607:
620:
709:
669:
739:
724:
656:
697:
295:
755:
681:
33:
168:
1763:
619:
1864:
256:
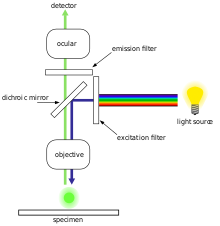
191:. Since most of the excitation light is transmitted through the specimen, only reflected excitatory light reaches the objective together with the emitted light and the epifluorescence method therefore gives a high signal-to-noise ratio. The dichroic beamsplitter acts as a wavelength specific filter, transmitting fluoresced light through to the eyepiece or detector, but reflecting any remaining excitation light back towards the source.
41:
1876:
1057:
492:. Photobleaching occurs as the fluorescent molecules accumulate chemical damage from the electrons excited during fluorescence. Photobleaching can severely limit the time over which a sample can be observed by fluorescence microscopy. Several techniques exist to reduce photobleaching such as the use of more robust fluorophores, by minimizing illumination, or by using photoprotective
147:(see figure below). The filters and the dichroic beamsplitter are chosen to match the spectral excitation and emission characteristics of the fluorophore used to label the specimen. In this manner, the distribution of a single fluorophore (color) is imaged at a time. Multi-color images of several types of fluorophores must be composed by combining several single-color images.
606:
723:
655:
539:
microscope as a confocal laser scanning fluorescence microscope where the light is focused ideally from all sides to a common focus which is used to scan the object by 'point-by-point' excitation combined with 'point-by-point' detection. However, the first experimental demonstration of the 4pi microscope took place in 1994.
258:
499:
Fluorescence microscopy with fluorescent reporter proteins has enabled analysis of live cells by fluorescence microscopy, however cells are susceptible to phototoxicity, particularly with short wavelength light. Furthermore, fluorescent molecules have a tendency to generate reactive chemical species
262:
261:
257:
503:
Unlike transmitted and reflected light microscopy techniques, fluorescence microscopy only allows observation of the specific structures which have been labeled for fluorescence. For example, observing a tissue sample prepared with a fluorescent DNA stain by fluorescence microscopy only reveals the
479:
and the techniques available for modifying DNA allow scientists to genetically modify proteins to also carry a fluorescent protein reporter. In biological samples this allows a scientist to directly make a protein of interest fluorescent. The protein location can then be directly tracked, including
507:
Computational techniques that propose to estimate the fluorescent signal from non-fluorescent images (such as brightfield) may reduce these concerns. In general, these approaches involve training a deep convolutional neural network on stained cells and then estimating the fluorescence on unstained
150:
Most fluorescence microscopes in use are epifluorescence microscopes, where excitation of the fluorophore and detection of the fluorescence are done through the same light path (i.e. through the objective). These microscopes are widely used in biology and are the basis for more advanced microscope
44:
538:
Several improvements in microscopy techniques have been invented in the 20th century and have resulted in increased resolution and contrast to some extent. However they did not overcome the diffraction limit. In 1978 first theoretical ideas have been developed to break this barrier by using a 4Pi
115:, causing them to emit light of longer wavelengths (i.e., of a different color than the absorbed light). The illumination light is separated from the much weaker emitted fluorescence through the use of a spectral emission filter. Typical components of a fluorescence microscope are a light source (
594:
could achieve similar results by relying on blinking or switching of single molecules, where the fraction of fluorescing molecules is very small at each time. This stochastic response of molecules on the applied light corresponds also to a highly nonlinear interaction, leading to subdiffraction
43:
263:
48:
47:
42:
566:
concept rely on a strong non-linear interaction between light and fluorescing molecules. The molecules are driven strongly between distinguishable molecular states at each specific location, so that finally light can be emitted at only a small fraction of space, hence an increased resolution.
708:
49:
587:(GFP) can be achieved by using a further development of SPDM the so-called SPDMphymod technology which makes it possible to detect and count two different fluorescent molecule types at the molecular level (this technology is referred to as two-color localization microscopy or 2CLM).
260:
50:
46:
696:
574:
stained with a fluorescent marker was achieved by development of SPDM localization microscopy and the structured laser illumination (spatially modulated illumination, SMI). Combining the principle of SPDM with SMI resulted in the development of the
321:
In order for a sample to be suitable for fluorescence microscopy it must be fluorescent. There are several methods of creating a fluorescent sample; the main techniques are labelling with fluorescent stains or, in the case of biological samples,
661:
Endothelial cells under the microscope. Nuclei are stained blue with DAPI, microtubules are marked green by an antibody bound to FITC and actin filaments are labeled red with phalloidin bound to TRITC. Bovine pulmonary artery endothelial (BPAE)
668:
554:
combines a fluorescence microscope with an electron microscope. This allows one to visualize ultrastructure and contextual information with the electron microscope while using the data from the fluorescence microscope as a labelling tool.
448:
in order to label specific proteins or other molecules within the cell. A sample is treated with a primary antibody specific for the molecule of interest. A fluorophore can be directly conjugated to the primary antibody. Alternatively a
534:
and "limits an optical microscope's resolution to approximately half of the wavelength of the light used." Fluorescence microscopy is central to many techniques which aim to reach past this limit by specialized optical configurations.
612:
A z-projection of an osteosarcoma cell, stained with phalloidin to visualise actin filaments. The image was taken on a confocal microscope, and the subsequent deconvolution was done using an experimentally derived point spread
680:
346:
Many fluorescent stains have been designed for a range of biological molecules. Some of these are small molecules which are intrinsically fluorescent and bind a biological molecule of interest. Major examples of these are
90:
substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a
1037:
Colin, S., Coelho, L.P., Sunagawa, S., Bowler, C., Karsenti, E., Bork, P., Pepperkok, R. and De Vargas, C. (2017) "Quantitative 3D-imaging for cell biology and ecology of environmental microbial eukaryotes".
738:
45:
367:
of cells. Others are drugs, toxins, or peptides which bind specific cellular structures and have been derivatised with a fluorescent reporter. A major example of this class of fluorescent stain is
754:
259:
1387:
729:
Super-resolution microscopy: Co-localization microscopy (2CLM) with GFP and RFP fusion proteins (nucleus of a bone cancer cell) 120.000 localized molecules in a wide-field area (470 μm)
1335:
334:) can be used. In the life sciences fluorescence microscopy is a powerful tool which allows the specific and sensitive staining of a specimen in order to detect the distribution of
1245:
S.W. Hell, E.H.K. Stelzer, S. Lindek, C. Cremer; Stelzer; Lindek; Cremer (1994). "Confocal microscopy with an increased detection aperture: type-B 4Pi confocal microscopy".
187:
emitted by the specimen is focused to the detector by the same objective that is used for the excitation which for greater resolution will need objective lens with higher
702:
Yeast cell membrane visualized by some membrane proteins fused with RFP and GFP fluorescent markers. Imposition of light from both of markers results in yellow color.
570:
As well in the 1990s another super resolution microscopy method based on wide field microscopy has been developed. Substantially improved size resolution of cellular
1117:
Kandel, Mikhail E.; He, Yuchen R.; Lee, Young Jae; Chen, Taylor Hsuan-Yu; Sullivan, Kathryn
Michele; Aydin, Onur; Saif, M. Taher A.; Kong, Hyunjoon; Sobh, Nahil;
508:
samples. Thus by decoupling the cells under investigation from the cells used to train the network, imaging can performed quicker and with reduced phototoxicity.
1197:
453:, conjugated to a fluorophore, which binds specifically to the first antibody can be used. For example, a primary antibody raised in a mouse which recognises
1428:
1806:
232:
156:
144:
36:
An upright fluorescence microscope (Olympus BX61) with the fluorescence filter cube turret above the objective lenses, coupled with a digital camera.
1298:
1504:
645:
is imaged separately using a different combination of excitation and emission filters, and the images are captured sequentially using a digital
1362:
1717:
288:
The animation starts by overlaying all available fluorescent channels, and then clarifies the visualisation by switching channels on and off
1645:
1388:"High-precision structural analysis of subnuclear complexes in fixed and live cells via spatially modulated illumination (SMI) microscopy"
714:
Super-resolution microscopy: Single YFP molecule detection in a human cancer cell. Typical distance measurements in the 15 nm range
1074:
Bidhendi, AJ; Chebli, Y; Geitmann, A (May 2020). "Fluorescence
Visualization of Cellulose and Pectin in the Primary Plant Cell Wall".
338:
or other molecules of interest. As a result, there is a diverse range of techniques for fluorescent staining of biological samples.
1313:
902:
674:
3D dual-color super-resolution microscopy with Her2 and Her3 in breast cells, standard dyes: Alexa 488, Alexa 568. LIMON microscopy
179:, are of the epifluorescence design shown in the diagram. Light of the excitation wavelength illuminates the specimen through the
17:
1912:
1849:
1844:
1712:
809:
591:
83:
912:
772:
551:
521:
1454:
1880:
1811:
359:(excited by UV wavelength light) and DRAQ5 and DRAQ7 (optimally excited by red light) which all bind the minor groove of
1747:
1727:
1336:"High-precision distance microscopy of 3D nanostructures by a spatially modulated excitation fluorescence microscope"
788:
687:
199:
Fluorescence microscopy requires intense, near-monochromatic, illumination which some widespread light sources, like
313:
illuminated by blue light in an epifluorescence microscope. The SYBR green in the sample binds to the herring sperm
1791:
1787:
1638:
1220:
544:
403:. The quest for fluorescent probes with a high specificity that also allow live imaging of plant cells is ongoing.
1589:
1570:
1123:"Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments"
686:
Human lymphocyte nucleus stained with DAPI with chromosome 13 (green) and 21 (red) centromere probes hybridized (
243:
arrays into the illumination path of a widefield epifluorescence microscope, highly uniform illumination with a
1816:
426:, which can be chemically linked to a different molecule which binds the target of interest within the sample.
872:
380:
1618:
animations and explanations on various types of microscopes including fluorescent and confocal microscopes
1608:
1386:
Reymann, J; Baddeley, D; Gunkel, M; Lemmer, P; Stadter, W; Jegou, T; Rippe, K; Cremer, C; Birk, U (2008).
847:
1868:
1631:
1118:
760:
Fluorescence microscopy images of sun flares pathology in a blood cell showing the affected areas in red.
517:
1834:
580:
1801:
1707:
1543:
793:
584:
294:
457:
combined with a secondary anti-mouse antibody derivatised with a fluorophore could be used to label
1267:
780:, pioneer of fluorescence microscopy techniques for visualization of bacterial subcellular proteins
244:
1503:
Gunkel, M; Erdel, F; Rippe, K; Lemmer, P; Kaufmann, R; Hörmann, C; Amberger, R; Cremer, C (2009).
1244:
1795:
1692:
543:
maximizes the amount of available focusing directions by using two opposing objective lenses or
1737:
1262:
504:
organization of the DNA within the cells and reveals nothing else about the cell morphologies.
1697:
526:
The wave nature of light limits the size of the spot to which light can be focused due to the
239:
excitation filter are commonly used for widefield epifluorescence microscopes. By placing two
1907:
1902:
1702:
1292:
957:
493:
323:
1584:
1917:
1254:
1144:
991:"Flat-top illumination profile in an epi-fluorescence microscope by dual micro lens arrays"
880:
783:
75:
8:
1782:
1722:
488:
Fluorophores lose their ability to fluoresce as they are illuminated in a process called
470:
327:
228:
152:
92:
1258:
1148:
918:
1673:
1535:
1485:
1420:
1354:
1198:"Considerations on a laser-scanning-microscope with high resolution and depth of field"
1178:
1165:
1134:
1122:
1099:
1020:
966:
941:
799:
450:
435:
227:. Lasers are most widely used for more complex fluorescence microscopy techniques like
208:
188:
180:
120:
96:
63:
32:
1527:
1477:
1412:
1334:
Hausmann, Michael; Schneider, Bernhard; Bradl, Joachim; Cremer, Christoph G. (1997),
1280:
1212:
1182:
1170:
1103:
1091:
1012:
971:
908:
527:
212:
132:
1539:
1489:
1424:
1358:
1024:
1752:
1519:
1469:
1402:
1346:
1272:
1160:
1152:
1083:
1047:
1002:
961:
953:
331:
744:
Fluorescence microscopy of DNA Expression in the Human Wild-Type and P239S Mutant
317:
and, once bound, fluoresces giving off green light when illuminated by blue light.
1839:
1612:
1341:, in Bigio, Irving J; Schneckenburger, Herbert; Slavik, Jan; et al. (eds.),
777:
559:
167:
140:
136:
625:
Epifluorescent imaging of the three components in a dividing human cancer cell.
1732:
1156:
1061:
988:
819:
540:
489:
440:
Immunofluorescence is a technique which uses the highly specific binding of an
364:
220:
204:
116:
1407:
1896:
1762:
900:
571:
423:
356:
176:
1617:
1314:"Correlative microscopy: Opening up worlds of information with fluorescence"
1051:
1531:
1523:
1481:
1473:
1416:
1284:
1174:
1095:
1016:
975:
814:
419:
411:
348:
200:
184:
67:
1455:"Nanostructure analysis using spatially modulated illumination microscopy"
280:(b) Cyan: - generic counterstain for visualising eukaryotic cell surfaces
1276:
1216:
1007:
990:
642:
638:
576:
458:
415:
407:
384:
112:
79:
1666:
1654:
804:
646:
558:
The first technique to really achieve a sub-diffraction resolution was
531:
368:
306:
278:(a) Green: - stains cellular membranes indicating the core cell bodies
175:
The majority of fluorescence microscopes, especially those used in the
108:
71:
1350:
1087:
1623:
396:
392:
240:
87:
203:
cannot provide. Four main types of light source are used, including
1139:
745:
476:
441:
236:
1060:
Material was copied from this source, which is available under a
630:
563:
562:, proposed in 1994. This method and all techniques following the
454:
445:
335:
310:
299:
1453:
Baddeley, D; Batram, C; Weiland, Y; Cremer, C; Birk, UJ (2003).
1742:
1605:
1599:
1505:"Dual color localization microscopy of cellular nanostructures"
634:
400:
376:
500:
when under illumination which enhances the phototoxic effect.
330:. Alternatively the intrinsic fluorescence of a sample (i.e.,
1056:
372:
302:
216:
128:
27:
Optical microscope that uses fluorescence and phosphorescence
1333:
989:
F.A.W. Coumans; E. van der Pol; L.W.M.M. Terstappen (2012).
1452:
1385:
352:
626:
360:
314:
224:
124:
54:
Fluorescence and confocal microscopes operating principle
1807:
Total internal reflection fluorescence microscopy (TIRF)
810:
Scanning electron microscope § Cathodoluminescence
530:. This limitation was described in the 19th century by
1502:
1073:
1062:
Creative
Commons Attribution 4.0 International License
901:
Juan Carlos
Stockert, Alfonso Blázquez-Castro (2017).
235:
while xenon lamps, and mercury lamps, and LEDs with a
107:
The specimen is illuminated with light of a specific
1845:
Photo-activated localization microscopy (PALM/STORM)
547:
using redshifted light and multi-photon excitation.
99:
to get better resolution of the fluorescence image.
598:
877:Microscopes—Help Scientists Explore Hidden Worlds
731:measured with a Vertico-SMI/SPDMphymod microscope
716:measured with a Vertico-SMI/SPDMphymod microscope
233:total internal reflection fluorescence microscopy
157:total internal reflection fluorescence microscope
1894:
579:microscope. Single molecule detection of normal
341:
276:Displays overlays from four fluorescent channels
1116:
123:are common; more advanced forms are high-power
1343:Optical Biopsies and Microscopic Techniques II
1748:Interference reflection microscopy (IRM/RICM)
1639:
649:, then superimposed to give a complete image.
511:
162:
1297:: CS1 maint: multiple names: authors list (
1195:
406:There are many fluorescent molecules called
395:is performed using stains or dyes that bind
1067:
1646:
1632:
942:"Super resolution fluorescence microscopy"
845:
282:(c) Blue: - stains DNA, identifies nuclei
111:(or wavelengths) which is absorbed by the
1406:
1266:
1164:
1138:
1006:
965:
848:"Introduction to Fluorescence Microscopy"
1718:Differential interference contrast (DIC)
1189:
1031:
958:10.1146/annurev.biochem.77.061906.092014
904:Fluorescence Microscopy in Life Sciences
841:
839:
837:
835:
293:
284:(d) Red: - resolves chloroplasts
254:
166:
86:, to study the properties of organic or
39:
31:
464:
391:collagen fibers. Staining of the plant
171:Schematic of a fluorescence microscope.
14:
1895:
1713:Quantitative phase-contrast microscopy
1653:
592:photoactivated localization microscopy
1627:
939:
832:
773:Correlative light-electron microscopy
522:Correlative Light-Electron Microscopy
429:
250:
1875:
1840:Stimulated emission depletion (STED)
379:cells. A new peptide, known as the
24:
1602:, the database of fluorescent dyes
1311:
25:
1929:
1812:Lightsheet microscopy (LSFM/SPIM)
1564:
789:Fluorescence in the life sciences
688:Fluorescent in situ hybridization
1874:
1863:
1862:
1761:
1055:
753:
737:
722:
707:
695:
679:
667:
654:
618:
605:
545:two-photon excitation microscopy
194:
1496:
1446:
1379:
1345:, vol. 3197, p. 217,
1327:
1305:
1238:
599:Fluorescence micrograph gallery
70:instead of, or in addition to,
1817:Lattice light-sheet microscopy
1728:Second harmonic imaging (SHIM)
1110:
982:
933:
907:. Bentham Science Publishers.
894:
865:
483:
383:, can also be conjugated with
13:
1:
1913:Optical microscopy techniques
1196:Cremer, C; Cremer, T (1978).
946:Annual Review of Biochemistry
873:"The Fluorescence Microscope"
826:
590:Alternatively, the advent of
342:Biological fluorescent stains
1590:Resources in other libraries
475:The modern understanding of
381:Collagen Hybridizing Peptide
102:
7:
765:
518:Super resolution microscopy
269:3D-animation of the diatom
10:
1934:
1606:Microscopy Resource Center
1157:10.1038/s41467-020-20062-x
515:
512:Sub-diffraction techniques
468:
433:
163:Epifluorescence microscopy
1858:
1825:
1770:
1759:
1683:
1661:
1585:Resources in your library
1408:10.1007/s10577-008-1238-2
794:Green fluorescent protein
585:green fluorescent protein
371:, which is used to stain
247:of 1-2% can be achieved.
846:Spring KR, Davidson MW.
245:coefficient of variation
223:sources, and high-power
1778:Fluorescence microscopy
1738:Structured illumination
1693:Bright-field microscopy
1611:22 October 2014 at the
1576:Fluorescence microscopy
1052:10.7554/eLife.26066.002
60:fluorescence microscope
18:Fluorescence microscopy
1850:Near-field (NSOM/SNOM)
1788:Multiphoton microscopy
1620:(Université Paris Sud)
1524:10.1002/biot.200900005
1474:10.1038/nprot.2007.399
940:Huang B (March 2010).
583:fluorescent dyes like
552:correlative microscopy
318:
291:
172:
55:
37:
1703:Dark-field microscopy
1512:Biotechnology Journal
1312:Baarle, Kaitlin van.
1127:Nature Communications
1076:Journal of Microscopy
297:
266:
170:
151:designs, such as the
141:dichroic beamsplitter
53:
35:
1771:Fluorescence methods
1277:10.1364/OL.19.000222
1008:10.1002/cyto.a.22029
881:The Nobel Foundation
784:Fluorescence imaging
465:Fluorescent proteins
363:, thus labeling the
1802:Image deconvolution
1783:Confocal microscopy
1723:Dispersion staining
1698:Köhler illumination
1395:Chromosome Research
1259:1994OptL...19..222H
1149:2020NatCo..11.6256K
629:is stained blue, a
471:Fluorescent protein
328:fluorescent protein
229:confocal microscopy
209:mercury-vapor lamps
153:confocal microscope
93:confocal microscope
1674:Optical microscopy
1655:Optical microscopy
800:Mercury-vapor lamp
637:is green, and the
451:secondary antibody
436:Immunofluorescence
430:Immunofluorescence
387:and used to stain
319:
292:
251:Sample preparation
189:numerical aperture
173:
121:mercury-vapor lamp
97:optical sectioning
64:optical microscope
56:
38:
1890:
1889:
1835:Diffraction limit
1571:Library resources
1351:10.1117/12.297969
1205:Microscopica Acta
1088:10.1111/jmi.12895
914:978-1-68108-519-7
852:Nikon MicroscopyU
732:
717:
528:diffraction limit
290:
264:
213:excitation filter
133:excitation filter
51:
16:(Redirected from
1925:
1878:
1877:
1866:
1865:
1828:limit techniques
1765:
1686:contrast methods
1684:Illumination and
1648:
1641:
1634:
1625:
1624:
1600:Fluorophores.org
1559:
1558:
1556:
1554:
1548:
1542:. Archived from
1509:
1500:
1494:
1493:
1462:Nature Protocols
1459:
1450:
1444:
1443:
1441:
1439:
1433:
1427:. Archived from
1410:
1392:
1383:
1377:
1376:
1375:
1373:
1367:
1361:, archived from
1340:
1331:
1325:
1324:
1322:
1320:
1309:
1303:
1302:
1296:
1288:
1270:
1242:
1236:
1235:
1233:
1231:
1225:
1219:. Archived from
1202:
1193:
1187:
1186:
1168:
1142:
1119:Popescu, Gabriel
1114:
1108:
1107:
1071:
1065:
1059:
1035:
1029:
1028:
1010:
995:Cytometry Part A
986:
980:
979:
969:
937:
931:
930:
928:
926:
917:. Archived from
898:
892:
891:
889:
887:
869:
863:
862:
860:
858:
843:
757:
741:
730:
726:
715:
711:
699:
683:
671:
658:
622:
609:
332:autofluorescence
286:
265:
52:
21:
1933:
1932:
1928:
1927:
1926:
1924:
1923:
1922:
1893:
1892:
1891:
1886:
1854:
1827:
1826:Sub-diffraction
1821:
1766:
1757:
1685:
1679:
1657:
1652:
1613:Wayback Machine
1596:
1595:
1594:
1579:
1578:
1574:
1567:
1562:
1552:
1550:
1549:on 4 March 2016
1546:
1507:
1501:
1497:
1457:
1451:
1447:
1437:
1435:
1434:on 4 March 2016
1431:
1390:
1384:
1380:
1371:
1369:
1368:on 4 March 2016
1365:
1338:
1332:
1328:
1318:
1316:
1310:
1306:
1290:
1289:
1243:
1239:
1229:
1227:
1226:on 4 March 2016
1223:
1200:
1194:
1190:
1115:
1111:
1072:
1068:
1036:
1032:
987:
983:
938:
934:
924:
922:
915:
899:
895:
885:
883:
871:
870:
866:
856:
854:
844:
833:
829:
824:
778:Elizabeth Harry
768:
761:
758:
749:
742:
733:
727:
718:
712:
703:
700:
691:
684:
675:
672:
663:
659:
650:
623:
614:
610:
601:
560:STED microscopy
524:
514:
486:
480:in live cells.
473:
467:
438:
432:
351:stains such as
344:
289:
285:
283:
281:
279:
277:
275:
255:
253:
205:xenon arc lamps
197:
165:
145:emission filter
137:dichroic mirror
105:
40:
28:
23:
22:
15:
12:
11:
5:
1931:
1921:
1920:
1915:
1910:
1905:
1888:
1887:
1885:
1884:
1872:
1859:
1856:
1855:
1853:
1852:
1847:
1842:
1837:
1831:
1829:
1823:
1822:
1820:
1819:
1814:
1809:
1804:
1799:
1785:
1780:
1774:
1772:
1768:
1767:
1760:
1758:
1756:
1755:
1750:
1745:
1740:
1735:
1733:4Pi microscope
1730:
1725:
1720:
1715:
1710:
1708:Phase contrast
1705:
1700:
1695:
1689:
1687:
1681:
1680:
1678:
1677:
1670:
1662:
1659:
1658:
1651:
1650:
1643:
1636:
1628:
1622:
1621:
1615:
1603:
1593:
1592:
1587:
1581:
1580:
1569:
1568:
1566:
1565:External links
1563:
1561:
1560:
1495:
1468:(10): 2640–6.
1445:
1378:
1326:
1304:
1268:10.1.1.501.598
1253:(3): 222–224.
1247:Optics Letters
1237:
1188:
1109:
1082:(3): 164–181.
1066:
1030:
1001:(4): 324–331.
981:
932:
921:on 14 May 2019
913:
893:
864:
830:
828:
825:
823:
822:
820:Xenon arc lamp
817:
812:
807:
802:
797:
791:
786:
781:
775:
769:
767:
764:
763:
762:
759:
752:
750:
743:
736:
734:
728:
721:
719:
713:
706:
704:
701:
694:
692:
685:
678:
676:
673:
666:
664:
660:
653:
651:
641:are red. Each
624:
617:
615:
611:
604:
600:
597:
572:nanostructures
541:4Pi microscopy
513:
510:
490:photobleaching
485:
482:
466:
463:
434:Main article:
431:
428:
343:
340:
287:
267:
252:
249:
221:supercontinuum
196:
193:
164:
161:
117:xenon arc lamp
104:
101:
26:
9:
6:
4:
3:
2:
1930:
1919:
1916:
1914:
1911:
1909:
1906:
1904:
1901:
1900:
1898:
1883:
1882:
1873:
1871:
1870:
1861:
1860:
1857:
1851:
1848:
1846:
1843:
1841:
1838:
1836:
1833:
1832:
1830:
1824:
1818:
1815:
1813:
1810:
1808:
1805:
1803:
1800:
1797:
1793:
1789:
1786:
1784:
1781:
1779:
1776:
1775:
1773:
1769:
1764:
1754:
1751:
1749:
1746:
1744:
1741:
1739:
1736:
1734:
1731:
1729:
1726:
1724:
1721:
1719:
1716:
1714:
1711:
1709:
1706:
1704:
1701:
1699:
1696:
1694:
1691:
1690:
1688:
1682:
1676:
1675:
1671:
1669:
1668:
1664:
1663:
1660:
1656:
1649:
1644:
1642:
1637:
1635:
1630:
1629:
1626:
1619:
1616:
1614:
1610:
1607:
1604:
1601:
1598:
1597:
1591:
1588:
1586:
1583:
1582:
1577:
1572:
1545:
1541:
1537:
1533:
1529:
1525:
1521:
1518:(6): 927–38.
1517:
1513:
1506:
1499:
1491:
1487:
1483:
1479:
1475:
1471:
1467:
1463:
1456:
1449:
1430:
1426:
1422:
1418:
1414:
1409:
1404:
1401:(3): 367–82.
1400:
1396:
1389:
1382:
1364:
1360:
1356:
1352:
1348:
1344:
1337:
1330:
1315:
1308:
1300:
1294:
1286:
1282:
1278:
1274:
1269:
1264:
1260:
1256:
1252:
1248:
1241:
1222:
1218:
1214:
1210:
1206:
1199:
1192:
1184:
1180:
1176:
1172:
1167:
1162:
1158:
1154:
1150:
1146:
1141:
1136:
1132:
1128:
1124:
1120:
1113:
1105:
1101:
1097:
1093:
1089:
1085:
1081:
1077:
1070:
1063:
1058:
1053:
1049:
1045:
1041:
1034:
1026:
1022:
1018:
1014:
1009:
1004:
1000:
996:
992:
985:
977:
973:
968:
963:
959:
955:
951:
947:
943:
936:
920:
916:
910:
906:
905:
897:
882:
878:
874:
868:
853:
849:
842:
840:
838:
836:
831:
821:
818:
816:
813:
811:
808:
806:
803:
801:
798:
795:
792:
790:
787:
785:
782:
779:
776:
774:
771:
770:
756:
751:
747:
740:
735:
725:
720:
710:
705:
698:
693:
689:
682:
677:
670:
665:
657:
652:
648:
644:
640:
636:
632:
628:
621:
616:
608:
603:
602:
596:
593:
588:
586:
582:
578:
573:
568:
565:
561:
556:
553:
548:
546:
542:
536:
533:
529:
523:
519:
509:
505:
501:
497:
495:
491:
481:
478:
472:
462:
460:
456:
452:
447:
443:
437:
427:
425:
421:
417:
413:
412:fluorochromes
409:
404:
402:
398:
394:
390:
386:
382:
378:
374:
370:
366:
362:
358:
354:
350:
339:
337:
333:
329:
325:
316:
312:
308:
305:stained with
304:
301:
296:
274:
272:
248:
246:
242:
238:
234:
230:
226:
222:
218:
214:
210:
206:
202:
201:halogen lamps
195:Light sources
192:
190:
186:
182:
178:
177:life sciences
169:
160:
158:
154:
148:
146:
142:
138:
134:
130:
126:
122:
118:
114:
110:
100:
98:
95:, which uses
94:
89:
85:
81:
77:
73:
69:
65:
61:
34:
30:
19:
1908:Cell imaging
1903:Fluorescence
1879:
1867:
1796:Three-photon
1777:
1672:
1665:
1575:
1551:. Retrieved
1544:the original
1515:
1511:
1498:
1465:
1461:
1448:
1436:. Retrieved
1429:the original
1398:
1394:
1381:
1370:, retrieved
1363:the original
1342:
1329:
1317:. Retrieved
1307:
1293:cite journal
1250:
1246:
1240:
1228:. Retrieved
1221:the original
1211:(1): 31–44.
1208:
1204:
1191:
1130:
1126:
1112:
1079:
1075:
1069:
1043:
1039:
1033:
998:
994:
984:
952:: 993–1016.
949:
945:
935:
923:. Retrieved
919:the original
903:
896:
886:28 September
884:. Retrieved
876:
867:
857:28 September
855:. Retrieved
851:
815:Stokes shift
639:microtubules
595:resolution.
589:
569:
557:
549:
537:
525:
506:
502:
498:
487:
474:
459:microtubules
439:
420:Alexa Fluors
408:fluorophores
405:
388:
385:fluorophores
349:nucleic acid
345:
320:
298:A sample of
270:
268:
198:
185:fluorescence
174:
149:
113:fluorophores
106:
68:fluorescence
59:
57:
29:
1918:Microscopes
1319:16 February
1133:(1): 6256.
925:17 December
643:fluorophore
577:Vertico SMI
550:Integrated
496:chemicals.
484:Limitations
461:in a cell.
424:DyLight 488
416:fluorescein
143:), and the
80:attenuation
1897:Categories
1792:Two-photon
1667:Microscope
1140:2002.08361
1046:: e26066.
827:References
805:Microscope
647:CCD camera
532:Ernst Abbe
516:See also:
469:See also:
393:cell walls
375:fibers in
369:phalloidin
324:expression
307:SYBR green
183:lens. The
109:wavelength
84:absorption
76:reflection
72:scattering
66:that uses
1553:12 August
1438:12 August
1372:12 August
1263:CiteSeerX
1230:12 August
1183:212725023
1104:215619998
613:function.
494:scavenger
397:cellulose
389:denatured
377:mammalian
271:Corethron
241:microlens
181:objective
103:Principle
88:inorganic
1869:Category
1609:Archived
1540:18162278
1532:19548231
1490:22042676
1482:17948007
1425:22811346
1417:18461478
1359:49339042
1285:19829598
1175:33288761
1121:(2020).
1096:32270489
1025:13812696
1017:22392641
976:19489737
766:See also
746:Palladin
581:blinking
477:genetics
442:antibody
414:such as
336:proteins
237:dichroic
211:with an
159:(TIRF).
155:and the
1881:Commons
1255:Bibcode
1166:7721808
1145:Bibcode
967:2835776
690:(FISH))
633:called
631:protein
564:RESOLFT
455:tubulin
446:antigen
444:to its
357:Hoechst
311:cuvette
300:herring
131:), the
1743:Sarfus
1573:about
1538:
1530:
1488:
1480:
1423:
1415:
1357:
1283:
1265:
1217:713859
1215:
1181:
1173:
1163:
1102:
1094:
1023:
1015:
974:
964:
911:
635:INCENP
401:pectin
365:nuclei
217:lasers
135:, the
129:lasers
78:, and
62:is an
1753:Raman
1547:(PDF)
1536:S2CID
1508:(PDF)
1486:S2CID
1458:(PDF)
1432:(PDF)
1421:S2CID
1391:(PDF)
1366:(PDF)
1355:S2CID
1339:(PDF)
1224:(PDF)
1201:(PDF)
1179:S2CID
1135:arXiv
1100:S2CID
1040:eLife
1021:S2CID
796:(GFP)
662:cells
422:, or
373:actin
326:of a
309:in a
303:sperm
1555:2013
1528:PMID
1478:PMID
1440:2013
1413:PMID
1374:2013
1321:2017
1299:link
1281:PMID
1232:2013
1213:PMID
1171:PMID
1092:PMID
1013:PMID
972:PMID
927:2017
909:ISBN
888:2008
859:2008
520:and
355:and
353:DAPI
231:and
225:LEDs
139:(or
127:and
125:LEDs
1520:doi
1470:doi
1403:doi
1347:doi
1273:doi
1161:PMC
1153:doi
1084:doi
1080:278
1048:doi
1003:doi
962:PMC
954:doi
627:DNA
410:or
399:or
361:DNA
315:DNA
273:sp.
207:or
119:or
82:or
1899::
1794:,
1534:.
1526:.
1514:.
1510:.
1484:.
1476:.
1464:.
1460:.
1419:.
1411:.
1399:16
1397:.
1393:.
1353:,
1295:}}
1291:{{
1279:.
1271:.
1261:.
1251:19
1249:.
1209:81
1207:.
1203:.
1177:.
1169:.
1159:.
1151:.
1143:.
1131:11
1129:.
1125:.
1098:.
1090:.
1078:.
1054:.
1042:,
1019:.
1011:.
999:81
997:.
993:.
970:.
960:.
950:78
948:.
944:.
879:.
875:.
850:.
834:^
418:,
219:,
215:,
74:,
58:A
1798:)
1790:(
1647:e
1640:t
1633:v
1557:.
1522::
1516:4
1492:.
1472::
1466:2
1442:.
1405::
1349::
1323:.
1301:)
1287:.
1275::
1257::
1234:.
1185:.
1155::
1147::
1137::
1106:.
1086::
1064:.
1050::
1044:6
1027:.
1005::
978:.
956::
929:.
890:.
861:.
748:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.