium-Catalyzed
Synthesis of 1,4-Dienes by Allylation of Alkenyalane: α-Farnesene".
3502:
1100:
Timmers, Peer H. A.; Welte, Cornelia U.; Koehorst, Jasper J.; Plugge, Caroline M.; Jetten, Mike S. M.; Stams, Alfons J. M. (2017).
1090:
Thauer, R. K., "Biochemistry of
Methanogenesis: a Tribute to Marjory Stephenson", Microbiology, 1998, volume 144, pages 2377-2406.
2654:
1457:
Rotondo JC, Bosi S, Bazzan E, Di
Domenico M, De Mattei M, Selvatici R, Patella A, Marci R, Tognon M, Martini F (December 2012).
286:
Biomethylation is the pathway for converting some heavy elements into more mobile or more lethal derivatives that can enter the
1544:
Rotondo JC, Borghi A, Selvatici R, Mazzoni E, Bononi I, Corazza M, Kussini J, Montinari E, Gafà R, Tognon M, Martini F (2018).
1266:
Tost J (2010). "DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker".
2484:
2180:
1962:
1754:
1057:
2500:
Lipsky, Sharon D.; Hall, Stan S. (1976). "Aromatic
Hydrocarbons from aromatic ketones and aldehydes: 1,1-Diphenylethane".
2633:
2647:
1000:
889:
163:
486:
428:
RNA stability and mRNA translation, and that abnormal RNA methylation contributes to etiology of human diseases.
2639:
2674:
671:
652:
2252:
Icke, Roland N.; Redemann, Ernst; Wisegarver, Burnett B.; Alles, Gordon A. (1949). "m-Methoxybenzaldehyde".
3324:
1596:"RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee"
548:
306:
is the methyl donor. The methanearsonates are the precursors to dimethylarsonates, again by the cycle of
454:
3497:
1040:
Ragsdale, Stephen W. (2008). "Catalysis of Methyl Group
Transfers Involving Tetrahydrofolate and B12".
687:
2775:
2670:
964:
955:– the biochemical method used to determine the presence or absence of methyl groups on a DNA sequence
568:
478:
830:
The method offers the advantage that the side products are easily removed from the product mixture.
311:
259:
3418:
861:
2436:
Icke, Roland N.; Wisegarver, Burnett B.; Alles, Gordon A. (1945). "β-Phenylethyldimethylamine".
1408:
Rotondo JC, Selvatici R, Di Domenico M, Marci R, Vesce F, Tognon M, Martini F (September 2013).
639:
1876:
1950:
3217:
474:
343:
310:(to methylarsonous acid) followed by a second methylation. Related pathways are found in the
171:
31:
1655:"Transcriptomic and Epigenomic Dynamics of Honey Bees in Response to Lethal Viral Infection"
1458:
1145:"The ribosome: A hot spot for the identification of new types of protein methyltransferases"
94:
3492:
3487:
2693:
2026:
1607:
1323:
952:
303:
8:
439:
368:
364:
247:
241:
194:
190:
186:
149:
2030:
1926:
1899:
1824:"N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics"
1611:
1327:
34:, or the substitution of an atom (or group) by a methyl group. Methylation is a form of
3023:
2873:
2607:
2582:
2140:
2113:
2088:
2061:
1848:
1823:
1798:
1771:
1681:
1654:
1630:
1595:
1570:
1545:
1434:
1409:
1344:
1311:
1291:
1248:
1171:
1144:
1068:
1020:
905:
619:
544:
424:
302:. Thus, trivalent inorganic arsenic compounds are methylated to give methanearsonate.
62:
2583:"Thermophoretic melting curves quantify the conformation and stability of RNA and DNA"
1049:
2977:
2863:
2822:
2612:
2480:
2198:
2176:
2145:
2093:
2042:
1999:
1958:
1931:
1853:
1803:
1750:
1686:
1635:
1575:
1526:
1490:
1439:
1390:
1349:
1283:
1240:
1176:
1073:
1053:
507:
372:
181:
Plants produce flavonoids and isoflavones with methylations on hydroxyl groups, i.e.
1295:
3280:
3258:
3157:
3076:
2967:
2944:
2868:
2840:
2602:
2594:
2563:
2536:
2509:
2472:
2445:
2418:
2387:
2360:
2332:
2289:
2261:
2234:
2207:
2168:
2135:
2125:
2083:
2073:
2034:
1991:
1921:
1911:
1843:
1835:
1793:
1783:
1742:
1717:
1676:
1666:
1625:
1615:
1565:
1561:
1557:
1522:
1518:
1480:
1470:
1429:
1421:
1380:
1339:
1331:
1275:
1252:
1232:
1204:
1166:
1156:
1123:
1113:
1063:
1045:
978:
925:
857:
540:
335:
263:
214:
16:
Chemical process in which a methyl (CH3) group is covalently attached to a molecule
2476:
1772:"Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases"
551:. Less common but more powerful (and more dangerous) methylating reagents include
114:. Sources of methyl groups include S-methylmethionine, methyl folate, methyl B12.
3391:
3091:
2982:
2858:
2817:
2669:
1015:
1005:
552:
331:
299:
210:
78:
1746:
1312:"Human DNA methylomes at base resolution show widespread epigenomic differences"
1221:
3253:
3147:
3124:
3066:
2916:
2898:
2785:
2751:
2172:
1916:
1010:
939:
897:
660:
363:, that are CG-rich (high cytosine and guanine content, made up of about 65% CG
360:
291:
122:
86:
2336:
2130:
1995:
1671:
1279:
185:. This 5-O-methylation affects the flavonoid's water solubility. Examples are
3481:
3272:
3199:
3175:
3114:
3109:
3061:
3056:
3005:
2962:
2883:
2827:
2731:
2567:
2540:
2513:
2449:
2391:
2364:
2293:
2265:
2238:
2211:
1788:
1161:
970:
845:
315:
182:
98:
39:
2527:
Grummitt, Oliver; Becker, Ernest I. (1950). "trans-1-Phenyl-1,3-butadiene".
2114:"Methylation deficiency disrupts biological rhythms from bacteria to humans"
2038:
1722:
1705:
1620:
1475:
1369:"DNA-binding factors shape the mouse methylome at distal regulatory regions"
820:{\displaystyle {\ce {RCO2H + tmsCHN2 + CH3OH -> RCO2CH3 + CH3Otms + N2}}}
3071:
2888:
2703:
2688:
2616:
2149:
2097:
2078:
2046:
2003:
1935:
1857:
1807:
1690:
1639:
1579:
1530:
1494:
1443:
1394:
1353:
1287:
1244:
1208:
1180:
1118:
1101:
1077:
901:
683:
631:
623:
560:
556:
530:
485:
to repress or activate gene expression. Protein methylation is one type of
255:
82:
74:
51:
47:
27:
1236:
3193:
3081:
3046:
3000:
2939:
2921:
2893:
2736:
2726:
2698:
2598:
2422:
1485:
839:
627:
572:
536:
482:
111:
1955:
Posttranslational Modification of Proteins: Expanding Nature's Inventory
1385:
1368:
1335:
3433:
3339:
3316:
3312:
3268:
3232:
3209:
2992:
2796:
2762:
2718:
2196:
Vyas, G. N.; Shah, N. M. (1951). "Quninacetophenone monomethyl ether".
990:
974:
511:
450:
287:
251:
167:
132:
35:
1839:
1425:
1128:
3462:
3375:
3185:
3104:
3051:
3015:
2954:
2931:
2878:
2809:
2741:
2463:
Shioiri T, Aoyama T, Snowden T (2001). "Trimethylsilyldiazomethane".
1980:"Regulation of heterochromatin by histone methylation and small RNAs"
379:
339:
271:
206:
66:
43:
23:
2406:
2378:
Neeman, M.; Johnson, William S. (1961). "Cholestanyl methyl ether".
1979:
3450:
3446:
3442:
3379:
3352:
3294:
3245:
3241:
3139:
3086:
2908:
2850:
2832:
933:
881:
844:
methyl reagents. Strongly nucleophilic methylating agents include
580:
443:
351:
347:
1652:
1102:"Reverse Methanogenesis and Respiration in Methanotrophic Archaea"
967:– a biophysical method to determine the methylisation state of DNA
3410:
3402:
3383:
3356:
3167:
1739:
Physiological and Molecular Mechanisms of Nutrition in Honey Bees
995:
599:
466:
461:(PRMTs). Lysine can be methylated once, twice, or three times by
355:
295:
159:
155:
55:
3441:
1407:
3454:
3406:
3348:
3290:
3033:
2805:
1741:. Advances in Insect Physiology. Vol. 49. pp. 25–58.
958:
663:, which occurs when amines are methylated with methyl halides.
603:
470:
447:
414:
236:
175:
70:
678:
3099:
2770:
1543:
656:
588:
576:
418:
410:
406:
367:), wherein none is methylated. These are associated with the
307:
140:
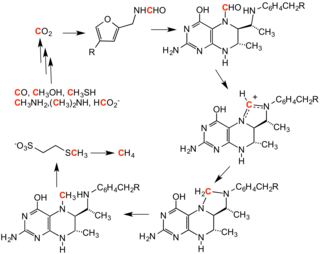
In reverse methanogenesis, methane is the methylating agent.
1456:
457:) or one on both nitrogens (symmetric dimethylarginine), by
2251:
1507:
1308:
1044:. Vitamins & Hormones. Vol. 79. pp. 293–324.
481:. Histones that are methylated on certain residues can act
402:
398:
394:
275:
93:
methylation of tissue samples is also a way to reduce some
2163:"Aromatic Substitution, Nucleophilic and Organometallic".
2062:"Natural history of S-adenosylmethionine-binding proteins"
1099:
281:
1706:"The Function of DNA Methylation Marks in Social Insects"
813:
794:
778:
765:
746:
730:
711:
350:(cytosine-phosphate-guanine sites—that is, sites where a
162:
derivatives. This process, catalyzed by such enzymes as
3392:
4-(p-hydroxybenzylidene)-5-imidazolinone (HBI) formation
690:
methylate carboxylic acids, phenols, and even alcohols:
2060:
Kozbial, Piotr Z; Mushegian, Arcady R (December 2005).
2016:
1366:
1193:
2580:
1592:
346:. In vertebrates, DNA methylation typically occurs at
73:; such methylation can be involved in modification of
2469:
e-EROS Encyclopedia of Reagents for Organic Synthesis
1873:"Methylation (MTHFR) Testing & Folate Deficiency"
699:
2462:
2110:
1957:. Roberts and Company Publishers. pp. 121–149.
1820:
915:
2435:
1897:
465:. Protein methylation has been most studied in the
3365:p-Hydroxybenzylidene-imidazolinone (HBI) formation
2349:
2306:
819:
2636:Detection of Methylations after Mass Spectrometry
2553:
3479:
2278:
2059:
646:
1600:Proceedings of the National Academy of Sciences
622:is a specific for the methylation at oxygen of
136:Cycle for methanogenesis, showing intermediates
2581:Wienken CJ, Baaske P, Duhr S, Braun D (2011).
2526:
2465:Encyclopedia of Reagents for Organic Synthesis
1736:
2655:
2411:Journal of the Chemical Society, Transactions
2377:
602:may be methylated on carbon to produce a new
575:may be methylated on oxygen to give a methyl
477:to histones is catalyzed by enzymes known as
3325:Tryptophan tryptophylquinone (TTQ) formation
2404:
2010:
1537:
1501:
1450:
1401:
973:, the reversible removal of methyl group in
524:
125:, the process that generates methane from CO
1978:Grewal, Shiv IS; Rice, Judd C (June 2004).
1770:Rana, Ajay K.; Ankri, Serge (6 June 2016).
1259:
984:
833:
679:Diazomethane and trimethylsilyldiazomethane
514:process used to describe the delivery of a
166:, is a key reaction in the biosynthesis of
2662:
2648:
2499:
2398:
1977:
1971:
1703:
529:Methylations are commonly performed using
178:, a major structural component of plants.
2606:
2139:
2129:
2087:
2077:
1925:
1915:
1847:
1828:Nature Structural & Molecular Biology
1797:
1787:
1769:
1721:
1680:
1670:
1629:
1619:
1569:
1484:
1474:
1433:
1384:
1343:
1302:
1170:
1160:
1127:
1117:
1067:
371:of 56% of mammalian genes, including all
321:
2574:
2195:
1737:Wang, Ying; Li-Byarlay, Hongmei (2015).
1039:
235:
131:
42:atom. These terms are commonly used in
3419:Methylidene-imidazolone (MIO) formation
2053:
1870:
298:compounds starts with the formation of
282:Heavy metals: arsenic, mercury, cadmium
258:(Hcy). The overall reaction transforms
143:
3480:
2104:
1265:
1142:
587:may be likewise methylated to give an
434:
240:The methylation reaction catalyzed by
231:
3303:Lysine tyrosylquinone (LTQ) formation
2643:
2224:
1948:
1871:Stewart, Kendal (15 September 2017).
1215:
838:Methylation sometimes involve use of
393:occurs in different RNA species viz.
334:is the conversion of the cytosine to
224:-methylhistidine, and two isomers of
2776:Glycosyl phosphatidylinositol (GPI)
1704:Li-Byarlay, Hongmei (19 May 2016).
1187:
459:protein arginine methyltransferases
13:
2405:Purdie, T.; Irvine, J. C. (1903).
2165:March's Advanced Organic Chemistry
1710:Frontiers in Ecology and Evolution
896:Milder methylating agents include
665:
385:
326:
38:, with a methyl group replacing a
14:
3514:
3087:Oxidative deamination to aldehyde
2627:
946:
659:. This method avoids the risk of
563:). These reagents all react via S
117:
2225:Hiers, G. S. (1929). "Anisole".
932:
918:
888:
884:(C=O) of ketones and aldehyde.:
638:
610:
164:caffeoyl-CoA O-methyltransferase
97:. The reverse of methylation is
3503:Post-translational modification
2675:posttranslational modifications
2547:
2520:
2493:
2456:
2429:
2371:
2343:
2300:
2272:
2245:
2218:
2189:
2156:
1984:Current Opinion in Cell Biology
1942:
1891:
1864:
1814:
1763:
1730:
1697:
1646:
1586:
1149:Journal of Biological Chemistry
655:is a method for methylation of
501:
487:post-translational modification
95:histological staining artifacts
1562:10.1001/jamadermatol.2018.1373
1523:10.1001/jamadermatol.2016.1336
1360:
1136:
1093:
1084:
1033:
880:will add methyl groups to the
752:
559:, and methyl fluorosulfonate (
158:undergo O-methylation to give
1:
2477:10.1002/047084289X.rt298.pub2
2407:"C.?The alkylation of sugars"
1050:10.1016/S0083-6729(08)00410-X
1026:
647:Eschweiler–Clarke methylation
338:. The formation of Me-CpG is
104:
674:is used to methylate amines.
549:tetramethylammonium chloride
492:
373:ubiquitously expressed genes
7:
2894:Topaquinone (TPQ) formation
1949:Walsh, Christopher (2006).
1747:10.1016/bs.aiip.2015.06.002
1001:Titanium–zinc methylenation
911:
455:asymmetric dimethylarginine
200:
10:
3519:
2173:10.1002/9780470084960.ch13
2167:. 2006. pp. 853–933.
1917:10.1186/s12864-023-09411-4
1143:Clarke, Steven G. (2018).
688:trimethylsilyldiazomethane
672:Eschweiler–Clarke reaction
653:Eschweiler–Clarke reaction
569:nucleophilic substitutions
479:histone methyltransferases
354:is directly followed by a
147:
3431:
3401:
3374:
3347:
3338:Crosslinks between three
3337:
3311:
3289:
3267:
3240:
3230:
3208:
3184:
3166:
3138:
3032:
3014:
2991:
2953:
2930:
2907:
2849:
2804:
2794:
2761:
2717:
2681:
2671:Protein primary structure
2337:10.1016/j.tet.2008.10.024
2131:10.1038/s42003-020-0942-0
1996:10.1016/j.ceb.2004.04.002
1672:10.3389/fgene.2020.566320
1280:10.1007/s12033-009-9216-2
965:Microscale thermophoresis
525:Electrophilic methylation
463:lysine methyltransferases
442:typically takes place on
3432:Crosslinks between four
2568:10.15227/orgsyn.062.0031
2541:10.15227/orgsyn.030.0075
2514:10.15227/orgsyn.055.0007
2450:10.15227/orgsyn.025.0089
2392:10.15227/orgsyn.041.0009
2365:10.15227/orgsyn.084.0295
2294:10.15227/orgsyn.076.0169
2266:10.15227/orgsyn.029.0063
2239:10.15227/orgsyn.009.0012
2212:10.15227/orgsyn.031.0090
1789:10.3389/fgene.2016.00099
1162:10.1074/jbc.AW118.003235
985:Organic chemistry topics
961:DNA Methylation Database
834:Nucleophilic methylation
506:The term methylation in
425:N6-methyladenosine (m6A)
260:5-methyltetrahydrofolate
3231:Crosslinks between two
2039:10.1126/science.1060118
1723:10.3389/fevo.2016.00057
1621:10.1073/pnas.1310735110
862:methylmagnesium bromide
686:and the safer analogue
535:methyl sources such as
26:, is the addition of a
2879:Porphyrin ring linkage
2587:Nucleic Acids Research
2118:Communications Biology
2079:10.1186/1472-6807-5-19
2066:BMC Structural Biology
1225:Archives of Toxicology
1209:10.1002/hlca.200390329
1197:Helvetica Chimica Acta
1042:Folic Acid and Folates
821:
675:
322:Epigenetic methylation
244:
137:
2940:Succinimide formation
1951:"Protein Methylation"
1776:Frontiers in Genetics
1659:Frontiers in Genetics
1476:10.1093/humrep/des319
1237:10.1007/s002040000134
822:
669:
475:S-adenosyl methionine
344:DNA methyltransferase
312:microbial methylation
239:
135:
2694:Protein biosynthesis
2423:10.1039/CT9038301021
1119:10.1155/2017/1654237
953:Bisulfite sequencing
697:
304:S-adenosylmethionine
144:O-methyltransferases
2331:(51): 11618–11624.
2031:2001Sci...292..110N
1612:2013PNAS..11012750L
1606:(31): 12750–12755.
1386:10.1038/nature11086
1336:10.1038/nature08514
1328:2009Natur.462..315L
1155:(27): 10438–10446.
815:
796:
780:
767:
748:
732:
713:
440:Protein methylation
435:Protein methylation
248:Methionine synthase
242:methionine synthase
232:Methionine synthase
195:5-O-methylquercetin
191:5-O-methylmyricetin
187:5-O-methylgenistein
150:O-methyltransferase
3024:Transglutamination
2599:10.1093/nar/gkr035
1879:on 12 October 2017
1463:Human Reproduction
906:trimethylaluminium
817:
803:
784:
768:
755:
736:
720:
701:
676:
620:Purdie methylation
545:dimethyl carbonate
469:. The transfer of
245:
154:A wide variety of
138:
63:biological systems
3498:Organic reactions
3475:
3474:
3471:
3470:
3427:
3426:
3333:
3332:
3226:
3225:
2978:Polyglutamylation
2864:Dephosphorylation
2823:Dephosphorylation
2556:Organic Syntheses
2529:Organic Syntheses
2502:Organic Syntheses
2486:978-0-471-93623-7
2438:Organic Syntheses
2380:Organic Syntheses
2353:Organic Syntheses
2282:Organic Syntheses
2254:Organic Syntheses
2227:Organic Syntheses
2199:Organic Syntheses
2182:978-0-471-72091-1
2025:(5514): 110–113.
1964:978-0-9747077-3-0
1840:10.1038/nsmb.3148
1756:978-0-12-802586-4
1426:10.4161/epi.25798
1203:(12): 3939–3954.
1059:978-0-12-374232-2
858:Grignard reagents
806:
799:
787:
771:
758:
751:
739:
723:
716:
704:
571:. For example, a
508:organic chemistry
228:-methylarginine.
220:, two isomers of
65:, methylation is
24:chemical sciences
3510:
3439:
3438:
3345:
3344:
3281:Sulfilimine bond
3259:ADP-ribosylation
3238:
3237:
3158:ADP-ribosylation
3077:ADP-ribosylation
2968:ADP-ribosylation
2945:ADP-ribosylation
2869:ADP-ribosylation
2841:ADP-ribosylation
2802:
2801:
2795:Single specific
2664:
2657:
2650:
2641:
2640:
2621:
2620:
2610:
2578:
2572:
2571:
2551:
2545:
2544:
2524:
2518:
2517:
2497:
2491:
2490:
2460:
2454:
2453:
2433:
2427:
2426:
2402:
2396:
2395:
2375:
2369:
2368:
2347:
2341:
2340:
2304:
2298:
2297:
2276:
2270:
2269:
2249:
2243:
2242:
2222:
2216:
2215:
2193:
2187:
2186:
2160:
2154:
2153:
2143:
2133:
2108:
2102:
2101:
2091:
2081:
2057:
2051:
2050:
2014:
2008:
2007:
1975:
1969:
1968:
1946:
1940:
1939:
1929:
1919:
1895:
1889:
1888:
1886:
1884:
1875:. Archived from
1868:
1862:
1861:
1851:
1818:
1812:
1811:
1801:
1791:
1767:
1761:
1760:
1734:
1728:
1727:
1725:
1701:
1695:
1694:
1684:
1674:
1650:
1644:
1643:
1633:
1623:
1590:
1584:
1583:
1573:
1550:JAMA Dermatology
1541:
1535:
1534:
1511:JAMA Dermatology
1505:
1499:
1498:
1488:
1478:
1454:
1448:
1447:
1437:
1405:
1399:
1398:
1388:
1364:
1358:
1357:
1347:
1322:(7271): 315–22.
1306:
1300:
1299:
1263:
1257:
1256:
1219:
1213:
1212:
1191:
1185:
1184:
1174:
1164:
1140:
1134:
1133:
1131:
1121:
1097:
1091:
1088:
1082:
1081:
1071:
1037:
979:5-methylcytosine
942:
937:
936:
928:
926:Chemistry portal
923:
922:
921:
892:
879:
872:). For example,
871:
855:
826:
824:
823:
818:
816:
814:
811:
804:
797:
795:
792:
785:
779:
776:
769:
766:
763:
756:
749:
747:
744:
737:
731:
728:
721:
714:
712:
709:
702:
642:
614:
597:
586:
541:dimethyl sulfate
520:
336:5-methylcytosine
300:methanearsonates
264:tetrahydrofolate
83:protein function
81:, regulation of
77:, regulation of
3518:
3517:
3513:
3512:
3511:
3509:
3508:
3507:
3478:
3477:
3476:
3467:
3423:
3397:
3370:
3329:
3307:
3285:
3263:
3222:
3218:C-mannosylation
3204:
3180:
3162:
3134:
3100:Imine formation
3028:
3010:
2987:
2983:Polyglycylation
2949:
2926:
2903:
2859:Phosphorylation
2845:
2818:Phosphorylation
2790:
2757:
2713:
2677:
2668:
2630:
2625:
2624:
2579:
2575:
2552:
2548:
2525:
2521:
2498:
2494:
2487:
2461:
2457:
2434:
2430:
2403:
2399:
2376:
2372:
2348:
2344:
2322:
2318:
2314:
2310:
2305:
2301:
2277:
2273:
2250:
2246:
2223:
2219:
2194:
2190:
2183:
2162:
2161:
2157:
2109:
2105:
2058:
2054:
2015:
2011:
1976:
1972:
1965:
1947:
1943:
1896:
1892:
1882:
1880:
1869:
1865:
1819:
1815:
1768:
1764:
1757:
1735:
1731:
1702:
1698:
1651:
1647:
1591:
1587:
1542:
1538:
1506:
1502:
1455:
1451:
1406:
1402:
1379:(7378): 490–5.
1365:
1361:
1307:
1303:
1264:
1260:
1220:
1216:
1192:
1188:
1141:
1137:
1098:
1094:
1089:
1085:
1060:
1038:
1034:
1029:
1021:Tebbe's reagent
1016:Wittig reaction
1006:Petasis reagent
987:
949:
938:
931:
924:
919:
917:
914:
877:
873:
869:
865:
853:
849:
836:
812:
807:
793:
788:
777:
772:
764:
759:
745:
740:
729:
724:
710:
705:
700:
698:
695:
694:
681:
649:
596:
592:
584:
566:
553:methyl triflate
527:
519:
515:
504:
495:
437:
391:RNA methylation
388:
386:RNA methylation
332:DNA methylation
329:
327:DNA methylation
324:
284:
262:(N-MeTHF) into
234:
218:-methylcysteine
211:phosphorylation
203:
152:
146:
128:
120:
107:
79:gene expression
17:
12:
11:
5:
3516:
3506:
3505:
3500:
3495:
3490:
3473:
3472:
3469:
3468:
3466:
3465:
3459:
3457:
3436:
3429:
3428:
3425:
3424:
3422:
3421:
3415:
3413:
3399:
3398:
3396:
3395:
3388:
3386:
3372:
3371:
3369:
3368:
3361:
3359:
3342:
3335:
3334:
3331:
3330:
3328:
3327:
3321:
3319:
3309:
3308:
3306:
3305:
3299:
3297:
3287:
3286:
3284:
3283:
3277:
3275:
3265:
3264:
3262:
3261:
3256:
3254:Disulfide bond
3250:
3248:
3235:
3228:
3227:
3224:
3223:
3221:
3220:
3214:
3212:
3206:
3205:
3203:
3202:
3197:
3190:
3188:
3182:
3181:
3179:
3178:
3172:
3170:
3164:
3163:
3161:
3160:
3155:
3150:
3148:Citrullination
3144:
3142:
3136:
3135:
3133:
3132:
3127:
3125:Propionylation
3122:
3117:
3112:
3107:
3102:
3097:
3095:-glycosylation
3089:
3084:
3079:
3074:
3069:
3067:Ubiquitination
3064:
3059:
3054:
3049:
3044:
3038:
3036:
3030:
3029:
3027:
3026:
3020:
3018:
3012:
3011:
3009:
3008:
3003:
2997:
2995:
2989:
2988:
2986:
2985:
2980:
2975:
2970:
2965:
2959:
2957:
2951:
2950:
2948:
2947:
2942:
2936:
2934:
2928:
2927:
2925:
2924:
2919:
2917:Palmitoylation
2913:
2911:
2905:
2904:
2902:
2901:
2899:Detyrosination
2896:
2891:
2889:Flavin linkage
2886:
2881:
2876:
2871:
2866:
2861:
2855:
2853:
2847:
2846:
2844:
2843:
2838:
2830:
2825:
2820:
2814:
2812:
2799:
2792:
2791:
2789:
2788:
2786:Detyrosination
2783:
2778:
2773:
2767:
2765:
2759:
2758:
2756:
2755:
2752:Myristoylation
2749:
2744:
2739:
2734:
2729:
2723:
2721:
2715:
2714:
2712:
2711:
2709:N–O acyl shift
2706:
2701:
2696:
2691:
2685:
2683:
2679:
2678:
2667:
2666:
2659:
2652:
2644:
2638:
2637:
2629:
2628:External links
2626:
2623:
2622:
2573:
2546:
2519:
2492:
2485:
2455:
2428:
2397:
2370:
2342:
2320:
2316:
2312:
2308:
2299:
2271:
2244:
2217:
2188:
2181:
2155:
2103:
2052:
2009:
1990:(3): 230–238.
1970:
1963:
1941:
1890:
1863:
1834:(2): 110–115.
1813:
1762:
1755:
1729:
1696:
1645:
1585:
1556:(7): 819–823.
1536:
1500:
1469:(12): 3632–8.
1449:
1400:
1359:
1301:
1268:Mol Biotechnol
1258:
1231:(6): 289–299.
1214:
1186:
1135:
1092:
1083:
1058:
1031:
1030:
1028:
1025:
1024:
1023:
1018:
1013:
1011:Nysted reagent
1008:
1003:
998:
993:
986:
983:
982:
981:
968:
962:
956:
948:
947:Biology topics
945:
944:
943:
940:Biology portal
929:
913:
910:
898:tetramethyltin
894:
893:
875:
867:
851:
835:
832:
828:
827:
810:
802:
791:
783:
775:
762:
754:
743:
735:
727:
719:
708:
680:
677:
661:quaternization
648:
645:
644:
643:
616:
615:
598:; or a ketone
594:
564:
526:
523:
517:
510:refers to the
503:
500:
494:
491:
483:epigenetically
436:
433:
387:
384:
342:by the enzyme
328:
325:
323:
320:
314:of mercury to
292:biomethylation
283:
280:
233:
230:
207:ubiquitination
202:
199:
148:Main article:
145:
142:
126:
123:Methanogenesis
119:
118:Methanogenesis
116:
106:
103:
87:RNA processing
15:
9:
6:
4:
3:
2:
3515:
3504:
3501:
3499:
3496:
3494:
3491:
3489:
3486:
3485:
3483:
3464:
3461:
3460:
3458:
3456:
3452:
3448:
3444:
3440:
3437:
3435:
3430:
3420:
3417:
3416:
3414:
3412:
3408:
3404:
3400:
3394:(chromophore)
3393:
3390:
3389:
3387:
3385:
3381:
3377:
3373:
3367:(chromophore)
3366:
3363:
3362:
3360:
3358:
3354:
3350:
3346:
3343:
3341:
3336:
3326:
3323:
3322:
3320:
3318:
3314:
3310:
3304:
3301:
3300:
3298:
3296:
3292:
3288:
3282:
3279:
3278:
3276:
3274:
3273:Hydroxylysine
3270:
3266:
3260:
3257:
3255:
3252:
3251:
3249:
3247:
3243:
3239:
3236:
3234:
3229:
3219:
3216:
3215:
3213:
3211:
3207:
3201:
3200:Adenylylation
3198:
3195:
3192:
3191:
3189:
3187:
3183:
3177:
3176:Hydroxylation
3174:
3173:
3171:
3169:
3165:
3159:
3156:
3154:
3151:
3149:
3146:
3145:
3143:
3141:
3137:
3131:
3128:
3126:
3123:
3121:
3118:
3116:
3115:Succinylation
3113:
3111:
3110:Carbamylation
3108:
3106:
3103:
3101:
3098:
3096:
3094:
3090:
3088:
3085:
3083:
3080:
3078:
3075:
3073:
3070:
3068:
3065:
3063:
3062:Hydroxylation
3060:
3058:
3057:Adenylylation
3055:
3053:
3050:
3048:
3045:
3043:
3040:
3039:
3037:
3035:
3031:
3025:
3022:
3021:
3019:
3017:
3013:
3007:
3006:Glycosylation
3004:
3002:
2999:
2998:
2996:
2994:
2990:
2984:
2981:
2979:
2976:
2974:
2971:
2969:
2966:
2964:
2963:Carboxylation
2961:
2960:
2958:
2956:
2952:
2946:
2943:
2941:
2938:
2937:
2935:
2933:
2929:
2923:
2920:
2918:
2915:
2914:
2912:
2910:
2906:
2900:
2897:
2895:
2892:
2890:
2887:
2885:
2884:Adenylylation
2882:
2880:
2877:
2875:
2872:
2870:
2867:
2865:
2862:
2860:
2857:
2856:
2854:
2852:
2848:
2842:
2839:
2837:
2835:
2831:
2829:
2828:Glycosylation
2826:
2824:
2821:
2819:
2816:
2815:
2813:
2811:
2807:
2803:
2800:
2798:
2793:
2787:
2784:
2782:
2781:O-methylation
2779:
2777:
2774:
2772:
2769:
2768:
2766:
2764:
2760:
2753:
2750:
2748:
2745:
2743:
2740:
2738:
2735:
2733:
2732:Carbamylation
2730:
2728:
2725:
2724:
2722:
2720:
2716:
2710:
2707:
2705:
2702:
2700:
2697:
2695:
2692:
2690:
2687:
2686:
2684:
2680:
2676:
2672:
2665:
2660:
2658:
2653:
2651:
2646:
2645:
2642:
2635:
2632:
2631:
2618:
2614:
2609:
2604:
2600:
2596:
2592:
2588:
2584:
2577:
2569:
2565:
2561:
2557:
2550:
2542:
2538:
2534:
2530:
2523:
2515:
2511:
2507:
2503:
2496:
2488:
2482:
2478:
2474:
2470:
2466:
2459:
2451:
2447:
2443:
2439:
2432:
2424:
2420:
2417:: 1021–1037.
2416:
2412:
2408:
2401:
2393:
2389:
2385:
2381:
2374:
2366:
2362:
2358:
2354:
2346:
2338:
2334:
2330:
2326:
2303:
2295:
2291:
2287:
2283:
2275:
2267:
2263:
2259:
2255:
2248:
2240:
2236:
2232:
2228:
2221:
2213:
2209:
2205:
2201:
2200:
2192:
2184:
2178:
2174:
2170:
2166:
2159:
2151:
2147:
2142:
2137:
2132:
2127:
2123:
2119:
2115:
2107:
2099:
2095:
2090:
2085:
2080:
2075:
2071:
2067:
2063:
2056:
2048:
2044:
2040:
2036:
2032:
2028:
2024:
2020:
2013:
2005:
2001:
1997:
1993:
1989:
1985:
1981:
1974:
1966:
1960:
1956:
1952:
1945:
1937:
1933:
1928:
1923:
1918:
1913:
1909:
1905:
1901:
1894:
1878:
1874:
1867:
1859:
1855:
1850:
1845:
1841:
1837:
1833:
1829:
1825:
1817:
1809:
1805:
1800:
1795:
1790:
1785:
1781:
1777:
1773:
1766:
1758:
1752:
1748:
1744:
1740:
1733:
1724:
1719:
1715:
1711:
1707:
1700:
1692:
1688:
1683:
1678:
1673:
1668:
1664:
1660:
1656:
1649:
1641:
1637:
1632:
1627:
1622:
1617:
1613:
1609:
1605:
1601:
1597:
1589:
1581:
1577:
1572:
1567:
1563:
1559:
1555:
1551:
1547:
1540:
1532:
1528:
1524:
1520:
1517:(8): 928–33.
1516:
1512:
1504:
1496:
1492:
1487:
1486:11392/1689715
1482:
1477:
1472:
1468:
1464:
1460:
1453:
1445:
1441:
1436:
1431:
1427:
1423:
1419:
1415:
1411:
1404:
1396:
1392:
1387:
1382:
1378:
1374:
1370:
1363:
1355:
1351:
1346:
1341:
1337:
1333:
1329:
1325:
1321:
1317:
1313:
1305:
1297:
1293:
1289:
1285:
1281:
1277:
1273:
1269:
1262:
1254:
1250:
1246:
1242:
1238:
1234:
1230:
1226:
1218:
1210:
1206:
1202:
1198:
1190:
1182:
1178:
1173:
1168:
1163:
1158:
1154:
1150:
1146:
1139:
1130:
1125:
1120:
1115:
1111:
1107:
1103:
1096:
1087:
1079:
1075:
1070:
1065:
1061:
1055:
1051:
1047:
1043:
1036:
1032:
1022:
1019:
1017:
1014:
1012:
1009:
1007:
1004:
1002:
999:
997:
994:
992:
989:
988:
980:
976:
972:
971:Remethylation
969:
966:
963:
960:
957:
954:
951:
950:
941:
935:
930:
927:
916:
909:
907:
903:
899:
891:
887:
886:
885:
883:
863:
859:
847:
846:methyllithium
843:
842:
831:
808:
800:
789:
781:
773:
760:
741:
733:
725:
717:
706:
693:
692:
691:
689:
685:
673:
668:
664:
662:
658:
654:
641:
637:
636:
635:
633:
629:
625:
624:carbohydrates
621:
613:
609:
608:
607:
605:
601:
590:
582:
578:
574:
570:
562:
558:
554:
550:
546:
542:
538:
534:
533:
532:electrophilic
522:
513:
509:
499:
490:
488:
484:
480:
476:
472:
468:
464:
460:
456:
452:
449:
445:
441:
432:
429:
426:
422:
420:
416:
412:
408:
404:
400:
396:
392:
383:
381:
376:
374:
370:
366:
362:
357:
353:
349:
345:
341:
337:
333:
319:
317:
316:methylmercury
313:
309:
305:
301:
297:
293:
289:
279:
277:
273:
267:
265:
261:
257:
253:
249:
243:
238:
229:
227:
223:
219:
217:
212:
208:
198:
197:(azaleatin).
196:
192:
188:
184:
183:methoxy bonds
179:
177:
173:
169:
165:
161:
157:
151:
141:
134:
130:
124:
115:
113:
102:
100:
99:demethylation
96:
92:
88:
84:
80:
76:
72:
68:
64:
59:
57:
53:
49:
45:
41:
37:
33:
29:
25:
21:
3152:
3130:Butyrylation
3092:
3041:
2972:
2833:
2746:
2704:Racemization
2689:Peptide bond
2590:
2586:
2576:
2559:
2555:
2549:
2532:
2528:
2522:
2505:
2501:
2495:
2468:
2464:
2458:
2441:
2437:
2431:
2414:
2410:
2400:
2383:
2379:
2373:
2356:
2352:
2345:
2328:
2324:
2302:
2285:
2281:
2274:
2257:
2253:
2247:
2230:
2226:
2220:
2203:
2197:
2191:
2164:
2158:
2121:
2117:
2106:
2069:
2065:
2055:
2022:
2018:
2012:
1987:
1983:
1973:
1954:
1944:
1907:
1904:BMC Genomics
1903:
1893:
1881:. Retrieved
1877:the original
1866:
1831:
1827:
1816:
1779:
1775:
1765:
1738:
1732:
1713:
1709:
1699:
1662:
1658:
1648:
1603:
1599:
1588:
1553:
1549:
1539:
1514:
1510:
1503:
1466:
1462:
1452:
1420:(9): 990–7.
1417:
1413:
1403:
1376:
1372:
1362:
1319:
1315:
1304:
1274:(1): 71–81.
1271:
1267:
1261:
1228:
1224:
1217:
1200:
1196:
1189:
1152:
1148:
1138:
1109:
1105:
1095:
1086:
1041:
1035:
902:dimethylzinc
895:
841:nucleophilic
840:
837:
829:
684:Diazomethane
682:
650:
632:silver oxide
617:
561:magic methyl
557:diazomethane
531:
528:
505:
502:In chemistry
496:
473:groups from
438:
430:
423:
390:
389:
377:
330:
285:
268:
256:homocysteine
250:regenerates
246:
225:
221:
215:
204:
180:
153:
139:
121:
108:
90:
75:heavy metals
60:
52:soil science
48:biochemistry
28:methyl group
19:
18:
3493:Epigenetics
3488:Methylation
3194:Diphthamide
3153:Methylation
3120:Lactylation
3082:Deamination
3072:Sumoylation
3047:Acetylation
3042:Methylation
3001:Deamidation
2973:Methylation
2922:Prenylation
2747:Methylation
2737:Formylation
2727:Acetylation
2699:Proteolysis
2634:deltaMasses
2325:Tetrahedron
1414:Epigenetics
628:iodomethane
573:carboxylate
537:iodomethane
361:CpG islands
254:(Met) from
205:Along with
112:epigenetics
20:Methylation
3482:Categories
3317:Tryptophan
3313:Tryptophan
3269:Methionine
3210:Tryptophan
2993:Asparagine
2763:C terminus
2719:N terminus
2593:(8): e52.
2124:(1): 211.
1910:(1): 315.
1883:11 October
1129:1822/47121
1027:References
991:Alkylation
975:methionine
512:alkylation
451:amino acid
380:honey bees
288:food chain
252:methionine
172:percursors
105:In biology
36:alkylation
3463:Desmosine
3376:Histidine
3196:formation
3186:Histidine
3105:Glycation
3052:Acylation
3016:Glutamine
2955:Glutamate
2932:Aspartate
2874:Sulfation
2810:Threonine
2771:Amidation
2742:Glycation
2072:(1): 19.
753:⟶
493:Evolution
369:promoters
348:CpG sites
340:catalyzed
308:reduction
272:cobalamin
67:catalyzed
44:chemistry
32:substrate
22:, in the
3451:Allysine
3447:Allysine
3443:Allysine
3380:Tyrosine
3353:Tyrosine
3295:Tyrosine
3246:Cysteine
3242:Cysteine
3140:Arginine
2909:Cysteine
2851:Tyrosine
2617:21297115
2150:32376902
2098:16225687
2047:11283354
2004:15145346
1936:37308882
1927:10258952
1858:26751643
1808:27375676
1691:33101388
1640:23852726
1580:29898214
1531:27223861
1495:23010533
1444:23975186
1395:22170606
1354:19829295
1296:20307488
1288:19842073
1245:11005674
1181:29743234
1112:: 1–22.
1078:18804699
912:See also
882:carbonyl
860:such as
581:alkoxide
467:histones
444:arginine
365:residues
352:cytosine
201:Proteins
91:In vitro
40:hydrogen
3411:Glycine
3403:Alanine
3384:Glycine
3357:Glycine
3168:Proline
2836:-GlcNAc
2682:General
2608:3082908
2359:: 295.
2288:: 169.
2141:7203018
2089:1282579
2027:Bibcode
2019:Science
1849:4826618
1799:4893491
1682:7546774
1631:3732956
1608:Bibcode
1571:6128494
1435:3883776
1345:2857523
1324:Bibcode
1253:1025140
1172:6036201
1106:Archaea
1069:3037834
996:Methoxy
600:enolate
521:group.
356:guanine
296:arsenic
168:lignols
160:anisole
156:phenols
71:enzymes
56:biology
3455:Lysine
3407:Serine
3349:Serine
3291:Lysine
3034:Lysine
2806:Serine
2615:
2605:
2562:: 31.
2535:: 75.
2483:
2444:: 89.
2260:: 63.
2233:: 12.
2206:: 90.
2179:
2148:
2138:
2096:
2086:
2045:
2002:
1961:
1934:
1924:
1856:
1846:
1806:
1796:
1782:: 99.
1753:
1689:
1679:
1638:
1628:
1578:
1568:
1529:
1493:
1442:
1432:
1393:
1373:Nature
1352:
1342:
1316:Nature
1294:
1286:
1251:
1243:
1179:
1169:
1076:
1066:
1056:
959:MethDB
904:, and
722:tmsCHN
657:amines
626:using
604:ketone
471:methyl
448:lysine
415:snoRNA
290:. The
193:, and
176:lignin
85:, and
54:, and
2754:(Gly)
2508:: 7.
2386:: 9.
2315:or Cs
1292:S2CID
1249:S2CID
856:) or
589:ether
583:salt
579:; an
577:ester
547:, or
419:miRNA
411:snRNA
407:tmRNA
30:on a
2673:and
2613:PMID
2481:ISBN
2177:ISBN
2146:PMID
2094:PMID
2043:PMID
2000:PMID
1959:ISBN
1932:PMID
1885:2017
1854:PMID
1804:PMID
1751:ISBN
1687:PMID
1636:PMID
1576:PMID
1527:PMID
1491:PMID
1440:PMID
1391:PMID
1350:PMID
1284:PMID
1241:PMID
1177:PMID
1110:2017
1074:PMID
1054:ISBN
977:and
798:Otms
670:The
651:The
630:and
618:The
593:ROCH
403:mRNA
399:rRNA
395:tRNA
276:zinc
209:and
3434:AAs
3340:AAs
3233:AAs
2797:AAs
2603:PMC
2595:doi
2564:doi
2537:doi
2510:doi
2473:doi
2446:doi
2419:doi
2388:doi
2361:doi
2333:doi
2323:".
2290:doi
2262:doi
2235:doi
2208:doi
2169:doi
2136:PMC
2126:doi
2084:PMC
2074:doi
2035:doi
2023:292
1992:doi
1922:PMC
1912:doi
1844:PMC
1836:doi
1794:PMC
1784:doi
1743:doi
1718:doi
1677:PMC
1667:doi
1626:PMC
1616:doi
1604:110
1566:PMC
1558:doi
1554:154
1519:doi
1515:152
1481:hdl
1471:doi
1430:PMC
1422:doi
1381:doi
1377:480
1340:PMC
1332:doi
1320:462
1276:doi
1233:doi
1205:doi
1167:PMC
1157:doi
1153:293
1124:hdl
1114:doi
1064:PMC
1046:doi
870:MgX
757:RCO
703:RCO
446:or
378:In
294:of
174:to
69:by
61:In
3484::
2611:.
2601:.
2591:39
2589:.
2585:.
2560:62
2558:.
2533:30
2531:.
2506:55
2504:.
2479:.
2471:.
2467:.
2442:25
2440:.
2415:83
2413:.
2409:.
2384:41
2382:.
2357:84
2355:.
2329:64
2327:.
2319:CO
2311:CO
2286:76
2284:.
2258:29
2256:.
2229:.
2204:31
2202:.
2175:.
2144:.
2134:.
2120:.
2116:.
2092:.
2082:.
2068:.
2064:.
2041:.
2033:.
2021:.
1998:.
1988:16
1986:.
1982:.
1953:.
1930:.
1920:.
1908:24
1906:.
1902:.
1852:.
1842:.
1832:23
1830:.
1826:.
1802:.
1792:.
1778:.
1774:.
1749:.
1716:.
1712:.
1708:.
1685:.
1675:.
1665:.
1663:11
1661:.
1657:.
1634:.
1624:.
1614:.
1602:.
1598:.
1574:.
1564:.
1552:.
1548:.
1525:.
1513:.
1489:.
1479:.
1467:27
1465:.
1461:.
1438:.
1428:.
1416:.
1412:.
1389:.
1375:.
1371:.
1348:.
1338:.
1330:.
1318:.
1314:.
1290:.
1282:.
1272:44
1270:.
1247:.
1239:.
1229:74
1227:.
1201:86
1199:.
1175:.
1165:.
1151:.
1147:.
1122:.
1108:.
1104:.
1072:.
1062:.
1052:.
908:.
900:,
878:Li
874:CH
866:CH
854:Li
850:CH
786:CH
770:CH
750:OH
738:CH
634:.
606:.
591:,
585:RO
567:2
555:,
543:,
539:,
516:CH
489:.
417:,
413:,
409:,
405:,
401:,
397:,
318:.
189:,
170:,
101:.
89:.
58:.
50:,
46:,
3453:–
3449:–
3445:–
3409:–
3405:–
3382:–
3378:–
3355:–
3351:–
3315:–
3293:–
3271:–
3244:–
3093:O
2834:O
2808:/
2663:e
2656:t
2649:v
2619:.
2597::
2570:.
2566::
2543:.
2539::
2516:.
2512::
2489:.
2475::
2452:.
2448::
2425:.
2421::
2394:.
2390::
2367:.
2363::
2339:.
2335::
2321:3
2317:2
2313:3
2309:2
2296:.
2292::
2268:.
2264::
2241:.
2237::
2231:9
2214:.
2210::
2185:.
2171::
2152:.
2128::
2122:3
2100:.
2076::
2070:5
2049:.
2037::
2029::
2006:.
1994::
1967:.
1938:.
1914::
1887:.
1860:.
1838::
1810:.
1786::
1780:7
1759:.
1745::
1726:.
1720::
1714:4
1693:.
1669::
1642:.
1618::
1610::
1582:.
1560::
1533:.
1521::
1497:.
1483::
1473::
1446:.
1424::
1418:8
1397:.
1383::
1356:.
1334::
1326::
1298:.
1278::
1255:.
1235::
1211:.
1207::
1183:.
1159::
1132:.
1126::
1116::
1080:.
1048::
876:3
868:3
864:(
852:3
848:(
809:2
805:N
801:+
790:3
782:+
774:3
761:2
742:3
734:+
726:2
718:+
715:H
707:2
595:3
565:N
518:3
226:N
222:N
216:S
127:2
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.