262:
concentration" of the reagents. This is understood when considering how increases in concentration leads to increases in reaction rate: essentially when the reactants are more concentrated, they collide more often and so react more often. In enzyme catalysis, the binding of the reagents to the enzyme restricts the conformational space of the reactants, holding them in the 'proper orientation' and close to each other, so that they collide more frequently, and with the correct geometry, to facilitate the desired reaction. The "effective concentration" is the concentration the reactant would have to be, free in solution, to experiences the same collisional frequency. Often such theoretical effective concentrations are unphysical and impossible to realize in reality – which is a testament to the great catalytic power of many enzymes, with massive rate increases over the uncatalyzed state.
145:
335:
lowering the barrier. A key feature of enzyme catalysis over many non-biological catalysis, is that both acid and base catalysis can be combined in the same reaction. In many abiotic systems, acids (large ) or bases ( large concentration H+ sinks, or species with electron pairs) can increase the rate of the reaction; but of course the environment can only have one overall pH (measure of acidity or basicity (alkalinity)). However, since enzymes are large molecules, they can position both acid groups and basic groups in their active site to interact with their substrates, and employ both modes independent of the bulk pH.
185:
2321:
1480:
1389:
22:
3140:
3076:
124:) increases the fraction of reactant molecules that can overcome this barrier and form the product. An important principle is that since they only reduce energy barriers between products and reactants, enzymes always catalyze reactions in both directions, and cannot drive a reaction forward or affect the equilibrium position – only the speed with which is it achieved. As with other catalysts, the enzyme is not consumed or changed by the reaction (as a substrate is) but is recycled such that a single enzyme performs many rounds of catalysis.
204:
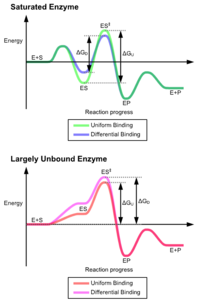
effect of uniform binding increases both substrate and transition state binding affinity, while differential binding increases only transition state binding affinity. Both are used by enzymes and have been evolutionarily chosen to minimize the activation energy of the reaction. Enzymes that are saturated, that is, have a high affinity substrate binding, require differential binding to reduce the energy of activation, whereas small substrate unbound enzymes may use either differential or uniform binding.
996:) due to the changes in the free energy content of every molecule, whether S or P, in water solution. This approach is in accordance with the following mechanism of muscle contraction. The final step of ATP hydrolysis in skeletal muscle is the product release caused by the association of myosin heads with actin. The closing of the actin-binding cleft during the association reaction is structurally coupled with the opening of the nucleotide-binding pocket on the myosin active site.
624:(TPP) to form covalent intermediates with reactant molecules. Such covalent intermediates function to reduce the energy of later transition states, similar to how covalent intermediates formed with active site amino acid residues allow stabilization, but the capabilities of cofactors allow enzymes to carryout reactions that amino acid side residues alone could not. Enzymes utilizing such cofactors include the PLP-dependent enzyme
3272:
24:
28:
27:
23:
29:
523:
distributions about the active sites are arranged so as to stabilize the transition states of the catalyzed reactions. In several enzymes, these charge distributions apparently serve to guide polar substrates toward their binding sites so that the rates of these enzymatic reactions are greater than their apparent diffusion-controlled limits.
516:
that the ionic transition states are stabilized by fixed dipoles. This is very different from transition state stabilization in water, where the water molecules must pay with "reorganization energy". In order to stabilize ionic and charged states. Thus, the catalysis is associated with the fact that the enzyme polar groups are preorganized
464:
catalyzed significantly, since it is not the rate determining barrier. Note that in the example shown, the histidine conjugate acid acts as a general acid catalyst for the subsequent loss of the amine from a tetrahedral intermediate. Evidence supporting this proposed mechanism (Figure 4 in Ref. 13) has, however been controverted.
26:
261:
of the reactants and thus makes addition or transfer reactions less unfavorable, since a reduction in the overall entropy when two reactants become a single product. However this is a general effect and is seen in non-addition or transfer reactions where it occurs due to an increase in the "effective
207:
These effects have led to most proteins using the differential binding mechanism to reduce the energy of activation, so most substrates have high affinity for the enzyme while in the transition state. Differential binding is carried out by the induced fit mechanism – the substrate first binds weakly,
648:
A metal ion in the active site participates in catalysis by coordinating charge stabilization and shielding. Because of a metal's positive charge, only negative charges can be stabilized through metal ions. However, metal ions are advantageous in biological catalysis because they are not affected by
455:
The initial step of the serine protease catalytic mechanism involves the histidine of the active site accepting a proton from the serine residue. This prepares the serine as a nucleophile to attack the amide bond of the substrate. This mechanism includes donation of a proton from serine (a base, pKa
203:
The advantages of the induced fit mechanism arise due to the stabilizing effect of strong enzyme binding. There are two different mechanisms of substrate binding: uniform binding, which has strong substrate binding, and differential binding, which has strong transition state binding. The stabilizing
640:
proposal of a covalent catalysis (where the barrier is lower than the corresponding barrier in solution) would require, for example, a partial covalent bond to the transition state by an enzyme group (e.g., a very strong hydrogen bond), and such effects do not contribute significantly to catalysis.
639:
for the reaction (via to the covalent intermediate) and so is distinct from true catalysis. For example, the energetics of the covalent bond to the serine molecule in chymotrypsin should be compared to the well-understood covalent bond to the nucleophile in the uncatalyzed solution reaction. A true
346:
groups, or to stabilize leaving groups. Many amino acids with acidic or basic groups are this employed in the active site, such as the glutamic and aspartic acid, histidine, cystine, tyrosine, lysine and arginine, as well as serine and threonine. In addition, the peptide backbone, with carbonyl and
661:
This is the principal effect of induced fit binding, where the affinity of the enzyme to the transition state is greater than to the substrate itself. This induces structural rearrangements which strain substrate bonds into a position closer to the conformation of the transition state, so lowering
522:
Binding of substrate usually excludes water from the active site, thereby lowering the local dielectric constant to that of an organic solvent. This strengthens the electrostatic interactions between the charged/polar substrates and the active sites. In addition, studies have shown that the charge
515:
Systematic computer simulation studies established that electrostatic effects give, by far, the largest contribution to catalysis. It can increase the rate of reaction by a factor of up to 10. In particular, it has been found that enzyme provides an environment which is more polar than water, and
463:
The modification of the pKa's is a pure part of the electrostatic mechanism. The catalytic effect of the above example is mainly associated with the reduction of the pKa of the oxyanion and the increase in the pKa of the histidine, while the proton transfer from the serine to the histidine is not
1011:
leads to transformation of a significant part of the free energy of ATP hydrolysis into the kinetic energy of the solvated phosphate, producing active streaming. This assumption of a local mechano-chemical transduction is in accord with Tirosh's mechanism of muscle contraction, where the muscle
729:
Quantum tunneling does not appear to provide a major catalytic advantage, since the tunneling contributions are similar in the catalyzed and the uncatalyzed reactions in solution. However, the tunneling contribution (typically enhancing rate constants by a factor of ~1000 compared to the rate of
572:
with residues in the enzyme active site or with a cofactor. This adds an additional covalent intermediate to the reaction, and helps to reduce the energy of later transition states of the reaction. The covalent bond must, at a later stage in the reaction, be broken to regenerate the enzyme. This
334:
may donate and accept protons in order to stabilize developing charges in the transition state. This is related to the overall principle of catalysis, that of reducing energy barriers, since in general transition states are high energy states, and by stabilizing them this high energy is reduced,
94:
is vital since many but not all metabolically essential reactions have very low rates when uncatalysed. One driver of protein evolution is the optimization of such catalytic activities, although only the most crucial enzymes operate near catalytic efficiency limits, and many enzymes are far from
870:
The crucial point for the verification of the present approach is that the catalyst must be a complex of the enzyme with the transfer group of the reaction. This chemical aspect is supported by the well-studied mechanisms of the several enzymatic reactions. Consider the reaction of peptide bond
311:
However, the situation might be more complex, since modern computational studies have established that traditional examples of proximity effects cannot be related directly to enzyme entropic effects. Also, the original entropic proposal has been found to largely overestimate the contribution of
702:
The substrate, on binding, is distorted from the half chair conformation of the hexose ring (because of the steric hindrance with amino acids of the protein forcing the equatorial c6 to be in the axial position) into the chair conformation, which is similar in shape to the transition state.
999:
Notably, the final steps of ATP hydrolysis include the fast release of phosphate and the slow release of ADP. The release of a phosphate anion from bound ADP anion into water solution may be considered as an exergonic reaction because the phosphate anion has low molecular mass.
856:
of the active enzyme appears in the product due to possibility of the exchange reaction inside enzyme to avoid both electrostatic inhibition and repulsion of atoms. So we represent the active enzyme as a powerful reactant of the enzymatic reaction. The reaction
127:
Enzymes are often highly specific and act on only certain substrates. Some enzymes are absolutely specific meaning that they act on only one substrate, while others show group specificity and can act on similar but not identical chemical groups such as the
718:). Some enzymes operate with kinetics which are faster than what would be predicted by the classical ΔG. In "through the barrier" models, a proton or an electron can tunnel through activation barriers. Quantum tunneling for protons has been observed in
963:) is the deacylation step. It is important to note that the group H+, initially found on the enzyme, but not in water, appears in the product before the step of hydrolysis, therefore it may be considered as an additional group of the enzymatic reaction.
972:) shows that the enzyme acts as a powerful reactant of the reaction. According to the proposed concept, the H transport from the enzyme promotes the first reactant conversion, breakdown of the first initial chemical bond (between groups P
983:
The proposed chemical mechanism does not depend on the concentration of the substrates or products in the medium. However, a shift in their concentration mainly causes free energy changes in the first and final steps of the reactions
25:
741:
The binding energy of the enzyme-substrate complex cannot be considered as an external energy which is necessary for the substrate activation. The enzyme of high energy content may firstly transfer some specific energetic group
1520:
Kuhn B, Kollman PA (2000). "QM-FE and
Molecular Dynamics Calculations on Catechol O-Methyltransferase: Free Energy of Activation in the Enzyme and in Aqueous Solution and Regioselectivity of the Enzyme-Catalyzed Reaction".
2929:
White HD, Belknap B, Webb MR (September 1997). "Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate".
225:
in the uncatalyzed reaction in water (without the enzyme). The induced fit only suggests that the barrier is lower in the closed form of the enzyme but does not tell us what the reason for the barrier reduction is.
665:
However, the strain effect is, in fact, a ground state destabilization effect, rather than transition state stabilization effect. Furthermore, enzymes are very flexible and they cannot apply large strain effect.
284:
1493:
Stanton RV, Peräkylä M, Bakowies D, Kollman PA (1998). "Combined ab initio and Free Energy
Calculations To Study Reactions in Enzymes and Solution: Amide Hydrolysis in Trypsin and Aqueous Solution".
548:
2965:
Tirosh R, Low WZ, Oplatka A (March 1990). "Translational motion of actin filaments in the presence of heavy meromyosin and MgATP as measured by
Doppler broadening of laser light scattering".
447:
695:
730:
reaction for the classical 'over the barrier' route) is likely crucial to the viability of biological organisms. This emphasizes the general importance of tunneling reactions in biology.
196:
interaction is the induced fit model. This model proposes that the initial interaction between enzyme and substrate is relatively weak, but that these weak interactions rapidly induce
750:
from the second bound reactant (or from the second group of the single reactant) must be transferred to active site to finish substrate conversion to product and enzyme regeneration.
2186:
Fife TH, Przystas TJ (1 February 1985). "Divalent metal ion catalysis in the hydrolysis of esters of picolinic acid. Metal ion promoted hydroxide ion and water catalyzed reactions".
245:
close to the chemical bonds in the substrate that will be altered in the reaction. After binding takes place, one or more mechanisms of catalysis lowers the energy of the reaction's
2292:
Warshel A, Levitt M (May 1976). "Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme".
257:
Enzyme-substrate interactions align the reactive chemical groups and hold them close together in an optimal geometry, which increases the rate of the reaction. This reduces the
249:, by providing an alternative chemical pathway for the reaction. There are six possible mechanisms of "over the barrier" catalysis as well as a "through the barrier" mechanism:
2413:
Olsson MH, Siegbahn PE, Warshel A (March 2004). "Simulations of the large kinetic isotope effect and the temperature dependence of the hydrogen atom transfer in lipoxygenase".
2451:
Masgrau L, Roujeinikova A, Johannissen LO, Hothi P, Basran J, Ranaghan KE, et al. (April 2006). "Atomic description of an enzyme reaction dominated by proton tunneling".
1754:
Fersht AR, Requena Y (December 1971). "Mechanism of the -chymotrypsin-catalyzed hydrolysis of amides. pH dependence of k c and K m . Kinetic detection of an intermediate".
366:
Many reaction mechanisms involving acid/base catalysis assume a substantially altered pKa. This alteration of pKa is possible through the local environment of the residue.
419:
pKa can also be influenced significantly by the surrounding environment, to the extent that residues which are basic in solution may act as proton donors, and vice versa.
211:
It is important to clarify, however, that the induced fit concept cannot be used to rationalize catalysis. That is, the chemical catalysis is defined as the reduction of
867:
remains inside enzyme. This approach as idea had formerly proposed relying on the hypothetical extremely high enzymatic conversions (catalytically perfect enzyme).
3438:
2749:
Hengge AC, Stein RL (January 2004). "Role of protein conformational mobility in enzyme catalysis: acylation of alpha-chymotrypsin by specific peptide substrates".
871:
hydrolysis catalyzed by a pure protein α-chymotrypsin (an enzyme acting without a cofactor), which is a well-studied member of the serine proteases family, see.
519:
The magnitude of the electrostatic field exerted by an enzyme's active site has been shown to be highly correlated with the enzyme's catalytic rate enhancement.
2819:
Holmes KC, Angert I, Kull FJ, Jahn W, Schröder RR (September 2003). "Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide".
3443:
59:. Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the
208:
then the enzyme changes conformation increasing the affinity to the transition state and stabilizing it, so reducing the activation energy to reach it.
82:). Many cofactors are vitamins, and their role as vitamins is directly linked to their use in the catalysis of biological process within metabolism.
2648:
Volkenshtein MV, Dogonadze RR, Madumarov AK, Urushadze ZD, Kharkats Yu I (1973). "Electronic and
Conformational Interactions in Enzyme Catalysis.".
714:
These traditional "over the barrier" mechanisms have been challenged in some cases by models and observations of "through the barrier" mechanisms (
3304:
669:
In addition to bond strain in the substrate, bond strain may also be induced within the enzyme itself to activate residues in the active site.
1145:
in the 2010s led to the observation that the movement of untethered enzymes increases with increasing substrate concentration and increasing
2359:
Garcia-Viloca M, Gao J, Karplus M, Truhlar DG (January 2004). "How enzymes work: analysis by modern rate theory and computer simulations".
1548:
Bruice TC, Lightstone FC (1999). "Ground State and
Transition State Contributions to the Rates of Intramolecular and Enzymatic Reactions".
2014:
2872:"ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle"
2586:
Olsson MH, Parson WW, Warshel A (May 2006). "Dynamical contributions to enzyme catalysis: critical tests of a popular hypothesis".
99:, orbital steering, entropic restriction, orientation effects (i.e. lock and key catalysis), as well as motional effects involving
1789:
Zeeberg B, Caswell M, Caplow M (April 1973). "Concerning a reported change in rate-determining step in chymotrypsin catalysis".
1634:
Warshel A, Parson WW (November 2001). "Dynamics of biochemical and biophysical reactions: insight from computer simulations".
3216:
2343:
2276:
2170:
2086:
2055:
1411:
1317:
1173:
556:
The tetrahedral intermediate is stabilised by a partial ionic bond between the Zn ion and the negative charge on the oxygen.
2102:
Piccirilli JA, Vyle JS, Caruthers MH, Cech TR (January 1993). "Metal ion catalysis in the
Tetrahymena ribozyme reaction".
229:
Induced fit may be beneficial to the fidelity of molecular recognition in the presence of competition and noise via the
3297:
74:. Enzymes often also incorporate non-protein components, such as metal ions or specialized organic molecules known as
3550:
3276:
2004:
Toney, M. D. "Reaction specificity in pyridoxal enzymes." Archives of biochemistry and biophysics (2005) 433: 279-287
512:. Metal ions are particularly effective and can reduce the pKa of water enough to make it an effective nucleophile.
3476:
1684:
Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MH (August 2006). "Electrostatic basis for enzyme catalysis".
2621:
Vol'kenshtein MV, Dogonadze RR, Madumarov AK, Urushadze ZD, Kharkats YI (1972). "The theory of enzyme catalysis".
1429:"Conformational proofreading: the impact of conformational changes on the specificity of molecular recognition"
635:
Rather than lowering the activation energy for a reaction pathway, covalent catalysis provides an alternative
3706:
3461:
3290:
2706:
Fogel AG (August 1982). "Cooperativity of enzymatic reactions and molecular aspects of energy transduction".
1849:"On the Theory of Electron-Transfer Reactions. VI. Unified Treatment for Homogeneous and Electrode Reactions"
1577:"Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect"
1239:
1146:
3471:
980:). The step of hydrolysis leads to a breakdown of the second chemical bond and regeneration of the enzyme.
723:
230:
2502:
Hwang JK, Warshel A (1996). "How important are quantum mechanical nuclear motions in enzyme catalysis".
3314:
1142:
1126:
1122:
1050:
1046:
1028:
636:
356:
746:
from catalytic site of the enzyme to the final place of the first bound reactant, then another group X
3807:
3794:
3781:
3768:
3755:
3742:
3729:
3691:
3433:
1118:
3139:
3075:
3002:"Ballistic protons and microwave-induced water solutions (solitons) in bioenergetic transformations"
2784:
Lymn RW, Taylor EW (December 1971). "Mechanism of adenosine triphosphate hydrolysis by actomyosin".
2320:
1479:
1388:
662:
the energy difference between the substrate and transition state and helping catalyze the reaction.
3701:
3655:
3598:
3359:
1261:"At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?"
1111:
1065:
1032:
321:
193:
1719:
Warshel A, Naray-Szabo G, Sussman F, Hwang JK (May 1989). "How do serine proteases really work?".
1020:
In reality, most enzyme mechanisms involve a combination of several different types of catalysis.
184:
3603:
3209:
Structure and
Mechanism in Protein Science : A Guide to Enzyme Catalysis and Protein Folding
3089:
Riedel C, Gabizon R, Wilson CA, Hamadani K, Tsekouras K, Marqusee S, et al. (January 2015).
2153:
Bender ML (1 January 1962). "Metal Ion
Catalysis of Nucleophilic Organic Reactions in Solution".
613:
505:
96:
75:
1333:
629:
625:
621:
476:(or partial ionic charge interactions) with the intermediate. These bonds can either come from
153:
79:
2223:"Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences"
472:
Stabilization of charged transition states can also be by residues in the active site forming
3624:
3543:
3497:
3411:
3328:
650:
197:
148:
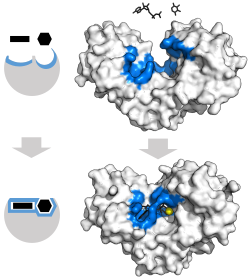
Enzyme changes shape by induced fit upon substrate binding to form enzyme-substrate complex.
110:
2078:
2072:
2047:
2041:
109:
in that the crucial factor is a reduction of energy barrier(s) separating the reactants (or
3696:
3507:
3492:
3343:
3102:
2883:
2828:
2542:
2460:
2368:
2111:
1961:
1902:
1863:
1848:
1588:
1440:
1349:
456:
14) to histidine (an acid, pKa 6), made possible due to the local environment of the bases.
71:
70:
are made predominantly of proteins, either a single protein chain or many such chains in a
2018:
8:
3660:
3405:
3282:
2663:
Foigel AG (June 2011). "Is the enzyme a powerful reactant of the biochemical reaction?".
1234:
617:
283:
3106:
2887:
2832:
2546:
2464:
2372:
2115:
1965:
1906:
1867:
1592:
1444:
1353:
105:
Mechanisms of enzyme catalysis vary, but are all similar in principle to other types of
3834:
3593:
3466:
3364:
3254:
3227:
3179:
3154:
3123:
3090:
3059:
3034:
2852:
2731:
2688:
2568:
2484:
2392:
2268:
2135:
1982:
1949:
1659:
1463:
1428:
1285:
1260:
1229:
1209:
1012:
force derives from an integrated action of active streaming created by ATP hydrolysis.
1003:
Thus, we arrive at the conclusion that the primary release of the inorganic phosphate H
2906:
2871:
1925:
1890:
1611:
1576:
1372:
1337:
3453:
3212:
3184:
3128:
3064:
2982:
2978:
2947:
2911:
2844:
2801:
2766:
2723:
2680:
2630:
2603:
2560:
2476:
2430:
2384:
2339:
2309:
2305:
2272:
2242:
2238:
2203:
2166:
2127:
2082:
2051:
1987:
1950:"Extreme electric fields power catalysis in the active site of ketosteroid isomerase"
1930:
1829:
1806:
1771:
1736:
1701:
1651:
1616:
1468:
1407:
1377:
1313:
1290:
1169:) can be calculated by using bond changes, reaction centres or substructure metrics (
715:
166:
114:
2735:
2692:
2488:
2396:
1663:
733:
In 1971-1972 the first quantum-mechanical model of enzyme catalysis was formulated.
132:
in different molecules. Many enzymes have stereochemical specificity and act on one
3829:
3639:
3634:
3536:
3502:
3428:
3338:
3249:
3239:
3174:
3166:
3118:
3110:
3054:
3046:
3013:
2974:
2939:
2901:
2891:
2856:
2836:
2793:
2758:
2715:
2672:
2595:
2550:
2511:
2468:
2422:
2376:
2301:
2234:
2195:
2158:
2139:
2119:
1977:
1969:
1920:
1910:
1871:
1798:
1763:
1728:
1693:
1643:
1606:
1596:
1557:
1530:
1502:
1458:
1448:
1367:
1357:
1280:
1272:
1214:
1199:
1150:
1096:
537:
481:
331:
246:
100:
2572:
2157:. Advances in Chemistry. Vol. 37. American Chemical Society. pp. 19–36.
1170:
957:) includes the formation of a covalent acyl-enzyme intermediate. The second step (
3686:
3670:
3583:
3512:
3400:
3380:
3354:
3091:"The heat released during catalytic turnover enhances the diffusion of an enzyme"
1453:
1204:
1189:
1177:
1088:
1072:
574:
437:
433:
133:
2967:
Biochimica et
Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology
3724:
3665:
2876:
Proceedings of the
National Academy of Sciences of the United States of America
2260:
2162:
1895:
Proceedings of the National Academy of Sciences of the United States of America
1581:
Proceedings of the National Academy of Sciences of the United States of America
1342:
Proceedings of the National Academy of Sciences of the United States of America
1154:
91:
34:
2676:
1647:
3823:
3629:
3588:
3395:
2207:
1833:
1092:
874:
We present the experimental results for this reaction as two chemical steps:
569:
547:
501:
497:
44:
2620:
2472:
2380:
1973:
1915:
694:
446:
3578:
3517:
3390:
3333:
3244:
3188:
3153:
Rahman SA, Cuesta SM, Furnham N, Holliday GL, Thornton JM (February 2014).
3132:
3068:
2848:
2770:
2684:
2607:
2564:
2480:
2434:
2388:
1991:
1705:
1655:
1601:
1472:
1381:
1294:
1219:
1194:
1166:
589:, where an acyl-enzyme intermediate is formed. An alternative mechanism is
582:
343:
129:
87:
2986:
2951:
2915:
2896:
2805:
2727:
2634:
2246:
2131:
1810:
1775:
1740:
1620:
276:
Similar reactions will occur far faster if the reaction is intramolecular.
3802:
3737:
3573:
2313:
2267:. McGraw-Hill series in advanced chemistry (reprint ed.). New York:
1934:
1362:
753:
We can present the whole enzymatic reaction as a two coupling reactions:
671:
590:
525:
473:
421:
339:
264:
242:
144:
60:
56:
3114:
2840:
2797:
2199:
1802:
1767:
1732:
1114:
1068:
3385:
3170:
3155:"EC-BLAST: a tool to automatically search and compare enzyme reactions"
2719:
2647:
1276:
1035:
719:
606:
485:
149:
3050:
3033:
Muddana HS, Sengupta S, Mallouk TE, Sen A, Butler PJ (February 2010).
2943:
2762:
2599:
2515:
2426:
1875:
1697:
1561:
1534:
1506:
649:
changes in pH. Metal ions can also act to ionize water by acting as a
176:
170:
3776:
3750:
3018:
3001:
2123:
352:
161:
106:
83:
2555:
2530:
2222:
1338:"Application of a Theory of Enzyme Specificity to Protein Synthesis"
2450:
1107:
1084:
684:
602:
578:
493:
863:) shows incomplete conversion of the substrate because its group X
683:
Substrate, bound substrate, and transition state conformations of
241:
These conformational changes also bring catalytic residues in the
3228:"Quantum catalysis in enzymes—beyond the transition state theory"
1076:
1061:
586:
348:
293:
258:
48:
3789:
3559:
3271:
1080:
1043:
1039:
598:
489:
157:
152:
has a large induced fit motion that closes over the substrates
67:
52:
2358:
1718:
1492:
568:
Covalent catalysis involves the substrate forming a transient
236:
3763:
1223:
1222:, whose ubiquity despite their catalytic inactivity suggests
594:
338:
Often general acid or base catalysis is employed to activate
653:. Metal ions may also be agents of oxidation and reduction.
3152:
3088:
2101:
2015:"Micronutrient Information Center, Oregon State University"
1130:
509:
477:
327:
3528:
3312:
1157:, resulting in a "recoil effect that propels the enzyme".
3032:
2336:
Fundamentals of Biochemistry: Life at the Molecular Level
2869:
1149:. Subsequent observations suggest that this increase in
2870:
Siemankowski RF, Wiseman MO, White HD (February 1985).
360:
95:
optimal. Important factors in enzyme catalysis include
3035:"Substrate catalysis enhances single-enzyme diffusion"
1683:
1038:) catalyses the reversible interconversion of the two
2412:
2818:
2650:
Konformatsionnie Izmenenia Biopolimerov v Rastvorakh
1788:
1153:
is driven by transient displacement of the enzyme's
296:
in the intramolecular reaction can be estimated as k
2652:. Moscow: Nauka Publishing House. pp. 153–157.
2585:
1015:
355:are very commonly involved, since they both have a
218:(when the system is already in the ES) relative to
363:and can therefore both accept and donate protons.
3444:Ultraviolet–visible spectroscopy of stereoisomers
160:. Binding sites in blue, substrates in black and
3821:
2964:
2928:
1947:
1547:
1401:
1310:Physical Chemistry with Biological Applications
1258:
3225:
1948:Fried SD, Bagchi S, Boxer SG (December 2014).
1427:Savir Y, Tlusty T (May 2007). Scalas E (ed.).
3544:
3298:
2446:
2444:
2333:
2291:
1753:
1633:
1023:
315:
188:The different mechanisms of substrate binding
2501:
2185:
1941:
252:
3026:
2748:
2614:
1519:
1426:
237:Mechanisms of an alternative reaction route
3551:
3537:
3305:
3291:
3082:
2783:
2441:
1679:
1677:
1675:
1673:
1574:
467:
16:Catalysis of chemical reactions by enzymes
3253:
3243:
3178:
3122:
3058:
3017:
2905:
2895:
2554:
1981:
1924:
1914:
1610:
1600:
1462:
1452:
1371:
1361:
1284:
3039:Journal of the American Chemical Society
2415:Journal of the American Chemical Society
2220:
2188:Journal of the American Chemical Society
2070:
2064:
2039:
1823:
1791:Journal of the American Chemical Society
1756:Journal of the American Chemical Society
1332:
1165:Similarity between enzymatic reactions (
183:
143:
20:
2408:
2406:
2338:(Fourth ed.). Hoboken, NJ: Wiley.
1888:
1670:
1307:
1252:
951:are products. The first chemical step (
200:in the enzyme that strengthen binding.
3822:
3206:
2999:
2662:
2259:
2152:
2077:. John Wiley & Sons Inc. pp.
2046:. John Wiley & Sons Inc. pp.
2033:
1846:
1160:
1091:to perform covalent catalysis, and an
643:
113:) from the products. The reduction of
3532:
3286:
2705:
2265:Catalysis in Chemistry and Enzymology
1136:
563:
3226:Sutcliffe M, Munro A (August 2006).
3146:
2528:
2403:
909:
876:
800:
755:
709:
612:Some enzymes utilize non-amino acid
2708:Molecular and Cellular Biochemistry
2665:Molecular and Cellular Biochemistry
2531:"Enzymes: by chance, or by design?"
2227:Free Radical Biology & Medicine
1259:Kamerlin SC, Warshel A (May 2010).
1095:to stabilise charge-buildup on the
13:
3199:
2334:Voet D, Voet JG, Pratt CW (2013).
1575:Page MI, Jencks WP (August 1971).
1312:. Benjamin/Cummings. p. 427.
347:amide N groups is often employed.
326:Proton donors and acceptors, i.e.
312:orientation entropy to catalysis.
14:
3846:
3439:NMR spectroscopy of stereoisomers
3264:
1404:Modern Physical Organic Chemistry
394:Adjacent residues of like charge
192:The classic model for the enzyme-
3477:Diastereomeric recrystallization
3270:
3138:
3074:
2319:
2155:Reactions of Coordinated Ligands
1960:(6216). New York, N.Y.: 1510–4.
1891:"Energetics of enzyme catalysis"
1478:
1402:Anslyn EV, Dougherty DA (2006).
1387:
1016:Examples of catalytic mechanisms
736:
693:
546:
445:
282:
2993:
2958:
2922:
2863:
2812:
2777:
2742:
2699:
2656:
2641:
2579:
2522:
2495:
2352:
2327:
2285:
2253:
2214:
2179:
2146:
2095:
2007:
1998:
1882:
1840:
1817:
1782:
1747:
1712:
1636:Quarterly Reviews of Biophysics
1627:
601:residue, as seen in the enzyme
292:The effective concentration of
97:general acid and base catalysis
2221:Stadtman ER (1 January 1990).
1568:
1541:
1513:
1486:
1420:
1395:
1326:
1301:
846:It may be seen from reaction (
656:
139:
1:
1245:
1240:Time resolved crystallography
1117:) catalyses the breakdown of
628:and the TPP-dependent enzyme
573:mechanism is utilised by the
3472:Chiral column chromatography
3232:Philosophical Transactions B
2979:10.1016/0167-4838(90)90025-b
2306:10.1016/0022-2836(76)90311-9
2294:Journal of Molecular Biology
2239:10.1016/0891-5849(90)90006-5
1454:10.1371/journal.pone.0000468
1406:. University Science Books.
724:aromatic amine dehydrogenase
701:
692:
681:
554:
545:
535:
453:
444:
431:
290:
281:
274:
7:
3558:
1889:Warshel A (November 1978).
1183:
1102:
992:
986:
968:
959:
953:
929:
900:
859:
848:
836:
791:
231:conformational proofreading
10:
3853:
3434:Chiral derivatizing agents
3315:enantioselective synthesis
3211:. New York: W.H. Freeman.
2163:10.1021/ba-1963-0037.ch002
1127:dihydroxyacetone phosphate
1123:glyceraldehyde 3-phosphate
1056:
1051:glyceraldehyde 3-phosphate
1047:dihydroxyacetone phosphate
1029:Triose phosphate isomerase
1024:Triose phosphate isomerase
405:Salt bridge (and hydrogen
319:
316:Proton donors or acceptors
3715:
3707:Michaelis–Menten kinetics
3679:
3648:
3617:
3566:
3485:
3452:
3421:
3373:
3321:
2677:10.1007/s11010-011-0742-4
2529:Ball P (September 2004).
1828:. John Wiley & Sons.
1648:10.1017/s0033583501003730
1119:fructose 1,6-bisphosphate
593:formation using the free
253:Proximity and orientation
3599:Diffusion-limited enzyme
3360:Supramolecular chirality
2071:Voet D, Voet JG (2004).
2040:Voet D, Voet JG (2004).
1824:Voet D, Voet JG (2011).
383:Hydrophobic environment
322:Protein pKa calculations
2473:10.1126/science.1126002
2381:10.1126/science.1088172
1974:10.1126/science.1259802
1916:10.1073/pnas.75.11.5250
1143:single-molecule studies
468:Electrostatic catalysis
43:is the increase in the
3245:10.1098/rstb.2006.1879
2629:(3). Moscow: 347–353.
1602:10.1073/pnas.68.8.1678
630:pyruvate dehydrogenase
626:aspartate transaminase
622:thiamine pyrophosphate
198:conformational changes
189:
181:
154:adenosine triphosphate
80:adenosine triphosphate
37:
3692:Eadie–Hofstee diagram
3625:Allosteric regulation
3498:Chiral pool synthesis
3412:Diastereomeric excess
2897:10.1073/pnas.82.3.658
484:side chains found on
187:
164:cofactor in yellow. (
147:
88:biochemical reactions
72:multi-subunit complex
32:
3702:Lineweaver–Burk plot
3508:Asymmetric catalysis
3493:Asymmetric induction
3279:at Wikimedia Commons
1363:10.1073/pnas.44.2.98
966:Thus, the reaction (
3406:Enantiomeric excess
3238:(1472): 1291–1455.
3115:10.1038/nature14043
3107:2015Natur.517..227R
2938:(39): 11828–11836.
2888:1985PNAS...82..658S
2841:10.1038/nature02005
2833:2003Natur.425..423H
2798:10.1021/bi00801a004
2547:2004Natur.431..396B
2510:(47): 11745–11751.
2465:2006Sci...312..237M
2373:2004Sci...303..186G
2200:10.1021/ja00290a048
2116:1993Natur.361...85P
1966:2014Sci...346.1510F
1907:1978PNAS...75.5250W
1868:1965JChPh..43..679M
1803:10.1021/ja00789a081
1768:10.1021/ja00754a066
1733:10.1021/bi00435a001
1593:1971PNAS...68.1678P
1445:2007PLoSO...2..468S
1354:1958PNAS...44...98K
1308:Laidler KJ (1978).
1235:The Proteolysis Map
1176:30 May 2019 at the
1161:Reaction similarity
943:is a polypeptide, P
644:Metal ion catalysis
618:pyridoxal phosphate
577:of enzymes such as
540:catalytic mechanism
504:or come from metal
57:biological molecule
3661:Enzyme superfamily
3594:Enzyme promiscuity
3503:Chiral auxiliaries
3467:Kinetic resolution
3365:Inherent chirality
3350:-symmetric ligands
3171:10.1038/nmeth.2803
2720:10.1007/bf00241567
2269:Dover Publications
1847:Marcus RA (1965).
1277:10.1002/prot.22654
1230:Quantum tunnelling
1210:Enzyme promiscuity
1137:Enzyme diffusivity
918:+ H−O−H → EH + P
852:) that the group X
564:Covalent catalysis
190:
182:
107:chemical catalysis
38:
3817:
3816:
3526:
3525:
3462:Recrystallization
3454:Chiral resolution
3275:Media related to
3218:978-0-7167-3268-6
3207:Fersht A (1998).
3101:(7533): 227–230.
3051:10.1021/ja908773a
3000:Tirosh R (2006).
2944:10.1021/bi970540h
2827:(6956): 423–427.
2792:(25): 4617–4624.
2763:10.1021/bi030222k
2623:Molecular Biology
2600:10.1021/cr040427e
2541:(7007): 396–397.
2516:10.1021/ja962007f
2459:(5771): 237–241.
2427:10.1021/ja037233l
2367:(5655): 186–195.
2345:978-0-470-54784-7
2278:978-0-486-65460-7
2172:978-0-8412-0038-8
2088:978-0-471-25090-6
2057:978-0-471-25090-6
1901:(11): 5250–5254.
1876:10.1063/1.1696792
1762:(25): 7079–7087.
1698:10.1021/cr0503106
1562:10.1021/ar960131y
1535:10.1021/ja992218v
1529:(11): 2586–2596.
1507:10.1021/ja972723x
1501:(14): 3448–3457.
1413:978-1-891389-31-3
1336:(February 1958).
1319:978-0-8053-5680-9
1147:reaction enthalpy
1097:transition states
1087:residues using a
1079:substrates after
937:
936:
908:
907:
844:
843:
799:
798:
716:quantum tunneling
710:Quantum tunneling
707:
706:
677:
561:
560:
531:
461:
460:
427:
417:
416:
359:close to neutral
309:
308:
270:
136:but not another.
115:activation energy
33:Visualization of
30:
3842:
3697:Hanes–Woolf plot
3640:Enzyme activator
3635:Enzyme inhibitor
3609:Enzyme catalysis
3553:
3546:
3539:
3530:
3529:
3429:Optical rotation
3374:Chiral molecules
3339:Planar chirality
3307:
3300:
3293:
3284:
3283:
3277:Enzyme catalysis
3274:
3259:
3257:
3247:
3222:
3193:
3192:
3182:
3150:
3144:
3143:
3142:
3136:
3126:
3086:
3080:
3079:
3078:
3072:
3062:
3045:(7): 2110–2111.
3030:
3024:
3023:
3021:
3019:10.3390/i7090320
3006:Int. J. Mol. Sci
2997:
2991:
2990:
2962:
2956:
2955:
2926:
2920:
2919:
2909:
2899:
2867:
2861:
2860:
2816:
2810:
2809:
2781:
2775:
2774:
2746:
2740:
2739:
2703:
2697:
2696:
2660:
2654:
2653:
2645:
2639:
2638:
2618:
2612:
2611:
2594:(5): 1737–1756.
2588:Chemical Reviews
2583:
2577:
2576:
2558:
2526:
2520:
2519:
2504:J. Am. Chem. Soc
2499:
2493:
2492:
2448:
2439:
2438:
2421:(9): 2820–2828.
2410:
2401:
2400:
2356:
2350:
2349:
2331:
2325:
2324:
2323:
2317:
2289:
2283:
2282:
2257:
2251:
2250:
2218:
2212:
2211:
2194:(4): 1041–1047.
2183:
2177:
2176:
2150:
2144:
2143:
2124:10.1038/361085a0
2099:
2093:
2092:
2068:
2062:
2061:
2037:
2031:
2030:
2028:
2026:
2021:on 21 March 2015
2017:. Archived from
2011:
2005:
2002:
1996:
1995:
1985:
1945:
1939:
1938:
1928:
1918:
1886:
1880:
1879:
1853:
1844:
1838:
1837:
1821:
1815:
1814:
1797:(8): 2734–2735.
1786:
1780:
1779:
1751:
1745:
1744:
1727:(9): 3629–3637.
1716:
1710:
1709:
1692:(8): 3210–3235.
1686:Chemical Reviews
1681:
1668:
1667:
1631:
1625:
1624:
1614:
1604:
1587:(8): 1678–1683.
1572:
1566:
1565:
1545:
1539:
1538:
1523:J. Am. Chem. Soc
1517:
1511:
1510:
1495:J. Am. Chem. Soc
1490:
1484:
1483:
1482:
1476:
1466:
1456:
1424:
1418:
1417:
1399:
1393:
1392:
1391:
1385:
1375:
1365:
1330:
1324:
1323:
1305:
1299:
1298:
1288:
1271:(6): 1339–1375.
1256:
1215:Protein dynamics
1200:Enzyme inhibitor
1121:(F-1,6-BP) into
931:
922:
910:
902:
893:
877:
838:
829:
801:
793:
784:
756:
697:
675:
672:
550:
538:Carboxypeptidase
529:
526:
449:
425:
422:
407:bond) formation
369:
368:
286:
268:
265:
247:transition state
179:
173:
101:protein dynamics
41:Enzyme catalysis
31:
3852:
3851:
3845:
3844:
3843:
3841:
3840:
3839:
3820:
3819:
3818:
3813:
3725:Oxidoreductases
3711:
3687:Enzyme kinetics
3675:
3671:List of enzymes
3644:
3613:
3584:Catalytic triad
3562:
3557:
3527:
3522:
3513:Organocatalysis
3481:
3448:
3417:
3401:Racemic mixture
3369:
3355:Axial chirality
3349:
3322:Chirality types
3317:
3311:
3267:
3262:
3219:
3202:
3200:Further reading
3197:
3196:
3151:
3147:
3137:
3087:
3083:
3073:
3031:
3027:
2998:
2994:
2963:
2959:
2927:
2923:
2868:
2864:
2817:
2813:
2782:
2778:
2747:
2743:
2704:
2700:
2661:
2657:
2646:
2642:
2619:
2615:
2584:
2580:
2556:10.1038/431396a
2527:
2523:
2500:
2496:
2449:
2442:
2411:
2404:
2357:
2353:
2346:
2332:
2328:
2318:
2290:
2286:
2279:
2258:
2254:
2219:
2215:
2184:
2180:
2173:
2151:
2147:
2110:(6407): 85–88.
2100:
2096:
2089:
2069:
2065:
2058:
2038:
2034:
2024:
2022:
2013:
2012:
2008:
2003:
1999:
1946:
1942:
1887:
1883:
1851:
1845:
1841:
1822:
1818:
1787:
1783:
1752:
1748:
1717:
1713:
1682:
1671:
1632:
1628:
1573:
1569:
1546:
1542:
1518:
1514:
1491:
1487:
1477:
1425:
1421:
1414:
1400:
1396:
1386:
1331:
1327:
1320:
1306:
1302:
1257:
1253:
1248:
1205:Enzyme kinetics
1190:Catalytic triad
1186:
1178:Wayback Machine
1163:
1139:
1105:
1089:catalytic triad
1073:serine protease
1059:
1026:
1018:
1010:
1006:
979:
975:
950:
946:
942:
921:
917:
913:
892:
888:
884:
880:
866:
855:
828:
824:
820:
816:
812:
808:
804:
783:
779:
775:
771:
767:
763:
759:
749:
745:
739:
712:
659:
646:
575:catalytic triad
566:
470:
438:serine protease
434:Catalytic triad
406:
324:
318:
304:= 2 x 10 Molar.
303:
299:
255:
239:
224:
217:
175:
165:
142:
123:
21:
17:
12:
11:
5:
3850:
3849:
3838:
3837:
3832:
3815:
3814:
3812:
3811:
3798:
3785:
3772:
3759:
3746:
3733:
3719:
3717:
3713:
3712:
3710:
3709:
3704:
3699:
3694:
3689:
3683:
3681:
3677:
3676:
3674:
3673:
3668:
3663:
3658:
3652:
3650:
3649:Classification
3646:
3645:
3643:
3642:
3637:
3632:
3627:
3621:
3619:
3615:
3614:
3612:
3611:
3606:
3601:
3596:
3591:
3586:
3581:
3576:
3570:
3568:
3564:
3563:
3556:
3555:
3548:
3541:
3533:
3524:
3523:
3521:
3520:
3515:
3510:
3505:
3500:
3495:
3489:
3487:
3483:
3482:
3480:
3479:
3474:
3469:
3464:
3458:
3456:
3450:
3449:
3447:
3446:
3441:
3436:
3431:
3425:
3423:
3419:
3418:
3416:
3415:
3409:
3403:
3398:
3393:
3388:
3383:
3377:
3375:
3371:
3370:
3368:
3367:
3362:
3357:
3352:
3347:
3341:
3336:
3331:
3325:
3323:
3319:
3318:
3310:
3309:
3302:
3295:
3287:
3281:
3280:
3266:
3265:External links
3263:
3261:
3260:
3223:
3217:
3203:
3201:
3198:
3195:
3194:
3165:(2): 171–174.
3159:Nature Methods
3145:
3081:
3025:
3012:(9): 320–345.
2992:
2973:(3): 274–280.
2957:
2921:
2882:(3): 658–662.
2862:
2811:
2776:
2757:(3): 742–747.
2741:
2698:
2671:(1–2): 87–89.
2655:
2640:
2613:
2578:
2521:
2494:
2440:
2402:
2351:
2344:
2326:
2300:(2): 227–249.
2284:
2277:
2252:
2233:(4): 315–325.
2213:
2178:
2171:
2145:
2094:
2087:
2063:
2056:
2032:
2006:
1997:
1940:
1881:
1862:(2): 679–701.
1839:
1816:
1781:
1746:
1711:
1669:
1642:(4): 563–679.
1626:
1567:
1556:(2): 127–136.
1550:Acc. Chem. Res
1540:
1512:
1485:
1419:
1412:
1394:
1325:
1318:
1300:
1250:
1249:
1247:
1244:
1243:
1242:
1237:
1232:
1227:
1217:
1212:
1207:
1202:
1197:
1192:
1185:
1182:
1162:
1159:
1155:center of mass
1141:The advent of
1138:
1135:
1104:
1101:
1058:
1055:
1025:
1022:
1017:
1014:
1008:
1004:
977:
973:
948:
944:
940:
935:
934:
925:
923:
919:
915:
906:
905:
896:
894:
890:
886:
882:
864:
853:
842:
841:
832:
830:
826:
822:
818:
814:
810:
806:
797:
796:
787:
785:
781:
777:
773:
769:
765:
761:
747:
743:
738:
735:
711:
708:
705:
704:
699:
698:
690:
689:
679:
678:
658:
655:
645:
642:
565:
562:
559:
558:
552:
551:
543:
542:
533:
532:
469:
466:
459:
458:
451:
450:
442:
441:
429:
428:
415:
414:
411:
408:
402:
401:
398:
395:
391:
390:
387:
384:
380:
379:
376:
373:
317:
314:
307:
306:
301:
297:
288:
287:
279:
278:
272:
271:
254:
251:
238:
235:
222:
215:
141:
138:
121:
35:ubiquitylation
15:
9:
6:
4:
3:
2:
3848:
3847:
3836:
3833:
3831:
3828:
3827:
3825:
3809:
3805:
3804:
3799:
3796:
3792:
3791:
3786:
3783:
3779:
3778:
3773:
3770:
3766:
3765:
3760:
3757:
3753:
3752:
3747:
3744:
3740:
3739:
3734:
3731:
3727:
3726:
3721:
3720:
3718:
3714:
3708:
3705:
3703:
3700:
3698:
3695:
3693:
3690:
3688:
3685:
3684:
3682:
3678:
3672:
3669:
3667:
3666:Enzyme family
3664:
3662:
3659:
3657:
3654:
3653:
3651:
3647:
3641:
3638:
3636:
3633:
3631:
3630:Cooperativity
3628:
3626:
3623:
3622:
3620:
3616:
3610:
3607:
3605:
3602:
3600:
3597:
3595:
3592:
3590:
3589:Oxyanion hole
3587:
3585:
3582:
3580:
3577:
3575:
3572:
3571:
3569:
3565:
3561:
3554:
3549:
3547:
3542:
3540:
3535:
3534:
3531:
3519:
3516:
3514:
3511:
3509:
3506:
3504:
3501:
3499:
3496:
3494:
3491:
3490:
3488:
3484:
3478:
3475:
3473:
3470:
3468:
3465:
3463:
3460:
3459:
3457:
3455:
3451:
3445:
3442:
3440:
3437:
3435:
3432:
3430:
3427:
3426:
3424:
3420:
3413:
3410:
3407:
3404:
3402:
3399:
3397:
3396:Meso compound
3394:
3392:
3389:
3387:
3384:
3382:
3379:
3378:
3376:
3372:
3366:
3363:
3361:
3358:
3356:
3353:
3351:
3346:
3342:
3340:
3337:
3335:
3332:
3330:
3327:
3326:
3324:
3320:
3316:
3308:
3303:
3301:
3296:
3294:
3289:
3288:
3285:
3278:
3273:
3269:
3268:
3256:
3251:
3246:
3241:
3237:
3233:
3229:
3224:
3220:
3214:
3210:
3205:
3204:
3190:
3186:
3181:
3176:
3172:
3168:
3164:
3160:
3156:
3149:
3141:
3134:
3130:
3125:
3120:
3116:
3112:
3108:
3104:
3100:
3096:
3092:
3085:
3077:
3070:
3066:
3061:
3056:
3052:
3048:
3044:
3040:
3036:
3029:
3020:
3015:
3011:
3007:
3003:
2996:
2988:
2984:
2980:
2976:
2972:
2968:
2961:
2953:
2949:
2945:
2941:
2937:
2933:
2925:
2917:
2913:
2908:
2903:
2898:
2893:
2889:
2885:
2881:
2877:
2873:
2866:
2858:
2854:
2850:
2846:
2842:
2838:
2834:
2830:
2826:
2822:
2815:
2807:
2803:
2799:
2795:
2791:
2787:
2780:
2772:
2768:
2764:
2760:
2756:
2752:
2745:
2737:
2733:
2729:
2725:
2721:
2717:
2713:
2709:
2702:
2694:
2690:
2686:
2682:
2678:
2674:
2670:
2666:
2659:
2651:
2644:
2636:
2632:
2628:
2624:
2617:
2609:
2605:
2601:
2597:
2593:
2589:
2582:
2574:
2570:
2566:
2562:
2557:
2552:
2548:
2544:
2540:
2536:
2532:
2525:
2517:
2513:
2509:
2505:
2498:
2490:
2486:
2482:
2478:
2474:
2470:
2466:
2462:
2458:
2454:
2447:
2445:
2436:
2432:
2428:
2424:
2420:
2416:
2409:
2407:
2398:
2394:
2390:
2386:
2382:
2378:
2374:
2370:
2366:
2362:
2355:
2347:
2341:
2337:
2330:
2322:
2315:
2311:
2307:
2303:
2299:
2295:
2288:
2280:
2274:
2270:
2266:
2262:
2256:
2248:
2244:
2240:
2236:
2232:
2228:
2224:
2217:
2209:
2205:
2201:
2197:
2193:
2189:
2182:
2174:
2168:
2164:
2160:
2156:
2149:
2141:
2137:
2133:
2129:
2125:
2121:
2117:
2113:
2109:
2105:
2098:
2090:
2084:
2080:
2076:
2075:
2067:
2059:
2053:
2049:
2045:
2044:
2036:
2020:
2016:
2010:
2001:
1993:
1989:
1984:
1979:
1975:
1971:
1967:
1963:
1959:
1955:
1951:
1944:
1936:
1932:
1927:
1922:
1917:
1912:
1908:
1904:
1900:
1896:
1892:
1885:
1877:
1873:
1869:
1865:
1861:
1857:
1856:J. Chem. Phys
1850:
1843:
1835:
1831:
1827:
1820:
1812:
1808:
1804:
1800:
1796:
1792:
1785:
1777:
1773:
1769:
1765:
1761:
1757:
1750:
1742:
1738:
1734:
1730:
1726:
1722:
1715:
1707:
1703:
1699:
1695:
1691:
1687:
1680:
1678:
1676:
1674:
1665:
1661:
1657:
1653:
1649:
1645:
1641:
1637:
1630:
1622:
1618:
1613:
1608:
1603:
1598:
1594:
1590:
1586:
1582:
1578:
1571:
1563:
1559:
1555:
1551:
1544:
1536:
1532:
1528:
1524:
1516:
1508:
1504:
1500:
1496:
1489:
1481:
1474:
1470:
1465:
1460:
1455:
1450:
1446:
1442:
1438:
1434:
1430:
1423:
1415:
1409:
1405:
1398:
1390:
1383:
1379:
1374:
1369:
1364:
1359:
1355:
1351:
1348:(2): 98–104.
1347:
1343:
1339:
1335:
1329:
1321:
1315:
1311:
1304:
1296:
1292:
1287:
1282:
1278:
1274:
1270:
1266:
1262:
1255:
1251:
1241:
1238:
1236:
1233:
1231:
1228:
1225:
1221:
1220:Pseudoenzymes
1218:
1216:
1213:
1211:
1208:
1206:
1203:
1201:
1198:
1196:
1193:
1191:
1188:
1187:
1181:
1179:
1175:
1172:
1168:
1158:
1156:
1152:
1148:
1144:
1134:
1132:
1128:
1124:
1120:
1116:
1113:
1109:
1100:
1098:
1094:
1093:oxyanion hole
1090:
1086:
1082:
1078:
1075:that cleaves
1074:
1070:
1067:
1063:
1054:
1052:
1048:
1045:
1041:
1037:
1034:
1030:
1021:
1013:
1001:
997:
995:
994:
989:
988:
981:
971:
970:
964:
962:
961:
956:
955:
933:
926:
924:
912:
911:
904:
897:
895:
879:
878:
875:
872:
868:
862:
861:
851:
850:
840:
833:
831:
803:
802:
795:
788:
786:
758:
757:
754:
751:
737:Active enzyme
734:
731:
727:
725:
722:oxidation by
721:
717:
700:
696:
691:
688:
686:
680:
674:
673:
670:
667:
663:
654:
652:
641:
638:
633:
631:
627:
623:
619:
615:
610:
608:
604:
600:
596:
592:
588:
584:
580:
576:
571:
570:covalent bond
557:
553:
549:
544:
541:
539:
534:
528:
527:
524:
520:
517:
513:
511:
507:
503:
502:glutamic acid
499:
498:aspartic acid
495:
491:
487:
483:
479:
475:
465:
457:
452:
448:
443:
440:
439:
435:
430:
424:
423:
420:
413:Increase pKa
412:
410:Decrease pKa
409:
404:
403:
400:Decrease pKa
399:
397:Increase pKa
396:
393:
392:
389:Decrease pKa
388:
386:Increase pKa
385:
382:
381:
377:
374:
371:
370:
367:
364:
362:
358:
354:
350:
345:
341:
336:
333:
329:
323:
313:
305:
295:
289:
285:
280:
277:
273:
267:
266:
263:
260:
250:
248:
244:
234:
232:
227:
221:
214:
209:
205:
201:
199:
195:
186:
178:
172:
168:
163:
159:
155:
151:
146:
137:
135:
131:
125:
120:
116:
112:
108:
103:
102:
98:
93:
89:
85:
81:
77:
73:
69:
64:
62:
58:
54:
50:
46:
42:
36:
19:
3803:Translocases
3800:
3787:
3774:
3761:
3748:
3738:Transferases
3735:
3722:
3608:
3579:Binding site
3518:Biocatalysis
3391:Diastereomer
3381:Stereoisomer
3344:
3334:Stereocenter
3313:Concepts in
3235:
3231:
3208:
3162:
3158:
3148:
3098:
3094:
3084:
3042:
3038:
3028:
3009:
3005:
2995:
2970:
2966:
2960:
2935:
2932:Biochemistry
2931:
2924:
2879:
2875:
2865:
2824:
2820:
2814:
2789:
2786:Biochemistry
2785:
2779:
2754:
2751:Biochemistry
2750:
2744:
2714:(1): 59–64.
2711:
2707:
2701:
2668:
2664:
2658:
2649:
2643:
2626:
2622:
2616:
2591:
2587:
2581:
2538:
2534:
2524:
2507:
2503:
2497:
2456:
2452:
2418:
2414:
2364:
2360:
2354:
2335:
2329:
2297:
2293:
2287:
2264:
2255:
2230:
2226:
2216:
2191:
2187:
2181:
2154:
2148:
2107:
2103:
2097:
2074:Biochemistry
2073:
2066:
2043:Biochemistry
2042:
2035:
2025:30 September
2023:. Retrieved
2019:the original
2009:
2000:
1957:
1953:
1943:
1898:
1894:
1884:
1859:
1855:
1842:
1826:Biochemistry
1825:
1819:
1794:
1790:
1784:
1759:
1755:
1749:
1724:
1721:Biochemistry
1720:
1714:
1689:
1685:
1639:
1635:
1629:
1584:
1580:
1570:
1553:
1549:
1543:
1526:
1522:
1515:
1498:
1494:
1488:
1436:
1432:
1422:
1403:
1397:
1345:
1341:
1328:
1309:
1303:
1268:
1264:
1254:
1226:implications
1195:Enzyme assay
1164:
1140:
1106:
1060:
1027:
1019:
1002:
998:
991:
985:
982:
967:
965:
958:
952:
938:
927:
898:
873:
869:
858:
847:
845:
834:
789:
752:
740:
732:
728:
713:
682:
676:For example:
668:
664:
660:
647:
634:
611:
583:chymotrypsin
567:
555:
536:
530:For example:
521:
518:
514:
471:
462:
454:
432:
426:For example:
418:
365:
344:electrophile
337:
325:
310:
291:
275:
269:For example:
256:
240:
228:
219:
212:
210:
206:
202:
191:
134:stereoisomer
130:peptide bond
126:
118:
104:
65:
40:
39:
18:
3574:Active site
1439:(5): e468.
1334:Koshland DE
1151:diffusivity
1042:phosphates
657:Bond strain
591:schiff base
486:amino acids
474:ionic bonds
372:Conditions
340:nucleophile
243:active site
233:mechanism.
140:Induced fit
61:active site
3824:Categories
3777:Isomerases
3751:Hydrolases
3618:Regulation
3386:Enantiomer
1246:References
885:+ EH → P
720:tryptamine
651:Lewis acid
607:glycolysis
320:See also:
150:Hexokinase
111:substrates
3835:Catalysis
3656:EC number
3486:Reactions
3329:Chirality
2263:(1987) .
2261:Jencks WP
2208:0002-7863
1834:808679090
620:(PLP) or
614:cofactors
579:proteases
506:cofactors
353:Histidine
194:substrate
174:,
84:Catalysis
3680:Kinetics
3604:Cofactor
3567:Activity
3422:Analysis
3189:24412978
3133:25487146
3069:20108965
2849:14508495
2771:14730979
2736:21790380
2693:11133081
2685:21318350
2608:16683752
2565:15385982
2489:27201250
2481:16614214
2435:14995199
2397:17498715
2389:14716003
1992:25525245
1706:16895325
1664:28961992
1656:11852595
1473:17520027
1433:PLOS ONE
1382:16590179
1295:20099310
1265:Proteins
1184:See also
1174:Archived
1171:EC-BLAST
1115:4.1.2.13
1108:Aldolase
1103:Aldolase
1085:arginine
1069:3.4.21.4
685:lysozyme
616:such as
603:aldolase
508:such as
494:arginine
488:such as
180:)
76:cofactor
3830:Enzymes
3790:Ligases
3560:Enzymes
3255:1647302
3180:4122987
3124:4363105
3103:Bibcode
3060:2832858
2987:2178685
2952:9305974
2916:3871943
2884:Bibcode
2857:2686184
2829:Bibcode
2806:4258719
2728:7132966
2635:4645409
2543:Bibcode
2461:Bibcode
2453:Science
2369:Bibcode
2361:Science
2247:2283087
2140:4326584
2132:8421499
2112:Bibcode
2079:604–606
2048:986–989
1983:4668018
1962:Bibcode
1954:Science
1903:Bibcode
1864:Bibcode
1811:4694533
1776:5133099
1741:2665806
1621:5288752
1589:Bibcode
1464:1868595
1441:Bibcode
1350:Bibcode
1286:2841229
1077:protein
1071:) is a
1062:Trypsin
1057:Trypsin
1044:isomers
1036:5.3.1.1
990:) and (
939:where S
637:pathway
605:during
597:from a
587:trypsin
349:Cystine
342:and/or
294:acetate
259:entropy
90:in the
68:enzymes
51:by an "
49:process
3764:Lyases
3252:
3215:
3187:
3177:
3131:
3121:
3095:Nature
3067:
3057:
2985:
2950:
2914:
2907:397104
2904:
2855:
2847:
2821:Nature
2804:
2769:
2734:
2726:
2691:
2683:
2633:
2606:
2573:228263
2571:
2563:
2535:Nature
2487:
2479:
2433:
2395:
2387:
2342:
2314:985660
2312:
2275:
2245:
2206:
2169:
2138:
2130:
2104:Nature
2085:
2054:
1990:
1980:
1935:281676
1933:
1926:392938
1923:
1832:
1809:
1774:
1739:
1704:
1662:
1654:
1619:
1612:389269
1609:
1471:
1461:
1410:
1380:
1373:335371
1370:
1316:
1293:
1283:
1081:lysine
1049:and D-
1040:triose
599:lysine
490:lysine
478:acidic
378:Bases
375:Acids
158:xylose
78:(e.g.
53:enzyme
3716:Types
2853:S2CID
2732:S2CID
2689:S2CID
2569:S2CID
2485:S2CID
2393:S2CID
2136:S2CID
1852:(PDF)
1660:S2CID
976:and P
947:and P
595:amine
581:like
482:basic
436:of a
328:acids
66:Most
55:", a
47:of a
3808:list
3801:EC7
3795:list
3788:EC6
3782:list
3775:EC5
3769:list
3762:EC4
3756:list
3749:EC3
3743:list
3736:EC2
3730:list
3723:EC1
3414:(de)
3408:(ee)
3213:ISBN
3185:PMID
3129:PMID
3065:PMID
2983:PMID
2971:1037
2948:PMID
2912:PMID
2845:PMID
2802:PMID
2767:PMID
2724:PMID
2681:PMID
2631:PMID
2604:PMID
2561:PMID
2477:PMID
2431:PMID
2385:PMID
2340:ISBN
2310:PMID
2273:ISBN
2243:PMID
2204:ISSN
2167:ISBN
2128:PMID
2083:ISBN
2052:ISBN
2027:2009
1988:PMID
1931:PMID
1830:OCLC
1807:PMID
1772:PMID
1737:PMID
1702:PMID
1652:PMID
1617:PMID
1469:PMID
1408:ISBN
1378:PMID
1314:ISBN
1291:PMID
1224:omic
1131:DHAP
1125:and
889:+ EP
825:+ EX
821:→ P
813:→ S
809:+ EP
780:+ EP
776:→ P
768:→ S
764:+ EX
585:and
510:zinc
351:and
332:base
330:and
177:2E2Q
171:2E2N
156:and
92:cell
45:rate
3250:PMC
3240:doi
3236:361
3175:PMC
3167:doi
3119:PMC
3111:doi
3099:517
3055:PMC
3047:doi
3043:132
3014:doi
2975:doi
2940:doi
2902:PMC
2892:doi
2837:doi
2825:425
2794:doi
2759:doi
2716:doi
2673:doi
2669:352
2596:doi
2592:106
2551:doi
2539:431
2512:doi
2508:118
2469:doi
2457:312
2423:doi
2419:126
2377:doi
2365:303
2302:doi
2298:103
2235:doi
2196:doi
2192:107
2159:doi
2120:doi
2108:361
1978:PMC
1970:doi
1958:346
1921:PMC
1911:doi
1872:doi
1799:doi
1764:doi
1729:doi
1694:doi
1690:106
1644:doi
1607:PMC
1597:doi
1558:doi
1531:doi
1527:122
1503:doi
1499:120
1459:PMC
1449:doi
1368:PMC
1358:doi
1281:PMC
1273:doi
1180:).
1133:).
1083:or
500:or
480:or
357:pKa
167:PDB
86:of
3826::
3248:.
3234:.
3230:.
3183:.
3173:.
3163:11
3161:.
3157:.
3127:.
3117:.
3109:.
3097:.
3093:.
3063:.
3053:.
3041:.
3037:.
3008:.
3004:.
2981:.
2969:.
2946:.
2936:36
2934:.
2910:.
2900:.
2890:.
2880:82
2878:.
2874:.
2851:.
2843:.
2835:.
2823:.
2800:.
2790:10
2788:.
2765:.
2755:43
2753:.
2730:.
2722:.
2712:47
2710:.
2687:.
2679:.
2667:.
2625:.
2602:.
2590:.
2567:.
2559:.
2549:.
2537:.
2533:.
2506:.
2483:.
2475:.
2467:.
2455:.
2443:^
2429:.
2417:.
2405:^
2391:.
2383:.
2375:.
2363:.
2308:.
2296:.
2271:.
2241:.
2229:.
2225:.
2202:.
2190:.
2165:.
2134:.
2126:.
2118:.
2106:.
2081:.
2050:.
1986:.
1976:.
1968:.
1956:.
1952:.
1929:.
1919:.
1909:.
1899:75
1897:.
1893:.
1870:.
1860:43
1858:.
1854:.
1805:.
1795:95
1793:.
1770:.
1760:93
1758:.
1735:.
1725:28
1723:.
1700:.
1688:.
1672:^
1658:.
1650:.
1640:34
1638:.
1615:.
1605:.
1595:.
1585:68
1583:.
1579:.
1554:32
1552:.
1525:.
1497:.
1467:.
1457:.
1447:.
1435:.
1431:.
1376:.
1366:.
1356:.
1346:44
1344:.
1340:.
1289:.
1279:.
1269:78
1267:.
1263:.
1167:EC
1112:EC
1099:.
1066:EC
1053:.
1033:EC
1007:PO
914:EP
817:EP
772:EX
726:.
632:.
609:.
496:,
492:,
361:pH
300:/k
169::
162:Mg
63:.
3810:)
3806:(
3797:)
3793:(
3784:)
3780:(
3771:)
3767:(
3758:)
3754:(
3745:)
3741:(
3732:)
3728:(
3552:e
3545:t
3538:v
3348:2
3345:C
3306:e
3299:t
3292:v
3258:.
3242::
3221:.
3191:.
3169::
3135:.
3113::
3105::
3071:.
3049::
3022:.
3016::
3010:7
2989:.
2977::
2954:.
2942::
2918:.
2894::
2886::
2859:.
2839::
2831::
2808:.
2796::
2773:.
2761::
2738:.
2718::
2695:.
2675::
2637:.
2627:6
2610:.
2598::
2575:.
2553::
2545::
2518:.
2514::
2491:.
2471::
2463::
2437:.
2425::
2399:.
2379::
2371::
2348:.
2316:.
2304::
2281:.
2249:.
2237::
2231:9
2210:.
2198::
2175:.
2161::
2142:.
2122::
2114::
2091:.
2060:.
2029:.
1994:.
1972::
1964::
1937:.
1913::
1905::
1878:.
1874::
1866::
1836:.
1813:.
1801::
1778:.
1766::
1743:.
1731::
1708:.
1696::
1666:.
1646::
1623:.
1599::
1591::
1564:.
1560::
1537:.
1533::
1509:.
1505::
1475:.
1451::
1443::
1437:2
1416:.
1384:.
1360::
1352::
1322:.
1297:.
1275::
1129:(
1110:(
1064:(
1031:(
1009:4
1005:2
993:2
987:1
984:(
978:2
974:1
969:3
960:4
954:3
949:2
945:1
941:1
932:)
930:4
928:(
920:2
916:2
903:)
901:3
899:(
891:2
887:1
883:1
881:S
865:2
860:2
857:(
854:1
849:1
839:)
837:2
835:(
827:2
823:2
819:2
815:2
811:2
807:2
805:S
794:)
792:1
790:(
782:2
778:1
774:1
770:1
766:1
762:1
760:S
748:2
744:1
742:X
687:.
302:1
298:2
223:a
220:E
216:a
213:E
122:a
119:E
117:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.