306:. There are different factors that can add variability to proteins. SAPs (single amino acid polymorphisms) and non-synonymous single nucleotide polymorphisms (nsSNPs) can lead to different "proteoforms" or "proteomorphs". Recent estimates have found ~135,000 validated nonsynonymous cSNPs currently housed within SwissProt. In dbSNP, there are 4.7 million candidate cSNPs, yet only ~670,000 cSNPs have been validated in the 1,000-genomes set as nonsynonymous cSNPs that change the identity of an amino acid in a protein.
135:
446:
366:, these projects seek to find and collect evidence for all predicted protein coding genes in the human genome. The Human Proteome Map currently (October 2020) claims 17,294 proteins and ProteomicsDB 15,479, using different criteria. On October 16, 2020, the HPP published a high-stringency blueprint covering more than 90% of the predicted protein coding genes. Proteins are identified from a wide range of fetal and adult tissues and cell types, including
379:
20:
142:
The proteome can be used in order to comparatively analyze different cancer cell lines. Proteomic studies have been used in order to identify the likelihood of metastasis in bladder cancer cell lines KK47 and YTS1 and were found to have 36 unregulated and 74 down regulated proteins. The differences
122:. Usually viral proteomes are predicted from the viral genome but some attempts have been made to determine all the proteins expressed from a virus genome, i.e. the viral proteome. More often, however, virus proteomics analyzes the changes of host proteins upon virus infection, so that in effect
514:
is an important tool in the study of the proteome. It allows for very sensitive separation of different kinds of proteins based on their affinity for a matrix. Some newer methods for the separation and identification of proteins include the use of monolithic capillary columns, high temperature
153:
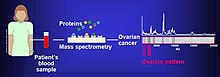
based proteomic analyses. The use of proteomics or the study of the proteome is a step forward in personalized medicine to tailor drug cocktails to the patient's specific proteomic and genomic profile. The analysis of ovarian cancer cell lines showed that putative biomarkers for ovarian cancer
1874:
Wilhelm, Mathias; Schlegl, Judith; Hahne, Hannes; Gholami, Amin
Moghaddas; Lieberenz, Marcus; Savitski, Mikhail M.; Ziegler, Emanuel; Butzmann, Lars; Gessulat, Siegfried; Marx, Harald; Mathieson, Toby; Lemeer, Simone; Schnatbaum, Karsten; Reimer, Ulf; Wenschuh, Holger; Mollenhauer, Martin;
621:
proteins. Because the range in protein contents in plasma is very large, it is difficult to detect proteins that tend to be scarce when compared to abundant proteins. This is an analytical limit that may possibly be a barrier for the detections of proteins with ultra low concentrations.
502:. This map was generated using high-resolution Fourier-transform mass spectrometry. This study profiled 30 histologically normal human samples resulting in the identification of proteins coded by 17,294 genes. This accounts for around 84% of the total annotated protein-coding genes.
397:
technology to make copies of a single protein. Numerous methods are available to study proteins, sets of proteins, or the whole proteome. In fact, proteins are often studied indirectly, e.g. using computational methods and analyses of genomes. Only a few examples are given below.
300:, DNA repair capability is positively related to genome information content and to genome size. “Proteomic constraint” proposes that modulators of mutation rates such as DNA repair genes are subject to selection pressure proportional to the amount of information in a genome.
598:
contains information about the human proteins in cells, tissues, and organs. All the data in the knowledge resource is open access to allow scientists both in academia and industry to freely access the data for exploration of the human proteome. The organization
206:, proteomic analyses were used in order to investigate how different proteins help each of these bacteria spores germinate after a prolonged period of dormancy. In order to better understand how to properly eliminate spores, proteomic analysis must be performed.
173:
Comparative proteomic analyses of 11 cell lines demonstrated the similarity between the metabolic processes of each cell line; 11,731 proteins were completely identified from this study. Housekeeping proteins tend to show greater variability between cell lines.
1635:
Acosta S, Carela M, Garcia-Gonzalez A, Gines M, Vicens L, Cruet R, Massey SE. DNA Repair Is
Associated with Information Content in Bacteria, Archaea, and DNA Viruses. J Hered. 2015 Sep-Oct;106(5):644-59. doi: 10.1093/jhered/esv055. Epub 2015 Aug 29. PMID:
392:
Analyzing proteins proves to be more difficult than analyzing nucleic acid sequences. While there are only 4 nucleotides that make up DNA, there are at least 20 different amino acids that can make up a protein. Additionally, there is currently no known
221:
in 1994 in a symposium on "2D Electrophoresis: from protein maps to genomes" held in Siena in Italy. It appeared in print in 1995, with the publication of part of his PhD thesis. Wilkins used the term to describe the entire complement of
182:. Studies of adenocarcinoma cell line LoVo demonstrated that 8 proteins were unregulated and 7 proteins were down-regulated. Proteins that showed a differential expression were involved in processes such as transcription,
1258:
Peng, Xing-Chen; Gong, Feng-Ming; Wei, Meng; Chen, Xi; Chen, Ye; Cheng, Ke; Gao, Feng; Xu, Feng; Bi, Feng; Liu, Ji-Yan (December 2010). "Proteomic analysis of cell lines to identify the irinotecan resistance proteins".
754:
Morgenstern, Marcel; Stiller, Sebastian B.; Lübbert, Philipp; Peikert, Christian D.; Dannenmaier, Stefan; Drepper, Friedel; Weill, Uri; Höß, Philipp; Feuerstein, Reinhild; Gebert, Michael; Bohnert, Maria (June 2017).
526:
can be used in order to quantify the abundance of certain proteins. By using antibodies specific to the protein of interest, it is possible to probe for the presence of specific proteins from a mixture of proteins.
1649:
Aebersold, Ruedi; Agar, Jeffrey N; Amster, I Jonathan; Baker, Mark S; Bertozzi, Carolyn R; Boja, Emily S; Costello, Catherine E; Cravatt, Benjamin F; Fenselau, Catherine; Garcia, Benjamin A; Ge, Ying (March 2018).
2043:
577:
provided predicted structures for over 200 million proteins from across the tree of life. Smaller projects have also used protein structure prediction to help map the proteome of individual organisms, for example
67:
While proteome generally refers to the proteome of an organism, multicellular organisms may have very different proteomes in different cells, hence it is important to distinguish proteomes in cells and organisms.
1414:
Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I (1995). "Progress with gene-product mapping of the
Mollicutes: Mycoplasma genitalium".
177:
Resistance to certain cancer drugs is still not well understood. Proteomic analysis has been used in order to identify proteins that may have anti-cancer drug properties, specifically for the colon cancer drug
2175:
Ponomarenko, Elena A.; Poverennaya, Ekaterina V.; Ilgisonis, Ekaterina V.; Pyatnitskiy, Mikhail A.; Kopylov, Arthur T.; Zgoda, Victor G.; Lisitsa, Andrey V.; Archakov, Alexander I. (2016).
1091:
An, Yao; Zhou, Li; Huang, Zhao; Nice, Edouard C.; Zhang, Haiyuan; Huang, Canhua (2019-05-04). "Molecular insights into cancer drug resistance from a proteomics perspective".
489:, on the other hand, can get sequence information from individual peptides by isolating them, colliding them with a non-reactive gas, and then cataloguing the fragment
2124:
Sommer, Markus J.; Cha, Sooyoung; Varabyou, Ales; Rincon, Natalia; Park, Sukhwan; Minkin, Ilia; Pertea, Mihaela; Steinegger, Martin; Salzberg, Steven L. (2022-12-15).
194:
Proteomic analyses have been performed in different kinds of bacteria to assess their metabolic reactions to different conditions. For example, in bacteria such as
55:, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions.
626:
1876:
90:, which can be conceptualized as the complete set of proteins from all of the various cellular proteomes. This is very roughly the protein equivalent of the
481:
identifies a protein by cleaving it into short peptides and then deduces the protein's identity by matching the observed peptide masses against a
382:
This image shows a two-dimensional gel with color-coded proteins. This is a way to visualize proteins based on their mass and isoelectric point.
1142:
Cruz, Isa N.; Coley, Helen M.; Kramer, Holger B.; Madhuri, Thumuluru
Kavitah; Safuwan, Nur a. M.; Angelino, Ana Rita; Yang, Min (2017-01-01).
272:(e.g. human proteome encodes about 20,000 proteins, but some estimates predicted 92,179 proteins out of which 71,173 are splicing variants).
569:
can be used to provide three-dimensional protein structure predictions of whole proteomes. In 2022, a large-scale collaboration between
2279:
159:
1831:
Altelaar, AF; Munoz, J; Heck, AJ (January 2013). "Next-generation proteomics: towards an integrative view of proteome dynamics".
535:
470:
is one of the key methods to study the proteome. Some important mass spectrometry methods include
Orbitrap Mass Spectrometry,
570:
1875:
Slotta-Huspenina, Julia; Boese, Joos-Hendrik; Bantscheff, Marcus; Gerstmair, Anja; Faerber, Franz; Kuster, Bernhard (2014).
2000:
Shi, Yang; Xiang, Rong; Horváth, Csaba; Wilkins, James A. (2004-10-22). "The role of liquid chromatography in proteomics".
254:
algorithms use certain cut-offs, such as 50 or 100 amino acids, so small proteins are often missed by such predictions. In
1026:"Quantitative Analysis of Differential Proteome Expression in Bladder Cancer vs. Normal Bladder Cells Using SILAC Method"
410:
603:
has selected the protein atlas as a core resource due to its fundamental importance for a wider life science community.
1201:"Comparative Proteomic Analysis of Eleven Common Cell Lines Reveals Ubiquitous but Varying Expression of Most Proteins"
1922:
1312:"Membrane Proteomes and Ion Transporters in Bacillus anthracis and Bacillus subtilis Dormant and Germinating Spores"
610:
250:(in viruses ranging from ~3 to ~1000, in bacteria ranging from about 500 proteins to about 10,000). However, most
2307:
1310:
Chen, Yan; Barat, Bidisha; Ray, W. Keith; Helm, Richard F.; Melville, Stephen B.; Popham, David L. (2019-03-15).
707:
Johnson, D. T.; Harris, R. A.; French, S.; Blair, P. V.; You, J.; Bemis, K. G.; Wang, M.; Balaban, R. S. (2006).
657:
539:
1024:
Yang, Ganglong; Xu, Zhipeng; Lu, Wei; Li, Xiang; Sun, Chengwen; Guo, Jia; Xue, Peng; Guan, Feng (2015-07-31).
246:
encode a relatively well-defined proteome as each protein can be predicted with high confidence, based on its
2272:
2247:
566:
1144:"Proteomics Analysis of Ovarian Cancer Cell Lines and Tissues Reveals Drug Resistance-associated Proteins"
478:
2237:
321:
417:, which resolves proteins on the basis of charge. In the second dimension, proteins are separated by
458:
214:
437:
to visualize the proteins. Spots on the gel are proteins that have migrated to specific locations.
486:
430:
367:
2265:
2004:. Bioanalytical Chemistry: Perspectives and Recent Advances with Recognition of Barry L. Karger.
662:
475:
394:
677:
558:. Pull-down assays are a method to determine the protein binding partners of a given protein.
409:, the study of the proteome, has largely been practiced through the separation of proteins by
2411:
2302:
2080:
1956:
1890:
1787:
1728:
1524:
1512:
1199:
Geiger, Tamar; Wehner, Anja; Schaab, Christoph; Cox, Juergen; Mann, Matthias (March 2012).
1037:
543:
414:
363:
269:
8:
2454:
2251:
687:
118:
2084:
1960:
1894:
1791:
1732:
1528:
1041:
2211:
2176:
2152:
2125:
2106:
1977:
1944:
1914:
1856:
1808:
1775:
1751:
1716:
1692:
1651:
1608:
1575:
1556:
1488:
1459:
1440:
1396:
1344:
1311:
1292:
1235:
1200:
1176:
1124:
1068:
1025:
1006:
946:
913:
840:
813:
789:
756:
736:
316:
coined by Perdigão and colleagues, defines regions of proteins that have no detectable
247:
2343:
2317:
2216:
2198:
2157:
2110:
2098:
2025:
2017:
1982:
1906:
1848:
1813:
1756:
1697:
1679:
1613:
1595:
1560:
1548:
1540:
1493:
1432:
1400:
1388:
1349:
1331:
1284:
1276:
1240:
1222:
1181:
1163:
1116:
1108:
1073:
1055:
998:
990:
951:
933:
894:
886:
845:
794:
776:
728:
482:
467:
462:
450:
325:
317:
163:
150:
1860:
1128:
1010:
971:"Viral proteomics: global evaluation of viruses and their interaction with the host"
740:
2421:
2379:
2374:
2369:
2206:
2188:
2147:
2137:
2088:
2009:
1972:
1964:
1918:
1898:
1881:
1840:
1803:
1795:
1746:
1736:
1687:
1671:
1663:
1603:
1587:
1532:
1483:
1475:
1444:
1424:
1380:
1339:
1323:
1296:
1268:
1230:
1212:
1171:
1155:
1100:
1063:
1045:
982:
941:
925:
876:
835:
825:
784:
768:
720:
498:
1104:
2327:
2312:
2013:
1536:
1050:
772:
614:
434:
251:
138:
The proteome can be used to determine the presence of different types of cancers.
134:
2069:"'The entire protein universe': AI predicts shape of nearly every known protein"
445:
2348:
2093:
2068:
1799:
724:
652:
511:
418:
76:
1272:
1143:
757:"Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale"
630:
359:
2448:
2406:
2401:
2202:
2021:
1683:
1599:
1544:
1428:
1335:
1280:
1226:
1167:
1112:
1059:
994:
986:
937:
890:
780:
708:
667:
490:
313:
106:
28:
1741:
1217:
582:
provides coverage of multiple protein isoforms for over 20,000 genes in the
347:
288:, which, in turn, is approximately related to the size of the proteome. In
79:
type under a particular set of environmental conditions such as exposure to
16:
Set of proteins that can be expressed by a genome, cell, tissue, or organism
2220:
2193:
2161:
2102:
2029:
1986:
1910:
1852:
1817:
1760:
1701:
1617:
1552:
1497:
1392:
1353:
1288:
1244:
1185:
1120:
1077:
1002:
970:
955:
898:
849:
830:
798:
732:
618:
583:
523:
346:. Currently, several projects aim to map the human proteome, including the
143:
in protein expression can help identify novel cancer signaling mechanisms.
2174:
1667:
1436:
929:
2426:
2416:
1675:
1591:
1479:
672:
546:
is the most popular of them but there are numerous variations, both used
530:
196:
2142:
1968:
1902:
1327:
607:
2353:
1159:
881:
864:
642:
406:
387:
281:
242:
179:
167:
56:
32:
1384:
378:
19:
2431:
2126:"Structure-guided isoform identification for the human transcriptome"
814:"Mitoproteomics: Tackling Mitochondrial Dysfunction in Human Disease"
329:
297:
255:
183:
146:
101:
has also been used to refer to the collection of proteins in certain
1844:
284:
capacity is positively correlated with the information content of a
2384:
574:
548:
426:
422:
337:
289:
202:
36:
2394:
2389:
2288:
1376:
682:
554:
333:
293:
261:
223:
80:
48:
753:
595:
496:
In May 2014, a draft map of the human proteome was published in
351:
1413:
647:
332:
and viruses was found to be "dark", compared with only ~14% in
285:
91:
52:
24:
709:"Tissue heterogeneity of the mammalian mitochondrial proteome"
471:
328:. For 546,000 Swiss-Prot proteins, 44–54% of the proteome in
236:
155:
600:
401:
2257:
2242:
265:
1873:
109:
proteome may consist of more than 3000 distinct proteins.
579:
413:. In the first dimension, the proteins are separated by
355:
1830:
1648:
706:
2123:
276:
Association of proteome size with DNA repair capability
1999:
1141:
865:"From ORFeomes to Protein Interaction Maps in Viruses"
531:
Protein complementation assays and interaction screens
2177:"The Size of the Human Proteome: The Width and Depth"
1877:"Mass-Spectrometry-Based Draft of the Human Proteome"
1198:
186:
and cell proliferation/differentiation among others.
1367:
Wilkins, Marc (Dec 2009). "Proteomics data mining".
969:
Viswanathan, Kasinath; Früh, Klaus (December 2007).
515:
chromatography and capillary electrochromatography.
260:
this becomes much more complicated as more than one
189:
75:
is the collection of proteins found in a particular
1776:"A high-stringency blueprint of the human proteome"
474:(Matrix Assisted Laser Desorption/Ionization), and
226:expressed by a genome, cell, tissue or organism.
2446:
1309:
633:are central resources for human proteomic data.
373:
86:It can also be useful to consider an organism's
23:General schema showing the relationships of the
1257:
1090:
968:
912:Maxwell, Karen L.; Frappier, Lori (June 2007).
911:
713:American Journal of Physiology. Cell Physiology
561:
126:proteomes (of virus and its host) are studied.
280:The concept of “proteomic constraint” is that
160:glyceraldehyde-3-phosphate dehydrogenase (G3P)
2273:
2181:International Journal of Analytical Chemistry
1631:
1629:
1627:
811:
162:, stress-70 protein, mitochondrial (GRP75),
2168:
2280:
2266:
1717:"Unexpected features of the dark proteome"
1624:
1023:
918:Microbiology and Molecular Biology Reviews
2210:
2192:
2151:
2141:
2092:
1976:
1807:
1750:
1740:
1691:
1607:
1487:
1457:
1343:
1234:
1216:
1175:
1067:
1049:
945:
880:
839:
829:
788:
402:Separation techniques and electrophoresis
2066:
1773:
1714:
1576:"UniProt: a hub for protein information"
444:
377:
133:
105:, such as organelles. For instance, the
18:
1652:"How many human proteoforms are there?"
1573:
1366:
536:Protein-fragment complementation assays
129:
2447:
2254:Web Archives (archived 2011-05-06)
1943:Kim, Min-Sik; et al. (May 2014).
1715:Perdigão, Nelson; et al. (2015).
1510:
1464:: proteome isoelectric point database"
2261:
1644:
1642:
62:
862:
589:
440:
229:
1945:"A draft map of the human proteome"
1942:
1205:Molecular & Cellular Proteomics
411:two dimensional gel electrophoresis
51:that is, or can be, expressed by a
13:
1639:
812:Gómez-Serrano, M (November 2018).
14:
2466:
2231:
1458:Kozlowski, LP (26 October 2016).
505:
190:The proteome in bacterial systems
1148:Cancer Genomics & Proteomics
360:The Human Proteome Project (HPP)
2308:Post-translational modification
2117:
2060:
2036:
1993:
1936:
1867:
1824:
1767:
1708:
1567:
1504:
1451:
1407:
1360:
1303:
1251:
1192:
658:List of omics topics in biology
617:contains information on 10,500
1135:
1084:
1017:
962:
905:
856:
805:
747:
700:
476:ESI (Electrospray Ionization).
59:is the study of the proteome.
1:
2067:Callaway, Ewen (2022-07-28).
1105:10.1080/14789450.2019.1601561
693:
374:Methods to study the proteome
149:of cancer have been found by
2287:
2014:10.1016/j.chroma.2004.07.044
1774:Adhikari, S (October 2020).
1574:Uniprot, Consortium (2014).
1537:10.1126/science.366.6463.296
1511:Leslie, Mitch (2019-10-18).
1051:10.1371/journal.pone.0134727
773:10.1016/j.celrep.2017.06.014
567:Protein structure prediction
562:Protein structure prediction
540:protein–protein interactions
166:, peroxiredoxin (PRDX2) and
7:
2002:Journal of Chromatography A
1369:Expert Review of Proteomics
1093:Expert Review of Proteomics
975:Expert Review of Proteomics
636:
518:
479:Peptide mass fingerprinting
453:commonly used in proteomics
322:three-dimensional structure
320:to other proteins of known
154:include "α-enolase (ENOA),
10:
2471:
2349:Protein structural domains
2094:10.1038/d41586-022-02083-2
1800:10.1038/s41467-020-19045-9
725:10.1152/ajpcell.00108.2006
456:
385:
264:can be produced from most
209:
164:apolipoprotein A-1 (APOA1)
2362:
2336:
2295:
1273:10.1007/s12038-010-0064-9
538:are often used to detect
459:Protein mass spectrometry
1429:10.1002/elps.11501601185
987:10.1586/14789450.4.6.815
608:Plasma Proteome database
487:Tandem mass spectrometry
431:Coomassie brilliant blue
324:and therefore cannot be
158:, mitochondrial (EFTU),
2044:"Pull-Down Assays - US"
1833:Nature Reviews Genetics
1742:10.1073/pnas.1508380112
1656:Nature Chemical Biology
1316:Journal of Bacteriology
1218:10.1074/mcp.M111.014050
863:Uetz, P. (2004-10-15).
663:Plant Proteome Database
1580:Nucleic Acids Research
1468:Nucleic Acids Research
1261:Journal of Biosciences
678:Human Proteome Project
544:yeast two-hybrid assay
454:
383:
139:
40:
2412:Photoreceptor protein
1780:Nature Communications
1668:10.1038/nchembio.2576
930:10.1128/MMBR.00042-06
448:
381:
137:
47:is the entire set of
22:
2303:Protein biosynthesis
2194:10.1155/2016/7436849
2048:www.thermofisher.com
831:10.1155/2018/1435934
818:Oxid Med Cell Longev
415:isoelectric focusing
364:human genome project
270:alternative splicing
156:elongation factor Tu
130:Importance in cancer
103:sub-cellular systems
2252:Library of Congress
2143:10.7554/eLife.82556
2085:2022Natur.608...15C
1969:10.1038/nature13302
1961:2014Natur.509..575K
1903:10.1038/nature13319
1895:2014Natur.509..582W
1792:2020NatCo..11.5301A
1733:2015PNAS..11215898P
1727:(52): 15898–15903.
1529:2019Sci...366..296L
1474:(D1): D1112–D1116.
1328:10.1128/JB.00662-18
1042:2015PLoSO..1034727Y
688:Human Protein Atlas
596:Human Protein Atlas
368:hematopoietic cells
326:modeled by homology
81:hormone stimulation
1592:10.1093/nar/gku989
1480:10.1093/nar/gkw978
1211:(3): M111.014050.
1160:10.21873/cgp.20017
914:"Viral proteomics"
882:10.1101/gr.2583304
875:(10b): 2029–2033.
625:Databases such as
613:2021-01-27 at the
455:
384:
348:Human Proteome Map
252:protein prediction
248:open reading frame
140:
112:The proteins in a
63:Types of proteomes
41:
2442:
2441:
2344:Protein structure
2318:Protein targeting
1586:(D1): D204–D212.
1523:(6463): 296–299.
1385:10.1586/epr.09.81
767:(13): 2836–2852.
590:Protein databases
483:sequence database
468:Mass spectrometry
463:Mass spectrometry
451:mass spectrometer
441:Mass spectrometry
318:sequence homology
230:Size and contents
151:mass spectrometry
88:complete proteome
73:cellular proteome
2462:
2422:Phycobiliprotein
2380:Globular protein
2375:Membrane protein
2370:List of proteins
2282:
2275:
2268:
2259:
2258:
2243:UniProt database
2225:
2224:
2214:
2196:
2172:
2166:
2165:
2155:
2145:
2121:
2115:
2114:
2096:
2064:
2058:
2057:
2055:
2054:
2040:
2034:
2033:
1997:
1991:
1990:
1980:
1955:(7502): 575–81.
1940:
1934:
1933:
1931:
1930:
1921:. Archived from
1871:
1865:
1864:
1828:
1822:
1821:
1811:
1771:
1765:
1764:
1754:
1744:
1712:
1706:
1705:
1695:
1646:
1637:
1633:
1622:
1621:
1611:
1571:
1565:
1564:
1513:"Outsize impact"
1508:
1502:
1501:
1491:
1455:
1449:
1448:
1411:
1405:
1404:
1364:
1358:
1357:
1347:
1307:
1301:
1300:
1255:
1249:
1248:
1238:
1220:
1196:
1190:
1189:
1179:
1139:
1133:
1132:
1088:
1082:
1081:
1071:
1053:
1021:
1015:
1014:
966:
960:
959:
949:
909:
903:
902:
884:
860:
854:
853:
843:
833:
809:
803:
802:
792:
751:
745:
744:
719:(2): c689–c697.
704:
524:Western blotting
419:molecular weight
362:. Much like the
217:coined the term
168:annexin A (ANXA)
116:can be called a
31:, proteome, and
2470:
2469:
2465:
2464:
2463:
2461:
2460:
2459:
2445:
2444:
2443:
2438:
2402:Fibrous protein
2358:
2332:
2328:Protein methods
2313:Protein folding
2291:
2286:
2234:
2229:
2228:
2173:
2169:
2122:
2118:
2079:(7921): 15–16.
2065:
2061:
2052:
2050:
2042:
2041:
2037:
1998:
1994:
1941:
1937:
1928:
1926:
1889:(7502): 582–7.
1872:
1868:
1845:10.1038/nrg3356
1829:
1825:
1772:
1768:
1713:
1709:
1647:
1640:
1634:
1625:
1572:
1568:
1509:
1505:
1456:
1452:
1417:Electrophoresis
1412:
1408:
1365:
1361:
1308:
1304:
1256:
1252:
1197:
1193:
1140:
1136:
1089:
1085:
1036:(7): e0134727.
1022:
1018:
967:
963:
910:
906:
869:Genome Research
861:
857:
810:
806:
752:
748:
705:
701:
696:
639:
615:Wayback Machine
592:
564:
533:
521:
508:
465:
457:Main articles:
443:
404:
395:high throughput
390:
376:
234:The genomes of
232:
212:
192:
132:
65:
17:
12:
11:
5:
2468:
2458:
2457:
2440:
2439:
2437:
2436:
2435:
2434:
2429:
2424:
2414:
2409:
2404:
2399:
2398:
2397:
2392:
2387:
2377:
2372:
2366:
2364:
2360:
2359:
2357:
2356:
2351:
2346:
2340:
2338:
2334:
2333:
2331:
2330:
2325:
2320:
2315:
2310:
2305:
2299:
2297:
2293:
2292:
2285:
2284:
2277:
2270:
2262:
2256:
2255:
2245:
2240:
2233:
2232:External links
2230:
2227:
2226:
2167:
2116:
2059:
2035:
1992:
1935:
1866:
1823:
1766:
1707:
1662:(3): 206–214.
1638:
1623:
1566:
1503:
1450:
1423:(1): 1090–94.
1406:
1359:
1302:
1267:(4): 557–564.
1250:
1191:
1134:
1099:(5): 413–429.
1083:
1016:
981:(6): 815–829.
961:
924:(2): 398–411.
904:
855:
804:
746:
698:
697:
695:
692:
691:
690:
685:
680:
675:
670:
665:
660:
655:
653:Bioinformatics
650:
645:
638:
635:
591:
588:
563:
560:
532:
529:
520:
517:
512:chromatography
507:
506:Chromatography
504:
442:
439:
425:. The gel is
403:
400:
386:Main article:
375:
372:
344:Human proteome
231:
228:
211:
208:
191:
188:
131:
128:
119:viral proteome
64:
61:
15:
9:
6:
4:
3:
2:
2467:
2456:
2453:
2452:
2450:
2433:
2430:
2428:
2425:
2423:
2420:
2419:
2418:
2415:
2413:
2410:
2408:
2407:Chromoprotein
2405:
2403:
2400:
2396:
2393:
2391:
2388:
2386:
2383:
2382:
2381:
2378:
2376:
2373:
2371:
2368:
2367:
2365:
2361:
2355:
2352:
2350:
2347:
2345:
2342:
2341:
2339:
2335:
2329:
2326:
2324:
2321:
2319:
2316:
2314:
2311:
2309:
2306:
2304:
2301:
2300:
2298:
2294:
2290:
2283:
2278:
2276:
2271:
2269:
2264:
2263:
2260:
2253:
2249:
2248:Pfam database
2246:
2244:
2241:
2239:
2236:
2235:
2222:
2218:
2213:
2208:
2204:
2200:
2195:
2190:
2186:
2182:
2178:
2171:
2163:
2159:
2154:
2149:
2144:
2139:
2135:
2131:
2127:
2120:
2112:
2108:
2104:
2100:
2095:
2090:
2086:
2082:
2078:
2074:
2070:
2063:
2049:
2045:
2039:
2031:
2027:
2023:
2019:
2015:
2011:
2007:
2003:
1996:
1988:
1984:
1979:
1974:
1970:
1966:
1962:
1958:
1954:
1950:
1946:
1939:
1925:on 2018-08-20
1924:
1920:
1916:
1912:
1908:
1904:
1900:
1896:
1892:
1888:
1884:
1883:
1878:
1870:
1862:
1858:
1854:
1850:
1846:
1842:
1838:
1834:
1827:
1819:
1815:
1810:
1805:
1801:
1797:
1793:
1789:
1785:
1781:
1777:
1770:
1762:
1758:
1753:
1748:
1743:
1738:
1734:
1730:
1726:
1722:
1718:
1711:
1703:
1699:
1694:
1689:
1685:
1681:
1677:
1676:1721.1/120977
1673:
1669:
1665:
1661:
1657:
1653:
1645:
1643:
1632:
1630:
1628:
1619:
1615:
1610:
1605:
1601:
1597:
1593:
1589:
1585:
1581:
1577:
1570:
1562:
1558:
1554:
1550:
1546:
1542:
1538:
1534:
1530:
1526:
1522:
1518:
1514:
1507:
1499:
1495:
1490:
1485:
1481:
1477:
1473:
1469:
1465:
1463:
1454:
1446:
1442:
1438:
1434:
1430:
1426:
1422:
1418:
1410:
1402:
1398:
1394:
1390:
1386:
1382:
1378:
1374:
1370:
1363:
1355:
1351:
1346:
1341:
1337:
1333:
1329:
1325:
1321:
1317:
1313:
1306:
1298:
1294:
1290:
1286:
1282:
1278:
1274:
1270:
1266:
1262:
1254:
1246:
1242:
1237:
1232:
1228:
1224:
1219:
1214:
1210:
1206:
1202:
1195:
1187:
1183:
1178:
1173:
1169:
1165:
1161:
1157:
1153:
1149:
1145:
1138:
1130:
1126:
1122:
1118:
1114:
1110:
1106:
1102:
1098:
1094:
1087:
1079:
1075:
1070:
1065:
1061:
1057:
1052:
1047:
1043:
1039:
1035:
1031:
1027:
1020:
1012:
1008:
1004:
1000:
996:
992:
988:
984:
980:
976:
972:
965:
957:
953:
948:
943:
939:
935:
931:
927:
923:
919:
915:
908:
900:
896:
892:
888:
883:
878:
874:
870:
866:
859:
851:
847:
842:
837:
832:
827:
823:
819:
815:
808:
800:
796:
791:
786:
782:
778:
774:
770:
766:
762:
758:
750:
742:
738:
734:
730:
726:
722:
718:
714:
710:
703:
699:
689:
686:
684:
681:
679:
676:
674:
671:
669:
668:Transcriptome
666:
664:
661:
659:
656:
654:
651:
649:
646:
644:
641:
640:
634:
632:
628:
623:
620:
616:
612:
609:
604:
602:
597:
587:
585:
581:
576:
572:
568:
559:
557:
556:
551:
550:
545:
541:
537:
528:
525:
516:
513:
503:
501:
500:
494:
492:
488:
484:
480:
477:
473:
469:
464:
460:
452:
447:
438:
436:
432:
428:
424:
420:
416:
412:
408:
399:
396:
389:
380:
371:
369:
365:
361:
357:
353:
349:
345:
341:
339:
335:
331:
327:
323:
319:
315:
314:dark proteome
311:
310:Dark proteome
307:
305:
301:
299:
295:
291:
287:
283:
278:
277:
273:
271:
267:
263:
259:
258:
253:
249:
245:
244:
239:
238:
227:
225:
220:
216:
207:
205:
204:
199:
198:
187:
185:
181:
175:
171:
169:
165:
161:
157:
152:
148:
144:
136:
127:
125:
121:
120:
115:
110:
108:
107:mitochondrial
104:
100:
95:
93:
89:
84:
82:
78:
74:
69:
60:
58:
54:
50:
46:
38:
34:
30:
29:transcriptome
26:
21:
2322:
2238:PIR database
2184:
2180:
2170:
2133:
2129:
2119:
2076:
2072:
2062:
2051:. Retrieved
2047:
2038:
2008:(1): 27–36.
2005:
2001:
1995:
1952:
1948:
1938:
1927:. Retrieved
1923:the original
1886:
1880:
1869:
1839:(1): 35–48.
1836:
1832:
1826:
1783:
1779:
1769:
1724:
1720:
1710:
1659:
1655:
1583:
1579:
1569:
1520:
1516:
1506:
1471:
1467:
1461:
1453:
1420:
1416:
1409:
1372:
1368:
1362:
1319:
1315:
1305:
1264:
1260:
1253:
1208:
1204:
1194:
1154:(1): 35–51.
1151:
1147:
1137:
1096:
1092:
1086:
1033:
1029:
1019:
978:
974:
964:
921:
917:
907:
872:
868:
858:
821:
817:
807:
764:
761:Cell Reports
760:
749:
716:
712:
702:
624:
619:blood plasma
605:
593:
584:human genome
565:
553:
547:
534:
522:
509:
497:
495:
466:
449:An Orbitrap
405:
391:
352:ProteomicsDB
343:
342:
309:
308:
303:
302:
279:
275:
274:
256:
241:
235:
233:
218:
215:Marc Wilkins
213:
201:
195:
193:
176:
172:
145:
141:
123:
117:
113:
111:
102:
98:
96:
87:
85:
72:
70:
66:
44:
42:
2427:Phytochrome
2417:Biliprotein
2187:: 7436849.
1786:(1): 5301.
1379:: 599–603.
824:: 1435934.
673:Interactome
312:. The term
304:Proteoforms
298:DNA viruses
243:prokaryotes
197:Clostridium
2455:Proteomics
2354:Proteasome
2337:Structures
2136:: e82556.
2053:2019-12-05
1929:2016-09-29
1460:"Proteome-
694:References
643:Metabolome
580:isoform.io
493:produced.
407:Proteomics
388:Proteomics
356:isoform.io
330:eukaryotes
282:DNA repair
257:eukaryotes
180:irinotecan
147:Biomarkers
57:Proteomics
33:metabolome
2432:Lipocalin
2296:Processes
2203:1687-8760
2111:251159714
2022:0021-9673
1684:1552-4450
1600:0305-1048
1561:204774732
1545:0036-8075
1401:207211912
1336:0021-9193
1281:0250-5991
1227:1535-9476
1168:1109-6535
1113:1478-9450
1060:1932-6203
995:1744-8387
938:1092-2172
891:1088-9051
781:2211-1247
184:apoptosis
97:The term
2449:Category
2385:Globulin
2323:Proteome
2289:Proteins
2221:27298622
2162:36519529
2103:35902752
2030:15543969
1987:24870542
1911:24870543
1861:10248311
1853:23207911
1818:33067450
1761:26578815
1702:29443976
1636:26320243
1618:25348405
1553:31624194
1498:27789699
1393:19929606
1354:30602489
1289:21289438
1245:22278370
1186:28031236
1129:88474614
1121:30925852
1078:26230496
1030:PLOS ONE
1011:25742649
1003:18067418
956:17554050
899:15489322
850:30533169
799:28658629
741:24412700
733:16928776
637:See also
627:neXtprot
611:Archived
575:DeepMind
571:EMBL-EBI
549:in vitro
519:Blotting
423:SDS-PAGE
338:bacteria
290:bacteria
224:proteins
219:proteome
203:Bacillus
99:proteome
49:proteins
45:proteome
37:lipidome
2395:Albumin
2390:Edestin
2250:at the
2212:4889822
2153:9812405
2081:Bibcode
1978:4403737
1957:Bibcode
1919:4467721
1891:Bibcode
1809:7568584
1788:Bibcode
1752:4702990
1729:Bibcode
1693:5837046
1609:4384041
1525:Bibcode
1517:Science
1489:5210655
1445:9269742
1437:7498152
1377:England
1345:6398275
1297:6082637
1236:3316730
1177:5267499
1069:4521931
1038:Bibcode
947:1899879
841:6250043
790:5494306
683:BioPlex
631:UniProt
555:in vivo
510:Liquid
427:stained
334:archaea
294:archaea
268:due to
262:protein
237:viruses
210:History
2219:
2209:
2201:
2160:
2150:
2109:
2101:
2073:Nature
2028:
2020:
1985:
1975:
1949:Nature
1917:
1909:
1882:Nature
1859:
1851:
1816:
1806:
1759:
1749:
1700:
1690:
1682:
1616:
1606:
1598:
1559:
1551:
1543:
1496:
1486:
1443:
1435:
1399:
1391:
1352:
1342:
1334:
1295:
1287:
1279:
1243:
1233:
1225:
1184:
1174:
1166:
1127:
1119:
1111:
1076:
1066:
1058:
1009:
1001:
993:
954:
944:
936:
897:
889:
848:
838:
797:
787:
779:
739:
731:
648:Cytome
601:ELIXIR
542:. The
499:Nature
435:silver
421:using
358:, and
286:genome
92:genome
53:genome
25:genome
2363:Types
2130:eLife
2107:S2CID
1915:S2CID
1857:S2CID
1557:S2CID
1441:S2CID
1397:S2CID
1375:(6).
1322:(6).
1293:S2CID
1125:S2CID
1007:S2CID
737:S2CID
472:MALDI
429:with
266:genes
114:virus
2217:PMID
2199:ISSN
2185:2016
2158:PMID
2099:PMID
2026:PMID
2018:ISSN
2006:1053
1983:PMID
1907:PMID
1849:PMID
1814:PMID
1757:PMID
1721:PNAS
1698:PMID
1680:ISSN
1614:PMID
1596:ISSN
1549:PMID
1541:ISSN
1494:PMID
1433:PMID
1389:PMID
1350:PMID
1332:ISSN
1285:PMID
1277:ISSN
1241:PMID
1223:ISSN
1182:PMID
1164:ISSN
1117:PMID
1109:ISSN
1074:PMID
1056:ISSN
999:PMID
991:ISSN
952:PMID
934:ISSN
895:PMID
887:ISSN
846:PMID
822:2018
795:PMID
777:ISSN
729:PMID
629:and
606:The
594:The
573:and
552:and
491:ions
461:and
336:and
296:and
240:and
200:and
77:cell
43:The
2207:PMC
2189:doi
2148:PMC
2138:doi
2089:doi
2077:608
2010:doi
1973:PMC
1965:doi
1953:509
1899:doi
1887:509
1841:doi
1804:PMC
1796:doi
1747:PMC
1737:doi
1725:112
1688:PMC
1672:hdl
1664:doi
1604:PMC
1588:doi
1533:doi
1521:366
1484:PMC
1476:doi
1425:doi
1381:doi
1340:PMC
1324:doi
1320:201
1269:doi
1231:PMC
1213:doi
1172:PMC
1156:doi
1101:doi
1064:PMC
1046:doi
983:doi
942:PMC
926:doi
877:doi
836:PMC
826:doi
785:PMC
769:doi
721:doi
717:292
485:.
433:or
170:".
124:two
2451::
2215:.
2205:.
2197:.
2183:.
2179:.
2156:.
2146:.
2134:11
2132:.
2128:.
2105:.
2097:.
2087:.
2075:.
2071:.
2046:.
2024:.
2016:.
1981:.
1971:.
1963:.
1951:.
1947:.
1913:.
1905:.
1897:.
1885:.
1879:.
1855:.
1847:.
1837:14
1835:.
1812:.
1802:.
1794:.
1784:11
1782:.
1778:.
1755:.
1745:.
1735:.
1723:.
1719:.
1696:.
1686:.
1678:.
1670:.
1660:14
1658:.
1654:.
1641:^
1626:^
1612:.
1602:.
1594:.
1584:43
1582:.
1578:.
1555:.
1547:.
1539:.
1531:.
1519:.
1515:.
1492:.
1482:.
1472:45
1470:.
1466:.
1462:pI
1439:.
1431:.
1421:16
1419:.
1395:.
1387:.
1371:.
1348:.
1338:.
1330:.
1318:.
1314:.
1291:.
1283:.
1275:.
1265:35
1263:.
1239:.
1229:.
1221:.
1209:11
1207:.
1203:.
1180:.
1170:.
1162:.
1152:14
1150:.
1146:.
1123:.
1115:.
1107:.
1097:16
1095:.
1072:.
1062:.
1054:.
1044:.
1034:10
1032:.
1028:.
1005:.
997:.
989:.
977:.
973:.
950:.
940:.
932:.
922:71
920:.
916:.
893:.
885:.
873:14
871:.
867:.
844:.
834:.
820:.
816:.
793:.
783:.
775:.
765:19
763:.
759:.
735:.
727:.
715:.
711:.
586:.
370:.
354:,
350:,
340:.
292:,
94:.
83:.
71:A
39:).
27:,
2281:e
2274:t
2267:v
2223:.
2191::
2164:.
2140::
2113:.
2091::
2083::
2056:.
2032:.
2012::
1989:.
1967::
1959::
1932:.
1901::
1893::
1863:.
1843::
1820:.
1798::
1790::
1763:.
1739::
1731::
1704:.
1674::
1666::
1620:.
1590::
1563:.
1535::
1527::
1500:.
1478::
1447:.
1427::
1403:.
1383::
1373:6
1356:.
1326::
1299:.
1271::
1247:.
1215::
1188:.
1158::
1131:.
1103::
1080:.
1048::
1040::
1013:.
985::
979:4
958:.
928::
901:.
879::
852:.
828::
801:.
771::
743:.
723::
35:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.