295:
171:
40:
1197:
1209:
1185:
590:, usually an antibiotic resistance gene, which confers on the bacteria an ability to survive and proliferate in a selective growth medium containing the particular antibiotics. The cells after transformation are exposed to the selective media, and only cells containing the plasmid may survive. In this way, the antibiotics act as a filter to select only the bacteria containing the plasmid DNA. The vector may also contain other
306:
287:
520:
162:
refer to genetic elements that reproduce autonomously. Later in 1968, it was decided that the term plasmid should be adopted as the term for extrachromosomal genetic element, and to distinguish it from viruses, the definition was narrowed to genetic elements that exist exclusively or predominantly outside of the chromosome and can replicate autonomously.
922:, extra-chromosomal linear or circular DNA molecules which have been considered to be plasmids. These can range from 0.7 kb to 20 kb in size. The plasmids have been generally classified into two categories- circular and linear. Circular plasmids have been isolated and found in many different plants, with those in
903:, yeast vectors that rely on integration into the host chromosome for survival and replication, and are usually used when studying the functionality of a solo gene or when the gene is toxic. Also connected with the gene URA3, that codes an enzyme related to the biosynthesis of pyrimidine nucleotides (T, C);
329:
Plasmids can also be classified into incompatibility groups. A microbe can harbour different types of plasmids, but different plasmids can only exist in a single bacterial cell if they are compatible. If two plasmids are not compatible, one or the other will be rapidly lost from the cell. Different
202:
boxes, and an adjacent AT-rich region. Smaller plasmids make use of the host replicative enzymes to make copies of themselves, while larger plasmids may carry genes specific for the replication of those plasmids. A few types of plasmids can also insert into the host chromosome, and these integrative
1085:
At a specified, low voltage, the migration rate of small linear DNA fragments is a function of their length. Large linear fragments (over 20 kb or so) migrate at a certain fixed rate regardless of length. This is because the molecules 'respirate', with the bulk of the molecule following the leading
858:
and used in biotechnical (fermentation) or biomedical (vaccine therapy) applications. Daughter cells that retain a copy of the plasmid survive, while a daughter cell that fails to inherit the plasmid dies or suffers a reduced growth-rate because of the lingering poison from the parent cell. Finally,
161:
to refer to "any extrachromosomal hereditary determinant." The term's early usage included any bacterial genetic material that exists extrachromosomally for at least part of its replication cycle, but because that description includes bacterial viruses, the notion of plasmid was refined over time to
330:
plasmids may therefore be assigned to different incompatibility groups depending on whether they can coexist together. Incompatible plasmids (belonging to the same incompatibility group) normally share the same replication or partition mechanisms and can thus not be kept together in a single cell.
325:
encoded by some of the transfer genes (see figure). Non-conjugative plasmids are incapable of initiating conjugation, hence they can be transferred only with the assistance of conjugative plasmids. An intermediate class of plasmids are mobilizable, and carry only a subset of the genes required for
761:
in the context of prokaryotes to refer to a plasmid that is capable of integrating into the chromosome. The integrative plasmids may be replicated and stably maintained in a cell through multiple generations, but at some stage, they will exist as an independent plasmid molecule. In the context of
640:
Another major use of plasmids is to make large amounts of proteins. In this case, researchers grow bacteria containing a plasmid harboring the gene of interest. Just as the bacterium produces proteins to confer its antibiotic resistance, it can also be induced to produce large amounts of proteins
494:
DNA structural instability can be defined as a series of spontaneous events that culminate in an unforeseen rearrangement, loss, or gain of genetic material. Such events are frequently triggered by the transposition of mobile elements or by the presence of unstable elements such as non-canonical
214:
Plasmids almost always carry at least one gene. Many of the genes carried by a plasmid are beneficial for the host cells, for example: enabling the host cell to survive in an environment that would otherwise be lethal or restrictive for growth. Some of these genes encode traits for antibiotic
963:
The function and origin of these plasmids remains largely unknown. It has been suggested that the circular plasmids share a common ancestor, some genes in the mitochondrial plasmid have counterparts in the nuclear DNA suggesting inter-compartment exchange. Meanwhile, the linear plasmids share
219:
that enable a bacterium to colonize a host and overcome its defences or have specific metabolic functions that allow the bacterium to utilize a particular nutrient, including the ability to degrade recalcitrant or toxic organic compounds. Plasmids can also provide bacteria with the ability to
782:, that can arise during artificial gene amplifications or in pathologic processes (e.g., cancer cell transformation). Episomes in eukaryotes behave similarly to plasmids in prokaryotes in that the DNA is stably maintained and replicated with the host cell. Cytoplasmic viral episomes (as in
1003:
In the latter, much larger volumes of bacterial suspension are grown from which a maxi-prep can be performed. In essence, this is a scaled-up miniprep followed by additional purification. This results in relatively large amounts (several hundred micrograms) of very pure plasmid DNA.
227:
Naturally occurring plasmids vary greatly in their physical properties. Their size can range from very small mini-plasmids of less than 1-kilobase pairs (kbp) to very large megaplasmids of several megabase pairs (Mbp). At the upper end, little differs between a megaplasmid and a
478:
particular genes. A wide variety of plasmids are commercially available for such uses. The gene to be replicated is normally inserted into a plasmid that typically contains a number of features for their use. These include a gene that confers resistance to particular antibiotics
964:
structural similarities such as invertrons with viral DNA and fungal plasmids, like fungal plasmids they also have low GC content, these observations have led to some hypothesizing that these linear plasmids have viral origins, or have ended up in plant mitochondria through
707:
Plasmids were historically used to genetically engineer the embryonic stem cells of rats to create rat genetic disease models. The limited efficiency of plasmid-based techniques precluded their use in the creation of more accurate human cell models. However, developments in
1081:
The rate of migration for small linear fragments is directly proportional to the voltage applied at low voltages. At higher voltages, larger fragments migrate at continuously increasing yet different rates. Thus, the resolution of a gel decreases with increased voltage.
499:
include direct, inverted, and tandem repeats, which are known to be conspicuous in a large number of commercially available cloning and expression vectors. Insertion sequences can also severely impact plasmid function and yield, by leading to
239:, and is determined by how the replication initiation is regulated and the size of the molecule. Larger plasmids tend to have lower copy numbers. Low-copy-number plasmids that exist only as one or a few copies in each bacterium are, upon
422:
and unencapsidated, which have been found in fungi and various plants, from algae to land plants. In many cases, however, it may be difficult or impossible to clearly distinguish RNA plasmids from RNA viruses and other infectious RNAs.
862:
In contrast, plasmids used in biotechnology, such as pUC18, pBR322 and derived vectors, hardly ever contain toxin-antitoxin addiction systems, and therefore need to be kept under antibiotic pressure to avoid plasmid loss.
598:
to facilitate selection of plasmids with cloned inserts. Bacteria containing the plasmid can then be grown in large amounts, harvested, and the plasmid of interest may then be isolated using various methods of
445:
similar to the chromosome, yet use a plasmid-type replication mechanism such as the low copy number RepABC. As a result, they have been variously classified as minichromosomes or megaplasmids in the past. In
75:. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances.
4053:
243:, in danger of being lost in one of the segregating bacteria. Such single-copy plasmids have systems that attempt to actively distribute a copy to both daughter cells. These systems, which include the
996:. The former can be used to quickly find out whether the plasmid is correct in any of several bacterial clones. The yield is a small amount of impure plasmid DNA, which is sufficient for analysis by
1963:
Bruhn, Matthias; Schindler, Daniel; Kemter, Franziska S.; Wiley, Michael R.; Chase, Kitty; Koroleva, Galina I.; Palacios, Gustavo; Sozhamannan, Shanmuga; Waldminghaus, Torsten (30 November 2018).
746:
in 1958 to refer to extra-chromosomal genetic material that may replicate autonomously or become integrated into the chromosome. Since the term was introduced, however, its use has changed, as
235:
Plasmids may be present in an individual cell in varying number, ranging from one to several hundreds. The normal number of copies of plasmid that may be found in a single cell is called the
224:. Some plasmids, however, have no observable effect on the phenotype of the host cell or its benefit to the host cells cannot be determined, and these plasmids are called cryptic plasmids.
4046:
313:
Plasmids may be classified in a number of ways. Plasmids can be broadly classified into conjugative plasmids and non-conjugative plasmids. Conjugative plasmids contain a set of
810:
that promote cancer cell proliferation. In cancers, these episomes passively replicate together with host chromosomes when the cell divides. When these viral episomes initiate
3223:
766:
is used to mean a non-integrated extrachromosomal closed circular DNA molecule that may be replicated in the nucleus. Viruses are the most common examples of this, such as
2083:"Evidence that the insertion events of IS2 transposition are biased towards abrupt compositional shifts in target DNA and modulated by a diverse set of culture parameters"
4039:
3034:
1090:
are frequently used to analyse purified plasmids. These enzymes specifically break the DNA at certain short sequences. The resulting linear fragments form 'bands' after
194:. A typical bacterial replicon may consist of a number of elements, such as the gene for plasmid-specific replication initiation protein (Rep), repeating units called
130:, plasmids are "naked" DNA and do not encode genes necessary to encase the genetic material for transfer to a new host; however, some classes of plasmids encode the
495:(non-B) structures. Accessory regions pertaining to the bacterial backbone may engage in a wide range of structural instability phenomena. Well-known catalysts of
232:. Plasmids are generally circular, but examples of linear plasmids are also known. These linear plasmids require specialized mechanisms to replicate their ends.
516:
backbone sequences would pointedly reduce the propensity for such events to take place, and consequently, the overall recombinogenic potential of the plasmid.
981:
Plasmids are often used to purify a specific sequence, since they can easily be purified away from the rest of the genome. For their use as vectors, and for
3007:
570:
Plasmids are the most-commonly used bacterial cloning vectors. These cloning vectors contain a site that allows DNA fragments to be inserted, for example a
909:, which transport a sequence of chromosomal DNA that includes an origin of replication. These plasmids are less stable, as they can be lost during budding.
2497:
474:. These plasmids serve as important tools in genetics and biotechnology labs, where they are commonly used to clone and amplify (make many copies of) or
890:
750:
has become the preferred term for autonomously replicating extrachromosomal DNA. At a 1968 symposium in London some participants suggested that the term
3216:
2795:
1154:
Many plasmids have been created over the years and researchers have given out plasmids to plasmid databases such as the non-profit organisations
2980:
932:
being the most studied and whose mechanism of replication is known. The circular plasmids can replicate using the θ model of replication (as in
1575:
1754:
Stes, Elisabeth; Vandeputte, Olivier; Jaziri, Mondher; Holsters, Marcelle; Vereecke, Danny (2011). "A Successful
Bacterial Coup d'État: How
3209:
1060:) DNA is fully intact with both strands uncut, and with an integral twist, resulting in a compact form. This can be modeled by twisting an
1370:
4609:
3072:"Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector"
1019:. The conformations are listed below in order of electrophoretic mobility (speed for a given applied voltage) from slowest to fastest:
803:
1094:. It is possible to purify certain fragments by cutting the bands out of the gel and dissolving the gel to release the DNA fragments.
174:
There are two types of plasmid integration into a host bacteria: Non-integrating plasmids replicate as with the top instance, whereas
3106:
1077:, but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation.
2707:
Gualberto, José M.; Mileshina, Daria; Wallet, Clémentine; Niazi, Adnan Khan; Weber-Lotfi, Frédérique; Dietrich, André (May 2014).
1928:
Harrison, PW; Lower, RP; Kim, NK; Young, JP (April 2010). "Introducing the bacterial 'chromid': not a chromosome, not a plasmid".
1038:(supercoils removed). This can be modeled by letting a twisted extension cord unwind and relax and then plugging it into itself.
4176:
1162:. One can find and request plasmids from those databases for research. Researchers also often upload plasmid sequences to the
4684:
2917:
2894:
2461:
Van
Craenenbroeck K, Vanhoenacker P, Haegeman G (September 2000). "Episomal vectors for gene expression in mammalian cells".
2286:
2261:
1904:
1847:
1691:
1558:
1477:
1322:
1297:
798:
plasmids). In either case, episomes remain physically separate from host cell chromosomes. Several cancer viruses, including
3134:
Haase R, Argyros O, Wong SP, Harbottle RP, Lipps HJ, Ogris M, Magnusson T, Vizoso Pinto MG, Haas J, Baiker A (March 2010).
1106:
1313:
Wickner RB, Hinnebusch A, Lambowitz AM, Gunsalus IC, Hollaender A, eds. (1987). "Mitochondrial and
Chloroplast Plasmids".
1007:
Many commercial kits have been created to perform plasmid extraction at various scales, purity, and levels of automation.
4542:
2445:
2418:
2391:
2232:
1818:
1611:
1352:
3062:
1015:
Plasmid DNA may appear in one of five conformations, which (for a given size) run at different speeds in a gel during
186:
In order for plasmids to replicate independently within a cell, they must possess a stretch of DNA that can act as an
4689:
4599:
879:
naturally harbour various plasmids. Notable among them are 2 μm plasmids—small circular plasmids often used for
854:. Several types of plasmid addiction systems (toxin/ antitoxin, metabolism-based, ORT systems) were described in the
641:
from the inserted gene. This is a cheap and easy way of mass-producing the protein the gene codes for, for example,
3196:
4674:
4604:
1883:
1097:
Because of its tight conformation, supercoiled DNA migrates faster through a gel than linear or open-circular DNA.
441:
and a plasmid, found in about 10% of bacterial species sequenced by 2009. These elements carry core genes and have
1629:"Shooting hoops: globetrotting plasmids spreading more than just antimicrobial resistance genes across One Health"
59:
and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in
1760:
1222:
619:
259:
1709:"Active stable maintenance functions in low copy-number plasmids of Gram-positive bacteria I. Partition systems"
4527:
3877:
3472:
3022:
2530:
1146:, and WebDSV. These pieces of software help conduct entire experiments in silico before doing wet experiments.
2748:"Unique Features of the Mitochondrial Rolling Circle-Plasmid mp1 from the Higher Plant Chenopodium Album (L.)"
993:
4515:
4013:
2082:
717:
487:
to allow the bacterial cells to replicate the plasmid DNA, and a suitable site for cloning (referred to as a
452:, the bacterium synchronizes the replication of the chromosome and chromid by a conserved genome size ratio.
2938:"A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells"
2496:
Colosimo A, Goncz KK, Holmes AR, Kunzelmann K, Novelli G, Malone RW, Bennett MJ, Gruenert DC (August 2000).
3661:
3342:
3070:
Argyros O, Wong SP, Niceta M, Waddington SN, Howe SJ, Coutelle C, Miller AD, Harbottle RP (December 2008).
3008:"Replicating minicircles: Generation of nonviral episomes for the efficient modification of dividing cells"
2181:
2142:
579:
3347:
1338:
326:
transfer. They can parasitize a conjugative plasmid, transferring at high frequency only in its presence.
3581:
3337:
3191:
1069:
1016:
623:
582:. After the gene of interest is inserted, the plasmids are introduced into bacteria by a process called
4679:
1769:
1175:
937:
806:, are maintained as latent, chromosomally distinct episomes in cancer cells, where the viruses express
467:
461:
360:
or antibacterial agents. Historically known as R-factors, before the nature of plasmids was understood.
82:
2981:"The Hitchhiking principle: Optimizing episomal vectors for the use in gene therapy and biotechnology"
2657:"Transformation of Saccharomyces cerevisiae with linear DNA killer plasmids from Kluyveromyces lactis"
2034:"Analysis of DNA repeats in bacterial plasmids reveals the potential for recurrent instability events"
4031:
3882:
2995:
583:
395:
94:
4586:
4532:
3729:
3630:
3547:
2081:
Gonçalves GA, Oliveira PH, Gomes AG, Prather KL, Lewis LA, Prazeres DM, Monteiro GA (August 2014).
1507:
965:
688:
252:
119:
106:, units of DNA capable of replicating autonomously within a suitable host. However, plasmids, like
17:
114:. Plasmids are transmitted from one bacterium to another (even of another species) mostly through
4621:
3756:
3532:
2810:
2708:
1965:"Functionality of Two Origins of Replication in Vibrio cholerae Strains With a Single Chromosome"
654:
4669:
3984:
3300:
2979:
Bode J, Fetzer CP, Nehlsen K, Scinteie M, Hinrichsen BH, Baiker A, et al. (January 2001).
1502:
709:
294:
93:
sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via
2905:
2549:"Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells"
2435:
2408:
2220:
1808:
3744:
3717:
3332:
2789:
2249:
1835:
1679:
1599:
1130:, and to plan manipulations. Examples of software packages that handle plasmid maps are ApE,
713:
571:
488:
484:
346:
318:
187:
115:
72:
3320:
2379:
1863:
1546:
126:. Unlike viruses, which encase their genetic material in a protective protein coat called a
4631:
3688:
3683:
3559:
3136:"pEPito: a significantly improved non-viral episomal expression vector for mammalian cells"
2560:
1247:
1044:
DNA has free ends, either because both strands have been cut or because the DNA was linear
885:
794:(bacterial phage viruses). Others replicate through a bidirectional replication mechanism (
680:
274:
52:
1384:
Sinkovics J, Horvath J, Horak A (1998). "The origin and evolution of viruses (a review)".
8:
4206:
3924:
3766:
3722:
3678:
3416:
3126:
1127:
1091:
989:
919:
880:
778:, but some are plasmids. Other examples include aberrant chromosomal fragments, such as
691:. Plasmids encoding ZFN could help deliver a therapeutic gene to a specific site so that
684:
600:
501:
496:
471:
419:
415:
411:
298:
236:
191:
102:
78:
2564:
2143:"Marker-free plasmids for biotechnological applications – implications and perspectives"
1777:
1469:
1048:. This can be modeled with an electrical extension cord that is not plugged into itself.
4500:
4422:
3974:
3162:
3135:
2632:
2607:
2334:
2123:
2063:
1991:
1964:
1655:
1628:
1437:
1412:
1262:
1087:
997:
799:
3201:
2962:
2937:
2811:"Linear plasmids in plant mitochondria: Peaceful coexistences or malicious invasions?"
2772:
2747:
2681:
2656:
2583:
2548:
1896:
4478:
4394:
4296:
4066:
3969:
3673:
3167:
3118:
3093:
3054:
2967:
2913:
2890:
2830:
2777:
2728:
2686:
2637:
2623:
2588:
2522:
2478:
2474:
2441:
2414:
2387:
2361:
2326:
2282:
2257:
2228:
2201:
2162:
2115:
2055:
1996:
1945:
1910:
1900:
1843:
1814:
1789:
1781:
1736:
1728:
1687:
1660:
1607:
1554:
1520:
1473:
1442:
1411:
Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F (September 2010).
1393:
1318:
1293:
1288:
Esser K, Kück U, Lang-Hinrichs C, Lemke P, Osiewacz HD, Stahl U, Tudzynski P (1986).
1213:
1112:
928:
635:
587:
575:
221:
155:
86:
2338:
2067:
739:
317:
which promote sexual conjugation between different cells. In the complex process of
3705:
3599:
3232:
3157:
3147:
3083:
3046:
2957:
2949:
2822:
2767:
2759:
2720:
2676:
2668:
2627:
2619:
2578:
2568:
2512:
2470:
2318:
2193:
2182:"Structural instability of plasmid biopharmaceuticals: challenges and implications"
2154:
2127:
2105:
2097:
2045:
1986:
1976:
1937:
1892:
1773:
1720:
1650:
1640:
1512:
1465:
1432:
1424:
982:
842:
833:
827:
552:
216:
158:
2672:
2197:
2158:
1516:
679:. Plasmid vectors are one of many approaches that could be used for this purpose.
4338:
3962:
3656:
3635:
3564:
3367:
2724:
1842:. Methods in Microbiology. Vol. 29. Academic Press. pp. 51-96 (75-77).
1232:
1135:
896:
Other types of plasmids are often related to yeast cloning vectors that include:
815:
786:
infections) can also occur. Some episomes, such as herpesviruses, replicate in a
754:
be abandoned, although others continued to use the term with a shift in meaning.
696:
509:
505:
475:
90:
743:
663:
so that it may express the protein that is lacking in the cells. Some forms of
170:
4387:
4352:
4345:
4324:
4303:
4280:
4160:
3952:
3839:
3710:
3569:
3552:
3357:
3287:
3050:
2826:
2553:
Proceedings of the
National Academy of Sciences of the United States of America
2303:
1201:
1189:
1116:
1061:
952:
837:
787:
565:
528:
379:
229:
139:
43:
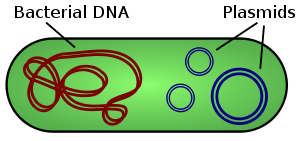
Illustration of a bacterium showing chromosomal DNA and plasmids (Not to scale)
2322:
2101:
2050:
2033:
1941:
606:
A plasmid cloning vector is typically used to clone DNA fragments of up to 15
418:. RNA plasmids are non-infectious extrachromosomal linear RNA replicons, both
350:
333:
Another way to classify plasmids is by function. There are five main classes:
322:
131:
4663:
4626:
4554:
4447:
4415:
4401:
4380:
4366:
4317:
4310:
4229:
4102:
3992:
3957:
3800:
3790:
3761:
3394:
3387:
3033:
Ehrhardt A, Haase R, Schepers A, Deutsch MJ, Lipps HJ, Baiker A (June 2008).
2256:. Methods in Molecular Biology. Vol. 235. Humana Press. pp. 19–26.
1981:
1785:
1732:
1227:
1131:
1052:
946:
791:
779:
767:
595:
513:
374:
Degradative plasmids, which enable the digestion of unusual substances, e.g.
356:
Resistance (R) plasmids, which contain genes that provide resistance against
314:
269:
248:
244:
240:
3152:
2573:
134:
necessary for their own transfer. Plasmids vary in size from 1 to over 400 k
4643:
4576:
4440:
4408:
4373:
4359:
4331:
4257:
4236:
3979:
3822:
3782:
3749:
3521:
3507:
3405:
3187:
International
Society for Plasmid Biology and other Mobile Genetic Elements
3171:
3122:
3097:
3058:
2953:
2834:
2763:
2732:
2641:
2608:"Plasmid addiction systems: perspectives and applications in biotechnology"
2526:
2482:
2365:
2330:
2205:
2166:
2119:
2059:
2000:
1949:
1793:
1740:
1708:
1664:
1553:. Methods in Molecular Biology. Vol. 235. Humana Press. pp. 1–5.
1524:
1446:
1159:
775:
771:
664:
660:
611:
544:
2971:
2912:(3rd ed.). Chichester, West Sussex, England: Wiley. pp. 101–11.
2781:
2690:
2592:
1914:
1724:
1645:
1428:
1397:
410:
Although most plasmids are double-stranded DNA molecules, some consist of
4505:
4250:
4003:
3869:
3817:
3812:
3325:
2352:
Jacob F, Wollman EL (1958), "Les épisomes, elements génétiques ajoutés",
2110:
811:
692:
591:
442:
364:
3088:
3071:
2606:
Kroll J, Klinter S, Schneider C, Voss I, Steinbüchel A (November 2010).
1163:
659:
Plasmids may also be used for gene transfer as a potential treatment in
30:
This article is about the DNA molecule. For the physics phenomenon, see
4636:
4583:
4170:
4083:
3911:
3906:
3859:
3852:
3847:
3832:
3827:
3668:
3603:
3352:
3294:
1257:
1237:
1143:
1034:
DNA is fully intact with both strands uncut but has been enzymatically
1024:
924:
855:
814:
to generate multiple virus particles, they generally activate cellular
672:
540:
480:
438:
390:
357:
301:
of a DNA fiber bundle, presumably of a single bacterial chromosome loop
208:
179:
4061:
2517:
1493:
Lederberg J (October 1952). "Cell genetics and hereditary symbiosis".
1196:
944:). Linear plasmids have been identified in some plant species such as
97:. Synthetic plasmids are available for procurement over the internet.
4243:
4222:
4124:
3916:
3901:
3888:
3640:
3428:
3259:
2460:
1686:(First ed.). Osney, Oxford OX: Wiley-Blackwell. pp. 21–22.
1252:
1139:
807:
607:
338:
190:. The self-replicating unit, in this case, the plasmid, is called a
135:
118:. This host-to-host transfer of genetic material is one mechanism of
68:
2277:
Kandavelou K, Chandrasegaran S (2008). "Plasmids for Gene
Therapy".
2180:
Oliveira PH, Prather KJ, Prazeres DM, Monteiro GA (September 2009).
309:
Electron micrograph of a bacterial DNA plasmid (chromosome fragment)
39:
4616:
4561:
4148:
4132:
4097:
3947:
3893:
3795:
3739:
3734:
3625:
3586:
3542:
3444:
3249:
2936:
Piechaczek C, Fetzer C, Baiker A, Bode J, Lipps HJ (January 1999).
1600:"Chapter 2 – Vectors for Gene Cloning: Plasmids and Bacteriophages"
1410:
1267:
1242:
1184:
1119:
851:
783:
386:
175:
123:
60:
31:
1312:
1208:
215:
resistance or resistance to heavy metal, while others may produce
4648:
4547:
4107:
4092:
3929:
3807:
3591:
3279:
3254:
2032:
Oliveira PH, Prather KJ, Prazeres DM, Monteiro GA (August 2010).
729:
642:
512:. Therefore, the reduction or complete elimination of extraneous
432:
402:
Plasmids can belong to more than one of these functional groups.
375:
368:
204:
64:
321:, plasmids may be transferred from one bacterium to another via
305:
4594:
4537:
4522:
4472:
4202:
4020:
3998:
3695:
3618:
3613:
3378:
3269:
3264:
2179:
2031:
1339:"GenBrick Gene Synthesis - Long DNA Sequences | GenScript"
847:
676:
615:
524:
448:
195:
127:
1166:, from which sequences of specific plasmids can be retrieved.
286:
4571:
4198:
4142:
4137:
3502:
3494:
3312:
3274:
2848:
2706:
2498:"Transfer and expression of foreign genes in mammalian cells"
2495:
2080:
956:
876:
840:(host killing/suppressor of killing) system of plasmid R1 in
107:
3105:
Wong SP, Argyros O, Coutelle C, Harbottle RP (August 2009).
3104:
3032:
1753:
4510:
4114:
4062:
4008:
3700:
3241:
3069:
1287:
668:
437:
Chromids are elements that exist at the boundary between a
199:
111:
56:
2935:
2709:"The plant mitochondrial genome: Dynamics and maintenance"
1155:
1126:
sequence of plasmid vectors, help to predict cut sites of
527:
plasmid, one of the first plasmids to be used widely as a
3133:
2978:
2605:
1962:
1627:
Smyth C, Leigh RJ, Delaney S, Murphy RA, Walsh F (2022).
1576:"Microbial Genomics: Standing on the Shoulders of Giants"
1123:
1100:
519:
55:
molecule within a cell that is physically separated from
2745:
2406:
2276:
2021:. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
3231:
3186:
1680:"Chapter 1 – The Function and Organization of Plasmids"
1626:
1547:"Chapter 1 – The Function and Organization of Plasmids"
960:, etc. but are rarer than their circular counterparts.
836:
or postsegregational killing system (PSK), such as the
531:. Shown on the plasmid diagram are the genes encoded (
2304:"What history tells us XIX. The notion of the episome"
1881:
Brown GG, Finnegan PM (January 1989). "RNA plasmids".
1927:
1383:
1290:
Plasmids of
Eukaryotes: fundamentals and Applications
1173:
547:
resistance respectively), its origin of replication (
2654:
2546:
2012:
2010:
1706:
385:
Virulence plasmids, which turn the bacterium into a
3107:"Strategies for the episomal modification of cells"
2875:
1460:Thomas CM, Summers D (2008). "Bacterial Plasmids".
716:, have enabled the creation of a new generation of
483:is most frequently used for bacterial strains), an
138:, and the number of identical plasmids in a single
2354:Comptes Rendus de l'Académie des Sciences de Paris
2884:
2702:
2700:
2254:E. Coli Plasmid Vectors: Methods and Applications
2140:
2007:
1551:E. Coli Plasmid Vectors: Methods and Applications
466:Artificially constructed plasmids may be used as
178:, the lower example, can integrate into the host
165:
71:. Plasmids often carry useful genes, such as for
4661:
3005:
1836:"Chapter 2: The Development of Plasmid Vectors."
1833:
1707:Dmowski, Michał; Jagura-Burdzy, Grazyna (2013).
1453:
363:Col plasmids, which contain genes that code for
2016:
918:The mitochondria of many higher plants contain
913:
2697:
2437:Introduction to Genetics: A Molecular Approach
2413:. CIBA Foundation Symposium. pp. 244–45.
2227:(2nd ed.). Wiley-Blackwell. p. 248.
1806:
1604:Gene Cloning and DNA Analysis: An Introduction
574:or polylinker which has several commonly used
4047:
3217:
1880:
1540:
1538:
1536:
1534:
1459:
1386:Acta Microbiologica et Immunologica Hungarica
1315:Extrachromosomal Elements in Lower Eukaryotes
63:; however, plasmids are sometimes present in
2794:: CS1 maint: multiple names: authors list (
2746:Backert, Meißner, Börner (1 February 1997).
2655:Gunge N, Murata K, Sakaguchi K (July 1982).
2547:Gerdes K, Rasmussen PB, Molin S (May 1986).
2351:
2279:Plasmids: Current Research and Future Trends
1813:(2nd ed.). Academic Cell. p. 795.
1317:. Boston, MA: Springer US. pp. 81–146.
859:the overall productivity could be enhanced.
832:Some plasmids or microbial hosts include an
818:defense mechanisms that kill the host cell.
683:(ZFNs) offer a way to cause a site-specific
281:
4610:Reverse transcriptase-related cellular gene
2386:. CIBA Foundation Symposium. pp. 4–8.
2141:Oliveira PH, Mairhofer J (September 2013).
1747:
4054:
4040:
3224:
3210:
2407:Wolstenholme GE, O'Connor M, eds. (1969).
2270:
1874:
1531:
1417:Microbiology and Molecular Biology Reviews
846:. This variant produces both a long-lived
122:, and plasmids are considered part of the
3161:
3151:
3111:Current Opinion in Molecular Therapeutics
3087:
2961:
2771:
2680:
2631:
2582:
2572:
2516:
2382:. In Wolstenholme GE, O'Connor M (eds.).
2218:
2109:
2049:
1990:
1980:
1654:
1644:
1506:
1492:
1436:
1142:, pDraw32, Serial Cloner, VectorFriends,
976:
2876:Klein DW, Prescott LM, Harley J (1999).
1593:
1591:
1589:
518:
304:
293:
285:
169:
38:
4591:Retroelements not elsewhere classified
2885:Moat AG, Foster JW, Spector MP (2002).
2301:
2250:"Chapter 2 – Choosing a Cloning Vector"
2247:
1840:Genetic Methods for Diverse Prokaryotes
1834:Radnedge L, Richards H (January 1999).
1677:
883:of yeast—and linear pGKL plasmids from
804:Kaposi's sarcoma-associated herpesvirus
154:was introduced in 1952 by the American
14:
4662:
2906:"Chapter 5: Manipulating Biomolecules"
2090:Applied Microbiology and Biotechnology
2038:Applied Microbiology and Biotechnology
2019:Molecular cloning: a laboratory manual
1149:
1111:The use of plasmids as a technique in
1101:Software for bioinformatics and design
985:, plasmids often need to be isolated.
821:
203:plasmids are sometimes referred to as
89:, serving to drive the replication of
4035:
3205:
2903:
2808:
2433:
2377:
2212:
1956:
1864:"Plasmids 101: Origin of Replication"
1597:
1586:
1544:
866:
667:require the insertion of therapeutic
629:
1107:List of genetic engineering software
994:miniprep to the maxiprep or bulkprep
971:
255:or partition function of a plasmid.
142:can range from one up to thousands.
4543:Integrative and conjugative element
3035:"Episomal vectors for gene therapy"
3006:Nehlsen K, Broll S, Bode J (2006).
2223:. In Streips UN, Yasbin RE (eds.).
1778:10.1146/annurev-phyto-072910-095217
1470:10.1002/9780470015902.a0000468.pub2
1371:"Invitrogen GeneArt Gene Synthesis"
24:
2988:Gene Therapy and Molecular Biology
2910:Elements of Molecular Neurobiology
2863:
1838:. In Smith MC, Sockett RE (eds.).
1573:
695:, cancer-causing mutations, or an
610:. To clone longer lengths of DNA,
523:A schematic representation of the
110:, are not generally classified as
25:
4701:
4600:Diversity-generating retroelement
3197:History of Plasmids with timeline
3180:
2511:(2): 314–18, 320–22, 324 passim.
2380:"What are episomes and plasmids?"
2252:. In Casali N, Preston A (eds.).
1606:(6th ed.). Wiley-Blackwell.
1134:, GeneConstructionKit, Geneious,
1064:and then plugging it into itself.
1000:and for some cloning techniques.
871:
702:
290:Overview of bacterial conjugation
4605:Telomerase reverse transcriptase
4177:Microbes with highly unusual DNA
2868:
2809:Handa, Hirokazu (January 2008).
2624:10.1111/j.1751-7915.2010.00170.x
2475:10.1046/j.1432-1327.2000.01645.x
2463:European Journal of Biochemistry
2440:. Garland Science. p. 238.
1884:International Review of Cytology
1549:. In Casali N, Presto A (eds.).
1207:
1195:
1183:
1010:
992:from bacteria, ranging from the
620:bacterial artificial chromosomes
504:and rearrangements, activation,
349:and result in the expression of
27:Small DNA molecule within a cell
2841:
2802:
2739:
2648:
2599:
2540:
2489:
2454:
2427:
2410:Bacterial Episomes and Plasmids
2400:
2384:Bacterial Episomes and Plasmids
2371:
2345:
2295:
2241:
2173:
2134:
2074:
2025:
2017:Russell DW, Sambrook J (2001).
1921:
1856:
1827:
1807:Clark DP, Pazdernik NJ (2012).
1800:
1761:Annual Review of Phytopathology
1700:
1671:
1620:
1567:
1223:Bacterial artificial chromosome
907:Yeast Replicative Plasmid (YRp)
901:Yeast integrative plasmid (YIp)
648:
508:or inactivation of neighboring
405:
260:Regulatory region of repBA gene
251:, are often referred to as the
4528:Defective interfering particle
3878:Last universal common ancestor
3473:Defective interfering particle
1758:Redirects Plant Development".
1713:Polish Journal of Microbiology
1486:
1404:
1377:
1363:
1345:
1331:
1306:
1281:
712:recombination techniques, and
675:target sites within the human
578:to which DNA fragments may be
166:Properties and characteristics
13:
1:
4516:Clonally transmissible cancer
4014:Clonally transmissible cancer
3450:Satellite-like nucleic acids
2928:
2673:10.1128/JB.151.1.462-464.1982
2198:10.1016/j.tibtech.2009.06.004
2159:10.1016/j.tibtech.2013.06.001
1897:10.1016/s0074-7696(08)61333-9
1517:10.1152/physrev.1952.32.4.403
1462:Encyclopedia of Life Sciences
1274:
988:There are several methods to
718:isogenic human disease models
614:with lysogeny genes deleted,
371:that can kill other bacteria.
4685:Molecular biology techniques
2725:10.1016/j.biochi.2013.09.016
1122:. These programs record the
1086:end through the gel matrix.
914:Plant mitochondrial plasmids
687:to the DNA genome and cause
624:yeast artificial chromosomes
7:
4069:, and comparable structures
2849:"Vector NTI feedback video"
2302:Morange M (December 2009).
1357:Integrated DNA Technologies
1353:"Gene synthesis | IDT"
1292:. Berlin: Springer-Verlag.
1169:
889:, that are responsible for
723:
586:. These plasmids contain a
426:
345:genes. They are capable of
10:
4706:
3570:Class II or DNA transposon
3565:Class I or retrotransposon
3051:10.2174/156652308784746440
2880:. Boston: WCB/McGraw-Hill.
2827:10.1016/j.mito.2007.10.002
2281:. Caister Academic Press.
1104:
1058:covalently closed-circular
938:rolling circle replication
825:
727:
652:
633:
563:
559:
462:Vector (molecular biology)
459:
455:
430:
257:
145:
29:
4491:
4465:
4432:
4288:
4274:
4267:
4214:
4197:
4190:
4158:
4123:
4081:
4074:
3940:
3883:Earliest known life forms
3868:
3781:
3757:Repeated sequences in DNA
3649:
3531:
3520:
3493:
3465:
3415:
3404:
3377:
3366:
3311:
3240:
2323:10.1007/s12038-009-0098-z
2225:Modern Microbial Genetics
2102:10.1007/s00253-014-5695-6
2051:10.1007/s00253-010-2671-7
1969:Frontiers in Microbiology
1942:10.1016/j.tim.2009.12.010
780:double minute chromosomes
396:Agrobacterium tumefaciens
282:Classifications and types
4690:Prokaryotic cell anatomy
4533:Endogenous viral element
3730:Endogenous viral element
3548:Horizontal gene transfer
3021:: 233–44. Archived from
2221:"Molecular Applications"
1982:10.3389/fmicb.2018.02932
1138:, LabGenius, Lasergene,
966:horizontal gene transfer
757:Today, some authors use
689:homologous recombination
120:horizontal gene transfer
100:Plasmids are considered
4675:Mobile genetic elements
3427:dsDNA satellite virus (
3153:10.1186/1472-6750-10-20
2994:: 33–46. Archived from
2661:Journal of Bacteriology
2612:Microbial Biotechnology
2574:10.1073/pnas.83.10.3116
2186:Trends in Biotechnology
2147:Trends in Biotechnology
1684:The Biology of Plasmids
1028:DNA has one strand cut.
968:from pathogenic fungi.
655:Vectors in gene therapy
268:form are unknown among
132:conjugative "sex" pilus
3985:Helper dependent virus
3301:Biological dark matter
2942:Nucleic Acids Research
2752:Nucleic Acids Research
2311:Journal of Biosciences
1930:Trends in Microbiology
1413:"Mobility of plasmids"
977:Plasmid DNA extraction
790:mechanism, similar to
710:adeno-associated virus
556:
310:
302:
291:
183:
44:
3745:Endogenous retrovirus
3718:Origin of replication
3434:ssDNA satellite virus
3424:ssRNA satellite virus
3192:What is Biotechnology
3129:on 17 September 2011.
3065:on 26 September 2011.
1725:10.33073/pjm-2013-001
1646:10.1099/mgen.0.000858
1495:Physiological Reviews
1429:10.1128/MMBR.00020-10
762:eukaryotes, the term
714:zinc finger nucleases
681:Zinc finger nucleases
572:multiple cloning site
522:
489:multiple cloning site
485:origin of replication
460:Further information:
308:
297:
289:
258:Further information:
188:origin of replication
173:
73:antibiotic resistance
42:
4632:Transposable element
4622:Spiegelman's Monster
3689:Secondary chromosome
3684:Extrachromosomal DNA
3560:Transposable element
3039:Current Gene Therapy
2954:10.1093/nar/27.2.426
2887:Microbial Physiology
2764:10.1093/nar/25.3.582
2219:Geoghegan T (2002).
1756:Rhodococcus fascians
1580:Microbiology Society
1248:Secondary chromosome
1025:Nicked open-circular
886:Kluyveromyces lactis
555:(indicated in blue).
275:Rhodococcus fascians
272:with one exception,
69:eukaryotic organisms
53:extrachromosomal DNA
3925:Model lipid bilayer
3767:Interspersed repeat
3089:10.1038/gt.2008.113
2565:1986PNAS...83.3116G
1678:Summers DK (1996).
1150:Plasmid collections
1128:restriction enzymes
1092:gel electrophoresis
1088:Restriction digests
990:isolate plasmid DNA
881:genetic engineering
822:Plasmid maintenance
685:double-strand break
601:plasmid preparation
497:genetic instability
472:genetic engineering
416:double-stranded RNA
414:, or predominantly
412:single-stranded DNA
299:Electron micrograph
237:plasmid copy number
156:molecular biologist
81:are widely used as
79:Artificial plasmids
4501:Bio-like structure
4423:Tolecusatellitidae
3235:organic structures
3015:Gene Ther Mol Biol
2248:Preston A (2003).
1633:Microbial Genomics
1263:Triparental mating
1073:DNA is similar to
998:restriction digest
867:Plasmids in nature
850:and a short-lived
800:Epstein-Barr virus
738:was introduced by
630:Protein production
557:
311:
303:
292:
184:
45:
4680:Molecular biology
4657:
4656:
4487:
4486:
4461:
4460:
4457:
4456:
4395:Portogloboviridae
4297:Alphasatellitidae
4191:Non-cellular life
4186:
4185:
4067:non-cellular life
4029:
4028:
3970:Non-cellular life
3777:
3776:
3516:
3515:
3489:
3488:
3443:ssRNA satellite (
3140:BMC Biotechnology
2919:978-0-470-85717-5
2904:Smith CU (2002).
2896:978-0-471-39483-9
2518:10.2144/00292rv01
2434:Brown TA (2011).
2288:978-1-904455-35-6
2263:978-1-58829-151-6
1906:978-0-12-364517-3
1849:978-0-12-652340-9
1810:Molecular Biology
1693:978-0-632-03436-9
1598:Brown TA (2010).
1560:978-1-58829-151-6
1479:978-0-470-01617-6
1324:978-1-4684-5251-8
1299:978-3-540-15798-4
1233:DNA recombination
1113:molecular biology
983:molecular cloning
972:Study of plasmids
929:Chenopodium album
891:killer phenotypes
812:lytic replication
636:Expression vector
588:selectable marker
576:restriction sites
553:restriction sites
217:virulence factors
87:molecular cloning
16:(Redirected from
4697:
4272:
4271:
4212:
4211:
4195:
4194:
4079:
4078:
4056:
4049:
4042:
4033:
4032:
3706:Gene duplication
3529:
3528:
3525:self-replication
3413:
3412:
3375:
3374:
3233:Self-replicating
3226:
3219:
3212:
3203:
3202:
3175:
3165:
3155:
3130:
3125:. Archived from
3101:
3091:
3082:(24): 1593–605.
3066:
3061:. Archived from
3029:
3027:
3012:
3002:
3000:
2985:
2975:
2965:
2923:
2900:
2881:
2857:
2856:
2845:
2839:
2838:
2806:
2800:
2799:
2793:
2785:
2775:
2743:
2737:
2736:
2704:
2695:
2694:
2684:
2652:
2646:
2645:
2635:
2603:
2597:
2596:
2586:
2576:
2544:
2538:
2537:
2536:on 24 July 2011.
2535:
2529:. Archived from
2520:
2502:
2493:
2487:
2486:
2458:
2452:
2451:
2431:
2425:
2424:
2404:
2398:
2397:
2378:Hayes W (1969).
2375:
2369:
2368:
2349:
2343:
2342:
2308:
2299:
2293:
2292:
2274:
2268:
2267:
2245:
2239:
2238:
2216:
2210:
2209:
2177:
2171:
2170:
2138:
2132:
2131:
2113:
2087:
2078:
2072:
2071:
2053:
2029:
2023:
2022:
2014:
2005:
2004:
1994:
1984:
1960:
1954:
1953:
1925:
1919:
1918:
1878:
1872:
1871:
1860:
1854:
1853:
1831:
1825:
1824:
1804:
1798:
1797:
1751:
1745:
1744:
1704:
1698:
1697:
1675:
1669:
1668:
1658:
1648:
1624:
1618:
1617:
1595:
1584:
1583:
1571:
1565:
1564:
1545:Hayes F (2003).
1542:
1529:
1528:
1510:
1490:
1484:
1483:
1457:
1451:
1450:
1440:
1408:
1402:
1401:
1381:
1375:
1374:
1367:
1361:
1360:
1349:
1343:
1342:
1335:
1329:
1328:
1310:
1304:
1303:
1285:
1212:
1211:
1200:
1199:
1188:
1187:
1179:
1115:is supported by
1032:Relaxed circular
920:self-replicating
843:Escherichia coli
834:addiction system
828:Addiction module
671:at pre-selected
341:, which contain
253:partition system
159:Joshua Lederberg
21:
4705:
4704:
4700:
4699:
4698:
4696:
4695:
4694:
4660:
4659:
4658:
4653:
4493:
4483:
4453:
4428:
4339:Finnlakeviridae
4284:
4263:
4205:
4201:
4182:
4154:
4119:
4070:
4060:
4030:
4025:
3975:Synthetic virus
3963:Artificial cell
3936:
3864:
3773:
3662:RNA replication
3657:DNA replication
3645:
3636:Group II intron
3534:
3524:
3512:
3503:Mammalian prion
3485:
3461:
3440:dsRNA satellite
3437:ssDNA satellite
3407:
3400:
3369:
3362:
3307:
3236:
3230:
3183:
3178:
3028:on 30 May 2009.
3025:
3010:
3001:on 30 May 2009.
2998:
2983:
2931:
2926:
2920:
2897:
2871:
2866:
2864:Further reading
2861:
2860:
2847:
2846:
2842:
2807:
2803:
2787:
2786:
2744:
2740:
2705:
2698:
2653:
2649:
2604:
2600:
2559:(10): 3116–20.
2545:
2541:
2533:
2500:
2494:
2490:
2469:(18): 5665–78.
2459:
2455:
2448:
2432:
2428:
2421:
2405:
2401:
2394:
2376:
2372:
2350:
2346:
2306:
2300:
2296:
2289:
2275:
2271:
2264:
2246:
2242:
2235:
2217:
2213:
2178:
2174:
2139:
2135:
2096:(15): 6609–19.
2085:
2079:
2075:
2030:
2026:
2015:
2008:
1961:
1957:
1926:
1922:
1907:
1879:
1875:
1862:
1861:
1857:
1850:
1832:
1828:
1821:
1805:
1801:
1752:
1748:
1705:
1701:
1694:
1676:
1672:
1625:
1621:
1614:
1596:
1587:
1572:
1568:
1561:
1543:
1532:
1491:
1487:
1480:
1458:
1454:
1409:
1405:
1392:(3–4): 349–90.
1382:
1378:
1369:
1368:
1364:
1351:
1350:
1346:
1337:
1336:
1332:
1325:
1311:
1307:
1300:
1286:
1282:
1277:
1272:
1218:
1206:
1194:
1182:
1174:
1172:
1152:
1136:Genome Compiler
1109:
1103:
1075:supercoiled DNA
1017:electrophoresis
1013:
979:
974:
916:
874:
869:
830:
824:
816:innate immunity
732:
726:
705:
697:immune response
657:
651:
638:
632:
568:
562:
551:), and various
510:gene expression
506:down-regulation
464:
458:
435:
429:
408:
284:
262:
168:
148:
91:recombinant DNA
57:chromosomal DNA
35:
28:
23:
22:
15:
12:
11:
5:
4703:
4693:
4692:
4687:
4682:
4677:
4672:
4655:
4654:
4652:
4651:
4646:
4641:
4640:
4639:
4629:
4624:
4619:
4614:
4613:
4612:
4607:
4602:
4597:
4589:
4581:
4580:
4579:
4569:
4564:
4559:
4550:
4545:
4540:
4535:
4530:
4525:
4520:
4519:
4518:
4513:
4503:
4497:
4495:
4489:
4488:
4485:
4484:
4482:
4481:
4476:
4469:
4467:
4463:
4462:
4459:
4458:
4455:
4454:
4452:
4451:
4444:
4436:
4434:
4430:
4429:
4427:
4426:
4419:
4412:
4405:
4398:
4391:
4388:Polydnaviridae
4384:
4377:
4370:
4363:
4356:
4353:Globuloviridae
4349:
4346:Fuselloviridae
4342:
4335:
4328:
4325:Bicaudaviridae
4321:
4314:
4307:
4304:Ampullaviridae
4300:
4292:
4290:
4286:
4285:
4281:Naldaviricetes
4278:
4276:
4269:
4265:
4264:
4262:
4261:
4254:
4247:
4240:
4233:
4226:
4218:
4216:
4209:
4192:
4188:
4187:
4184:
4183:
4181:
4180:
4174:
4166:
4164:
4161:Incertae sedis
4156:
4155:
4153:
4152:
4145:
4140:
4135:
4129:
4127:
4121:
4120:
4118:
4117:
4112:
4111:
4110:
4105:
4095:
4089:
4087:
4076:
4072:
4071:
4059:
4058:
4051:
4044:
4036:
4027:
4026:
4024:
4023:
4018:
4017:
4016:
4011:
4001:
3995:
3989:
3988:
3987:
3982:
3972:
3967:
3966:
3965:
3960:
3950:
3944:
3942:
3938:
3937:
3935:
3934:
3933:
3932:
3927:
3919:
3914:
3909:
3904:
3898:
3897:
3896:
3885:
3880:
3874:
3872:
3866:
3865:
3863:
3862:
3857:
3856:
3855:
3850:
3842:
3840:Kappa organism
3837:
3836:
3835:
3830:
3825:
3820:
3815:
3805:
3804:
3803:
3798:
3787:
3785:
3779:
3778:
3775:
3774:
3772:
3771:
3770:
3769:
3764:
3754:
3753:
3752:
3747:
3742:
3737:
3727:
3726:
3725:
3715:
3714:
3713:
3711:Non-coding DNA
3708:
3703:
3693:
3692:
3691:
3686:
3681:
3676:
3666:
3665:
3664:
3653:
3651:
3647:
3646:
3644:
3643:
3638:
3633:
3631:Group I intron
3628:
3623:
3622:
3621:
3611:
3610:
3609:
3606:
3597:
3594:
3589:
3584:
3574:
3573:
3572:
3567:
3557:
3556:
3555:
3553:Genomic island
3550:
3539:
3537:
3533:Mobile genetic
3526:
3518:
3517:
3514:
3513:
3511:
3510:
3505:
3499:
3497:
3491:
3490:
3487:
3486:
3484:
3483:
3482:
3481:
3478:
3469:
3467:
3463:
3462:
3460:
3459:
3458:
3457:
3454:
3448:
3441:
3438:
3435:
3432:
3425:
3421:
3419:
3410:
3402:
3401:
3399:
3398:
3391:
3383:
3381:
3372:
3364:
3363:
3361:
3360:
3358:dsDNA-RT virus
3355:
3353:ssRNA-RT virus
3350:
3348:(−)ssRNA virus
3345:
3343:(+)ssRNA virus
3340:
3335:
3330:
3329:
3328:
3317:
3315:
3309:
3308:
3306:
3305:
3304:
3303:
3298:
3288:Incertae sedis
3284:
3283:
3282:
3277:
3272:
3267:
3257:
3252:
3246:
3244:
3238:
3237:
3229:
3228:
3221:
3214:
3206:
3200:
3199:
3194:
3189:
3182:
3181:External links
3179:
3177:
3176:
3131:
3102:
3067:
3030:
3003:
2976:
2932:
2930:
2927:
2925:
2924:
2918:
2901:
2895:
2889:. Wiley-Liss.
2882:
2872:
2870:
2867:
2865:
2862:
2859:
2858:
2840:
2801:
2758:(3): 582–589.
2738:
2696:
2647:
2598:
2539:
2488:
2453:
2447:978-0815365099
2446:
2426:
2420:978-0700014057
2419:
2399:
2393:978-0700014057
2392:
2370:
2344:
2294:
2287:
2269:
2262:
2240:
2234:978-0471386650
2233:
2211:
2172:
2133:
2073:
2044:(6): 2157–67.
2024:
2006:
1955:
1920:
1905:
1873:
1855:
1848:
1826:
1820:978-0123785947
1819:
1799:
1770:Annual Reviews
1746:
1699:
1692:
1670:
1619:
1613:978-1405181730
1612:
1585:
1566:
1559:
1530:
1508:10.1.1.458.985
1485:
1478:
1452:
1403:
1376:
1362:
1344:
1330:
1323:
1305:
1298:
1279:
1278:
1276:
1273:
1271:
1270:
1265:
1260:
1255:
1250:
1245:
1240:
1235:
1230:
1225:
1219:
1217:
1216:
1204:
1192:
1171:
1168:
1151:
1148:
1117:bioinformatics
1105:Main article:
1102:
1099:
1079:
1078:
1065:
1062:extension cord
1049:
1039:
1029:
1012:
1009:
978:
975:
973:
970:
953:Brassica napus
936:) and through
915:
912:
911:
910:
904:
873:
872:Yeast plasmids
870:
868:
865:
826:Main article:
823:
820:
792:bacteriophages
788:rolling circle
776:polyomaviruses
740:François Jacob
728:Main article:
725:
722:
704:
703:Disease models
701:
653:Main article:
650:
647:
634:Main article:
631:
628:
596:reporter genes
584:transformation
566:Cloning vector
564:Main article:
561:
558:
529:cloning vector
457:
454:
431:Main article:
428:
425:
407:
404:
400:
399:
383:
380:salicylic acid
372:
361:
354:
315:transfer genes
283:
280:
270:phytopathogens
230:minichromosome
167:
164:
147:
144:
95:transformation
26:
9:
6:
4:
3:
2:
4702:
4691:
4688:
4686:
4683:
4681:
4678:
4676:
4673:
4671:
4670:Gene delivery
4668:
4667:
4665:
4650:
4647:
4645:
4642:
4638:
4635:
4634:
4633:
4630:
4628:
4627:Tandem repeat
4625:
4623:
4620:
4618:
4615:
4611:
4608:
4606:
4603:
4601:
4598:
4596:
4593:
4592:
4590:
4588:
4585:
4582:
4578:
4575:
4574:
4573:
4570:
4568:
4565:
4563:
4560:
4557:
4556:
4555:Nanobacterium
4551:
4549:
4546:
4544:
4541:
4539:
4536:
4534:
4531:
4529:
4526:
4524:
4521:
4517:
4514:
4512:
4509:
4508:
4507:
4504:
4502:
4499:
4498:
4496:
4490:
4480:
4477:
4474:
4471:
4470:
4468:
4464:
4450:
4449:
4448:Rhizidiovirus
4445:
4443:
4442:
4438:
4437:
4435:
4431:
4425:
4424:
4420:
4418:
4417:
4416:Thaspiviridae
4413:
4411:
4410:
4406:
4404:
4403:
4402:Pospiviroidae
4399:
4397:
4396:
4392:
4390:
4389:
4385:
4383:
4382:
4381:Plasmaviridae
4378:
4376:
4375:
4371:
4369:
4368:
4367:Halspiviridae
4364:
4362:
4361:
4357:
4355:
4354:
4350:
4348:
4347:
4343:
4341:
4340:
4336:
4334:
4333:
4329:
4327:
4326:
4322:
4320:
4319:
4318:Avsunviroidae
4315:
4313:
4312:
4311:Anelloviridae
4308:
4306:
4305:
4301:
4299:
4298:
4294:
4293:
4291:
4287:
4283:
4282:
4277:
4273:
4270:
4266:
4260:
4259:
4255:
4253:
4252:
4248:
4246:
4245:
4241:
4239:
4238:
4234:
4232:
4231:
4230:Duplodnaviria
4227:
4225:
4224:
4220:
4219:
4217:
4213:
4210:
4208:
4204:
4200:
4196:
4193:
4189:
4178:
4175:
4173:
4172:
4168:
4167:
4165:
4163:
4162:
4157:
4150:
4146:
4144:
4141:
4139:
4136:
4134:
4131:
4130:
4128:
4126:
4122:
4116:
4113:
4109:
4106:
4104:
4103:Mitochondrion
4101:
4100:
4099:
4096:
4094:
4091:
4090:
4088:
4085:
4080:
4077:
4075:Cellular life
4073:
4068:
4064:
4057:
4052:
4050:
4045:
4043:
4038:
4037:
4034:
4022:
4019:
4015:
4012:
4010:
4007:
4006:
4005:
4002:
4000:
3996:
3994:
3993:Nanobacterium
3990:
3986:
3983:
3981:
3978:
3977:
3976:
3973:
3971:
3968:
3964:
3961:
3959:
3958:Cell division
3956:
3955:
3954:
3951:
3949:
3946:
3945:
3943:
3939:
3931:
3928:
3926:
3923:
3922:
3920:
3918:
3915:
3913:
3910:
3908:
3905:
3903:
3899:
3895:
3892:
3891:
3890:
3886:
3884:
3881:
3879:
3876:
3875:
3873:
3871:
3867:
3861:
3858:
3854:
3851:
3849:
3846:
3845:
3843:
3841:
3838:
3834:
3831:
3829:
3826:
3824:
3821:
3819:
3816:
3814:
3811:
3810:
3809:
3806:
3802:
3801:Hydrogenosome
3799:
3797:
3794:
3793:
3792:
3791:Mitochondrion
3789:
3788:
3786:
3784:
3783:Endosymbiosis
3780:
3768:
3765:
3763:
3762:Tandem repeat
3760:
3759:
3758:
3755:
3751:
3748:
3746:
3743:
3741:
3738:
3736:
3733:
3732:
3731:
3728:
3724:
3721:
3720:
3719:
3716:
3712:
3709:
3707:
3704:
3702:
3699:
3698:
3697:
3694:
3690:
3687:
3685:
3682:
3680:
3677:
3675:
3672:
3671:
3670:
3667:
3663:
3660:
3659:
3658:
3655:
3654:
3652:
3650:Other aspects
3648:
3642:
3639:
3637:
3634:
3632:
3629:
3627:
3624:
3620:
3617:
3616:
3615:
3612:
3607:
3605:
3601:
3598:
3595:
3593:
3590:
3588:
3585:
3583:
3580:
3579:
3578:
3575:
3571:
3568:
3566:
3563:
3562:
3561:
3558:
3554:
3551:
3549:
3546:
3545:
3544:
3541:
3540:
3538:
3536:
3530:
3527:
3523:
3519:
3509:
3506:
3504:
3501:
3500:
3498:
3496:
3492:
3479:
3476:
3475:
3474:
3471:
3470:
3468:
3464:
3455:
3452:
3451:
3449:
3446:
3442:
3439:
3436:
3433:
3430:
3426:
3423:
3422:
3420:
3418:
3414:
3411:
3409:
3403:
3397:
3396:
3395:Avsunviroidae
3392:
3390:
3389:
3388:Pospiviroidae
3385:
3384:
3382:
3380:
3376:
3373:
3371:
3365:
3359:
3356:
3354:
3351:
3349:
3346:
3344:
3341:
3339:
3336:
3334:
3331:
3327:
3324:
3323:
3322:
3319:
3318:
3316:
3314:
3310:
3302:
3299:
3297:
3296:
3292:
3291:
3290:
3289:
3285:
3281:
3278:
3276:
3273:
3271:
3268:
3266:
3263:
3262:
3261:
3258:
3256:
3253:
3251:
3248:
3247:
3245:
3243:
3242:Cellular life
3239:
3234:
3227:
3222:
3220:
3215:
3213:
3208:
3207:
3204:
3198:
3195:
3193:
3190:
3188:
3185:
3184:
3173:
3169:
3164:
3159:
3154:
3149:
3145:
3141:
3137:
3132:
3128:
3124:
3120:
3117:(4): 433–41.
3116:
3112:
3108:
3103:
3099:
3095:
3090:
3085:
3081:
3077:
3073:
3068:
3064:
3060:
3056:
3052:
3048:
3045:(3): 147–61.
3044:
3040:
3036:
3031:
3024:
3020:
3016:
3009:
3004:
2997:
2993:
2989:
2982:
2977:
2973:
2969:
2964:
2959:
2955:
2951:
2948:(2): 426–28.
2947:
2943:
2939:
2934:
2933:
2921:
2915:
2911:
2907:
2902:
2898:
2892:
2888:
2883:
2879:
2874:
2873:
2869:General works
2854:
2850:
2844:
2836:
2832:
2828:
2824:
2820:
2816:
2815:Mitochondrion
2812:
2805:
2797:
2791:
2783:
2779:
2774:
2769:
2765:
2761:
2757:
2753:
2749:
2742:
2734:
2730:
2726:
2722:
2718:
2714:
2710:
2703:
2701:
2692:
2688:
2683:
2678:
2674:
2670:
2667:(1): 462–64.
2666:
2662:
2658:
2651:
2643:
2639:
2634:
2629:
2625:
2621:
2618:(6): 634–57.
2617:
2613:
2609:
2602:
2594:
2590:
2585:
2580:
2575:
2570:
2566:
2562:
2558:
2554:
2550:
2543:
2532:
2528:
2524:
2519:
2514:
2510:
2506:
2505:BioTechniques
2499:
2492:
2484:
2480:
2476:
2472:
2468:
2464:
2457:
2449:
2443:
2439:
2438:
2430:
2422:
2416:
2412:
2411:
2403:
2395:
2389:
2385:
2381:
2374:
2367:
2363:
2360:(1): 154–56,
2359:
2355:
2348:
2340:
2336:
2332:
2328:
2324:
2320:
2317:(6): 845–48.
2316:
2312:
2305:
2298:
2290:
2284:
2280:
2273:
2265:
2259:
2255:
2251:
2244:
2236:
2230:
2226:
2222:
2215:
2207:
2203:
2199:
2195:
2192:(9): 503–11.
2191:
2187:
2183:
2176:
2168:
2164:
2160:
2156:
2153:(9): 539–47.
2152:
2148:
2144:
2137:
2129:
2125:
2121:
2117:
2112:
2111:1721.1/104375
2107:
2103:
2099:
2095:
2091:
2084:
2077:
2069:
2065:
2061:
2057:
2052:
2047:
2043:
2039:
2035:
2028:
2020:
2013:
2011:
2002:
1998:
1993:
1988:
1983:
1978:
1974:
1970:
1966:
1959:
1951:
1947:
1943:
1939:
1935:
1931:
1924:
1916:
1912:
1908:
1902:
1898:
1894:
1890:
1886:
1885:
1877:
1869:
1865:
1859:
1851:
1845:
1841:
1837:
1830:
1822:
1816:
1812:
1811:
1803:
1795:
1791:
1787:
1783:
1779:
1775:
1771:
1767:
1763:
1762:
1757:
1750:
1742:
1738:
1734:
1730:
1726:
1722:
1718:
1714:
1710:
1703:
1695:
1689:
1685:
1681:
1674:
1666:
1662:
1657:
1652:
1647:
1642:
1638:
1634:
1630:
1623:
1615:
1609:
1605:
1601:
1594:
1592:
1590:
1581:
1577:
1570:
1562:
1556:
1552:
1548:
1541:
1539:
1537:
1535:
1526:
1522:
1518:
1514:
1509:
1504:
1501:(4): 403–30.
1500:
1496:
1489:
1481:
1475:
1471:
1467:
1463:
1456:
1448:
1444:
1439:
1434:
1430:
1426:
1423:(3): 434–52.
1422:
1418:
1414:
1407:
1399:
1395:
1391:
1387:
1380:
1372:
1366:
1358:
1354:
1348:
1340:
1334:
1326:
1320:
1316:
1309:
1301:
1295:
1291:
1284:
1280:
1269:
1266:
1264:
1261:
1259:
1256:
1254:
1251:
1249:
1246:
1244:
1241:
1239:
1236:
1234:
1231:
1229:
1228:Bacteriophage
1226:
1224:
1221:
1220:
1215:
1210:
1205:
1203:
1198:
1193:
1191:
1186:
1181:
1180:
1177:
1167:
1165:
1164:NCBI database
1161:
1157:
1147:
1145:
1141:
1137:
1133:
1132:Clone Manager
1129:
1125:
1121:
1118:
1114:
1108:
1098:
1095:
1093:
1089:
1083:
1076:
1072:
1071:
1066:
1063:
1059:
1055:
1054:
1050:
1047:
1043:
1040:
1037:
1033:
1030:
1027:
1026:
1022:
1021:
1020:
1018:
1011:Conformations
1008:
1005:
1001:
999:
995:
991:
986:
984:
969:
967:
961:
959:
958:
954:
949:
948:
947:Beta vulgaris
943:
939:
935:
931:
930:
926:
921:
908:
905:
902:
899:
898:
897:
894:
892:
888:
887:
882:
878:
864:
860:
857:
853:
849:
845:
844:
839:
835:
829:
819:
817:
813:
809:
805:
801:
797:
793:
789:
785:
781:
777:
773:
769:
768:herpesviruses
765:
760:
755:
753:
749:
745:
741:
737:
731:
721:
719:
715:
711:
700:
698:
694:
690:
686:
682:
678:
674:
670:
666:
662:
656:
646:
644:
637:
627:
625:
621:
617:
613:
609:
604:
602:
597:
593:
589:
585:
581:
577:
573:
567:
554:
550:
546:
542:
538:
534:
530:
526:
521:
517:
515:
511:
507:
503:
498:
492:
490:
486:
482:
477:
473:
469:
463:
453:
451:
450:
444:
440:
434:
424:
421:
417:
413:
403:
398:
397:
392:
388:
384:
381:
377:
373:
370:
366:
362:
359:
355:
352:
348:
344:
340:
336:
335:
334:
331:
327:
324:
320:
316:
307:
300:
296:
288:
279:
277:
276:
271:
267:
261:
256:
254:
250:
249:parMRC system
246:
245:parABS system
242:
241:cell division
238:
233:
231:
225:
223:
218:
212:
210:
206:
201:
197:
193:
189:
181:
177:
172:
163:
160:
157:
153:
143:
141:
137:
133:
129:
125:
121:
117:
113:
109:
105:
104:
98:
96:
92:
88:
84:
80:
76:
74:
70:
66:
62:
58:
54:
50:
41:
37:
33:
19:
4644:Transpoviron
4577:Fungal prion
4566:
4553:
4446:
4441:Dinodnavirus
4439:
4421:
4414:
4409:Spiraviridae
4407:
4400:
4393:
4386:
4379:
4374:Ovaliviridae
4372:
4365:
4360:Guttaviridae
4358:
4351:
4344:
4337:
4332:Clavaviridae
4330:
4323:
4316:
4309:
4302:
4295:
4279:
4258:Varidnaviria
4256:
4249:
4242:
4237:Monodnaviria
4235:
4228:
4221:
4169:
4159:
3980:Viral vector
3823:Gerontoplast
3750:Transpoviron
3576:
3522:Nucleic acid
3508:Fungal prion
3406:Helper-virus
3393:
3386:
3293:
3286:
3143:
3139:
3127:the original
3114:
3110:
3079:
3076:Gene Therapy
3075:
3063:the original
3042:
3038:
3023:the original
3018:
3014:
2996:the original
2991:
2987:
2945:
2941:
2909:
2886:
2878:Microbiology
2877:
2852:
2843:
2821:(1): 15–25.
2818:
2814:
2804:
2790:cite journal
2755:
2751:
2741:
2716:
2712:
2664:
2660:
2650:
2615:
2611:
2601:
2556:
2552:
2542:
2531:the original
2508:
2504:
2491:
2466:
2462:
2456:
2436:
2429:
2409:
2402:
2383:
2373:
2357:
2353:
2347:
2314:
2310:
2297:
2278:
2272:
2253:
2243:
2224:
2214:
2189:
2185:
2175:
2150:
2146:
2136:
2093:
2089:
2076:
2041:
2037:
2027:
2018:
1972:
1968:
1958:
1936:(4): 141–8.
1933:
1929:
1923:
1888:
1882:
1876:
1867:
1858:
1839:
1829:
1809:
1802:
1765:
1759:
1755:
1749:
1716:
1712:
1702:
1683:
1673:
1636:
1632:
1622:
1603:
1579:
1569:
1550:
1498:
1494:
1488:
1461:
1455:
1420:
1416:
1406:
1389:
1385:
1379:
1365:
1356:
1347:
1333:
1314:
1308:
1289:
1283:
1153:
1110:
1096:
1084:
1080:
1074:
1068:Supercoiled
1067:
1057:
1051:
1045:
1041:
1035:
1031:
1023:
1014:
1006:
1002:
987:
980:
962:
951:
945:
941:
933:
923:
917:
906:
900:
895:
884:
875:
861:
841:
831:
795:
772:adenoviruses
763:
758:
756:
751:
747:
744:Élie Wollman
735:
733:
706:
699:is avoided.
665:gene therapy
661:gene therapy
658:
649:Gene therapy
639:
612:lambda phage
605:
592:marker genes
569:
548:
545:tetracycline
536:
532:
493:
465:
447:
436:
420:encapsidated
409:
406:RNA plasmids
401:
394:
365:bacteriocins
342:
332:
328:
312:
273:
265:
264:Plasmids of
263:
234:
226:
222:fix nitrogen
213:
185:
151:
149:
101:
99:
77:
51:is a small,
48:
46:
36:
4587:microsphere
4506:Cancer cell
4251:Ribozyviria
4004:Cancer cell
3870:Abiogenesis
3818:Chromoplast
3813:Chloroplast
3596:Degradative
3338:dsRNA virus
3333:ssDNA virus
3326:Giant virus
3321:dsDNA virus
2853:The DNA Lab
2719:: 107–120.
1868:addgene.org
1719:(1): 3–16.
1639:(8): 1–10.
1053:Supercoiled
693:cell damage
673:chromosomal
443:codon usage
358:antibiotics
347:conjugation
319:conjugation
209:prokaryotes
116:conjugation
4664:Categories
4637:Retroposon
4584:Proteinoid
4494:structures
4492:Comparable
4268:Unassigned
4171:Parakaryon
4084:Prokaryota
3912:Proteinoid
3907:Coacervate
3860:Nitroplast
3853:Trophosome
3848:Bacteriome
3833:Apicoplast
3828:Leucoplast
3669:Chromosome
3587:Resistance
3295:Parakaryon
1574:Falkow S.
1275:References
1258:Transposon
1238:Plasmidome
1214:Technology
1144:Vector NTI
934:Vicia faba
925:Vicia faba
856:literature
796:Theta type
626:are used.
541:ampicillin
481:ampicillin
439:chromosome
391:Ti plasmid
339:F-plasmids
337:Fertility
180:chromosome
4244:Riboviria
4223:Adnaviria
4207:Satellite
4125:Eukaryota
3921:Research
3902:Protocell
3641:Retrozyme
3600:Virulence
3582:Fertility
3429:Virophage
3417:Satellite
3408:dependent
3260:Eukaryota
2713:Biochimie
1786:0066-4286
1772:: 69–86.
1733:1733-1331
1503:CiteSeerX
1253:Segrosome
1160:BCCM/LMBP
1140:MacVector
1070:denatured
808:oncogenes
734:The term
514:noncoding
502:deletions
150:The term
103:replicons
4617:Ribozyme
4562:Phagemid
4289:Families
4149:Protista
4133:Animalia
4098:Bacteria
3948:Organism
3941:See also
3917:Sulphobe
3894:Ribozyme
3889:RNA life
3796:Mitosome
3740:Prophage
3735:Provirus
3723:Replicon
3679:Circular
3626:Phagemid
3543:Mobilome
3535:elements
3445:Virusoid
3368:Subviral
3280:Protista
3265:Animalia
3250:Bacteria
3172:20230618
3123:19649988
3098:18633447
3059:18537590
2929:Episomes
2835:18326073
2733:24075874
2642:21255361
2527:10948433
2483:10971576
2366:13561654
2339:11367145
2331:20093737
2206:19656584
2167:23830144
2120:24769900
2068:19780633
2060:20496146
2001:30559732
1975:: 2932.
1950:20080407
1891:: 1–56.
1794:21495844
1741:23829072
1665:35960657
1525:13003535
1447:20805406
1268:VectorDB
1243:Provirus
1170:See also
1120:software
957:Zea mays
852:antidote
784:poxvirus
724:Episomes
427:Chromids
387:pathogen
369:proteins
351:sex pili
323:sex pili
205:episomes
192:replicon
176:episomes
124:mobilome
61:bacteria
32:plasmoid
18:Plasmids
4649:Xenobot
4567:Plasmid
4548:Jeewanu
4479:Obelisk
4275:Classes
4143:Plantae
4108:Plastid
4093:Archaea
3930:Jeewanu
3844:Organs
3808:Plastid
3608:Cryptic
3577:Plasmid
3275:Plantae
3255:Archaea
3163:2847955
2972:9862961
2782:9016599
2691:7045080
2633:3815339
2593:3517851
2561:Bibcode
2128:9826684
1992:6284228
1915:2684889
1656:9484753
1438:2937521
1398:9873943
1202:Science
1190:Biology
1176:Portals
1156:Addgene
1046:in vivo
1036:relaxed
942:C.album
940:(as in
838:hok/sok
764:episome
759:episome
752:episome
748:plasmid
736:episome
730:Episome
643:insulin
616:cosmids
580:ligated
560:Cloning
476:express
468:vectors
456:Vectors
433:Chromid
389:. e.g.
376:toluene
196:iterons
152:plasmid
146:History
108:viruses
83:vectors
65:archaea
49:plasmid
4595:Retron
4538:Fosmid
4523:Cosmid
4473:Nanobe
4433:Genera
4215:Realms
4203:Viroid
4021:Virome
3999:Nanobe
3696:Genome
3674:Linear
3619:Fosmid
3614:Cosmid
3379:Viroid
3370:agents
3170:
3160:
3146:: 20.
3121:
3096:
3057:
2970:
2963:148196
2960:
2916:
2893:
2833:
2780:
2773:146482
2770:
2731:
2689:
2682:220260
2679:
2640:
2630:
2591:
2584:323463
2581:
2525:
2481:
2444:
2417:
2390:
2364:
2337:
2329:
2285:
2260:
2231:
2204:
2165:
2126:
2118:
2066:
2058:
1999:
1989:
1948:
1913:
1903:
1846:
1817:
1792:
1784:
1739:
1731:
1690:
1663:
1653:
1610:
1557:
1523:
1505:
1476:
1445:
1435:
1396:
1321:
1296:
1042:Linear
877:Yeasts
848:poison
774:, and
677:genome
525:pBR322
449:Vibrio
266:linear
128:capsid
4572:Prion
4466:Other
4199:Virus
4138:Fungi
3495:Prion
3466:Other
3313:Virus
3270:Fungi
3026:(PDF)
3011:(PDF)
2999:(PDF)
2984:(PDF)
2534:(PDF)
2501:(PDF)
2335:S2CID
2307:(PDF)
2124:S2CID
2086:(PDF)
2064:S2CID
1768:(1).
669:genes
622:, or
4511:HeLa
4115:LUCA
4063:Life
4009:HeLa
3953:Cell
3701:Gene
3168:PMID
3119:PMID
3094:PMID
3055:PMID
2968:PMID
2914:ISBN
2891:ISBN
2831:PMID
2796:link
2778:PMID
2729:PMID
2687:PMID
2638:PMID
2589:PMID
2523:PMID
2479:PMID
2442:ISBN
2415:ISBN
2388:ISBN
2362:PMID
2327:PMID
2283:ISBN
2258:ISBN
2229:ISBN
2202:PMID
2163:PMID
2116:PMID
2056:PMID
1997:PMID
1946:PMID
1911:PMID
1901:ISBN
1844:ISBN
1815:ISBN
1790:PMID
1782:ISSN
1737:PMID
1729:ISSN
1688:ISBN
1661:PMID
1608:ISBN
1555:ISBN
1521:PMID
1474:ISBN
1443:PMID
1394:PMID
1319:ISBN
1294:ISBN
1158:and
1056:(or
927:and
802:and
742:and
543:and
539:for
535:and
378:and
247:and
200:DnaA
140:cell
112:life
67:and
4475:(?)
4179:(?)
3592:Col
3480:DNA
3477:RNA
3456:DNA
3453:RNA
3158:PMC
3148:doi
3084:doi
3047:doi
2958:PMC
2950:doi
2823:doi
2768:PMC
2760:doi
2721:doi
2717:100
2677:PMC
2669:doi
2665:151
2628:PMC
2620:doi
2579:PMC
2569:doi
2513:doi
2471:doi
2467:267
2358:247
2319:doi
2194:doi
2155:doi
2106:hdl
2098:doi
2046:doi
1987:PMC
1977:doi
1938:doi
1893:doi
1889:117
1774:doi
1721:doi
1651:PMC
1641:doi
1513:doi
1466:doi
1433:PMC
1425:doi
1124:DNA
608:kbp
594:or
549:ori
537:tet
533:amp
491:).
470:in
393:in
343:tra
207:in
85:in
4666::
4065:,
3604:Ti
3166:.
3156:.
3144:10
3142:.
3138:.
3115:11
3113:.
3109:.
3092:.
3080:15
3078:.
3074:.
3053:.
3041:.
3037:.
3019:10
3017:.
3013:.
2990:.
2986:.
2966:.
2956:.
2946:27
2944:.
2940:.
2908:.
2851:.
2829:.
2817:.
2813:.
2792:}}
2788:{{
2776:.
2766:.
2756:25
2754:.
2750:.
2727:.
2715:.
2711:.
2699:^
2685:.
2675:.
2663:.
2659:.
2636:.
2626:.
2614:.
2610:.
2587:.
2577:.
2567:.
2557:83
2555:.
2551:.
2521:.
2509:29
2507:.
2503:.
2477:.
2465:.
2356:,
2333:.
2325:.
2315:34
2313:.
2309:.
2200:.
2190:27
2188:.
2184:.
2161:.
2151:31
2149:.
2145:.
2122:.
2114:.
2104:.
2094:98
2092:.
2088:.
2062:.
2054:.
2042:87
2040:.
2036:.
2009:^
1995:.
1985:.
1971:.
1967:.
1944:.
1934:18
1932:.
1909:.
1899:.
1887:.
1866:.
1788:.
1780:.
1766:49
1764:.
1735:.
1727:.
1717:62
1715:.
1711:.
1682:.
1659:.
1649:.
1635:.
1631:.
1602:.
1588:^
1578:.
1533:^
1519:.
1511:.
1499:32
1497:.
1472:.
1464:.
1441:.
1431:.
1421:74
1419:.
1415:.
1390:45
1388:.
1355:.
955:,
950:,
893:.
770:,
720:.
645:.
618:,
603:.
367:,
278:.
211:.
198:,
136:bp
47:A
4558:"
4552:"
4151:'
4147:'
4086:"
4082:"
4055:e
4048:t
4041:v
3997:?
3991:?
3900:†
3887:?
3602:/
3447:)
3431:)
3225:e
3218:t
3211:v
3174:.
3150::
3100:.
3086::
3049::
3043:8
2992:6
2974:.
2952::
2922:.
2899:.
2855:.
2837:.
2825::
2819:8
2798:)
2784:.
2762::
2735:.
2723::
2693:.
2671::
2644:.
2622::
2616:3
2595:.
2571::
2563::
2515::
2485:.
2473::
2450:.
2423:.
2396:.
2341:.
2321::
2291:.
2266:.
2237:.
2208:.
2196::
2169:.
2157::
2130:.
2108::
2100::
2070:.
2048::
2003:.
1979::
1973:9
1952:.
1940::
1917:.
1895::
1870:.
1852:.
1823:.
1796:.
1776::
1743:.
1723::
1696:.
1667:.
1643::
1637:8
1616:.
1582:.
1563:.
1527:.
1515::
1482:.
1468::
1449:.
1427::
1400:.
1373:.
1359:.
1341:.
1327:.
1302:.
1178::
479:(
382:.
353:.
182:.
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.