355:
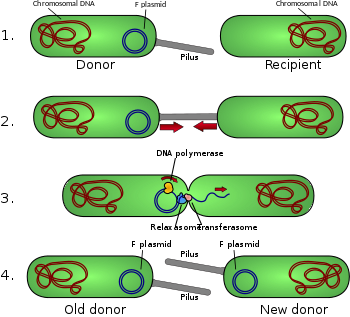
chromosome is being replicated. 5.The pilus detaches from the recipient cell and retracts. The Hfr cell ideally wants to transfer its entire genome to the recipient cell. However, due to its large size and inability to keep in contact with the recipient cell, it is not able to do so. 6.a. The F- cell remains F- because the entire F factor sequence was not received. Since no homologous recombination occurred, the DNA that was transferred is degraded by enzymes. b. In very rare cases, the F factor will be completely transferred and the F- cell will become an Hfr cell.
485:
system for exchange of DNA) and Ted (Thermoproteales system for exchange of DNA), appears to be responsible for the transfer of cellular DNA between members of the same species. It has been suggested that in these archaea the conjugation machinery has been fully domesticated for promoting DNA repair through homologous recombination rather than spread of mobile genetic elements. In addition to the VirB2-like conjugative pilus, the Ced and Ted systems include components for the VirB6-like transmembrane mating pore and the VirB4-like ATPase.
37:
335:
494:
168:
373:
A cell culture that contains in its population cells with non-integrated F-plasmids usually also contains a few cells that have accidentally integrated their plasmids. It is these cells that are responsible for the low-frequency chromosomal gene transfers that occur in such cultures. Some strains of
359:
If the F-plasmid that is transferred has previously been integrated into the donor's genome (producing an Hfr strain ) some of the donor's chromosomal DNA may also be transferred with the plasmid DNA. The amount of chromosomal DNA that is transferred depends on how long the two conjugating bacteria
484:
encode pili structurally similar to the bacterial conjugative pili. However, unlike in bacteria, where conjugation apparatus typically mediates the transfer of mobile genetic elements, such as plasmids or transposons, the conjugative machinery of hyperthermophilic archaea, called Ced (Crenarchaeal
280:
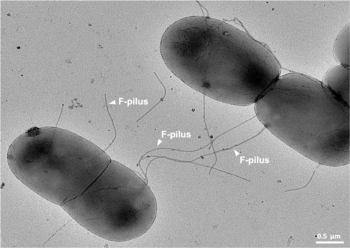
that attach themselves to the surface of F bacteria and initiate conjugation. Though there is some debate on the exact mechanism of conjugation it seems that the pili are the structures through which DNA exchange occurs. The F-pili are extremely resistant to mechanical and thermochemical stress,
354:
and creates an Hfr (high frequency of recombination) cell. 2.The Hfr cell forms a pilus and attaches to a recipient F- cell. 3.A nick in one strand of the Hfr cell's chromosome is created. 4.DNA begins to be transferred from the Hfr cell to the recipient cell while the second strand of its
471:
all regions of the chromosome are transferred with comparable efficiencies. The lengths of the donor segments vary widely, but have an average length of 44.2kb. Since a mean of 13 tracts are transferred, the average total of transferred DNA per genome is 575kb. This process is referred to as
399:
was originally mapped by interrupted mating experiments in which various Hfr cells in the process of conjugation were sheared from recipients after less than 100 minutes (initially using a Waring blender). The genes that were transferred were then investigated.
1224:
Beltran, Leticia C.; Cvirkaite-Krupovic, Virginija; Miller, Jessalyn; Wang, Fengbin; Kreutzberger, Mark A. B.; Patkowski, Jonasz B.; Costa, Tiago R. D.; Schouten, Stefan; Levental, Ilya; Conticello, Vincent P.; Egelman, Edward H.; Krupovic, Mart (2023-02-07).
1361:
Pan, Shen Q.; Jin, Shouguang; Boulton, Margaret I.; Hawes, Martha; Gordon, Milton P.; Nester, Eugene W. (July 1995). "An
Agrobacterium virulence factor encoded by a Ti plasmid gene or a chromosomal gene is required for T-DNA transfer into plants".
407:
chromosome is a rare spontaneous occurrence, and since the numerous genes promoting DNA transfer are in the plasmid genome rather than in the bacterial genome, it has been argued that conjugative bacterial gene transfer, as it occurs in the
319:, is then unwound from the unbroken strand and transferred to the recipient cell in a 5'-terminus to 3'-terminus direction. The remaining strand is replicated either independent of conjugative action (vegetative replication beginning at the
472:"Distributive conjugal transfer." Gray et al. found substantial blending of the parental genomes as a result of conjugation and regarded this blending as reminiscent of that seen in the meiotic products of sexual reproduction.
331:). Conjugative replication may require a second nick before successful transfer can occur. A recent report claims to have inhibited conjugation with chemicals that mimic an intermediate step of this second nicking event.
1025:
Patkowski, Jonasz B.; Dahlberg, Tobias; Amin, Himani; Gahlot, Dharmender K.; Vijayrajratnam, Sukhithasri; Vogel, Joseph P.; Francis, Matthew S.; Baker, Joseph L.; Andersson, Magnus; Costa, Tiago R. D. (5 April 2023).
459:, requires stable and extended contact between a donor and a recipient strain, is DNase resistant, and the transferred DNA is incorporated into the recipient chromosome by homologous recombination. However, unlike
1558:
Karas, Bogumil J.; Diner, Rachel E.; Lefebvre, Stephane C.; McQuaid, Jeff; Phillips, Alex P.R.; Noddings, Chari M.; Brunson, John K.; Valas, Ruben E.; Deerinck, Thomas J. (2015-04-21).
46:
undergoing bacterial conjugation using F-pili. These long and extremely robust extracellular appendages serve as physical conduits for translocation of DNA. Adapted from
315:. In the F-plasmid system the relaxase enzyme is called TraI and the relaxosome consists of TraI, TraY, TraM and the integrated host factor IHF. The nicked strand, or
194:
Both cells synthesize a complementary strand to produce a double stranded circular plasmid and also reproduce pili; both cells are now viable donor for the F-factor.
374:
bacteria with an integrated F-plasmid can be isolated and grown in pure culture. Because such strains transfer chromosomal genes very efficiently they are called
863:"Distributive Conjugal Transfer in Mycobacteria Generates Progeny with Meiotic-Like Genome-Wide Mosaicism, Allowing Mapping of a Mating Identity Locus"
289:
locus seem to open a channel between the bacteria and it is thought that the traD enzyme, located at the base of the pilus, initiates membrane fusion.
366:
the transfer of the entire bacterial chromosome takes about 100 minutes. The transferred DNA can then be integrated into the recipient genome via
1702:
1674:
77:
since it involves the exchange of genetic material. However, it is not sexual reproduction, since no exchange of gamete occurs, and indeed no
1859:
230:. There can only be one copy of the F-plasmid in a given bacterium, either free or integrated, and bacteria that possess a copy are called
531:
contain genes that are capable of transferring to plant cells. The expression of these genes effectively transforms the plant cells into
1405:
Heinemann, Jack A.; Sprague, George F. (July 1989). "Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast".
578:
to a variety of targets. In laboratories, successful transfers have been reported from bacteria to yeast, plants, mammalian cells,
1028:"The F-pilus biomechanical adaptability accelerates conjugative dissemination of antimicrobial resistance and biofilm formation"
667:
794:
755:
721:
745:
1695:
1168:
427:, a form of conjugation referred to as spontaneous zygogenesis (Z-mating for short) is observed in certain strains of
1844:
681:
1456:
Kunik, Tayla; Tzfira, Tzvi; Kapulnik, Yoram; Gafni, Yedidya; Dingwall, Colin; Citovsky, Vitaly (13 February 2001).
412:
Hfr system, is not an evolutionary adaptation of the bacterial host, nor is it likely ancestral to eukaryotic sex.
921:"Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria"
463:
Hfr conjugation, mycobacterial conjugation is chromosome rather than plasmid based. Furthermore, in contrast to
1688:
1680:
586:. Conjugation has advantages over other forms of genetic transfer including minimal disruption of the target's
211:
1075:
Lujan, Scott A.; Guogas, Laura M.; Ragonese, Heather; Matson, Steven W.; Redinbo, Matthew R. (24 July 2007).
535:-producing factories. Opines are used by the bacteria as sources of nitrogen and energy. Infected cells form
26:
by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a
861:
Gray, Todd A.; Krywy, Janet A.; Harold, Jessica; Palumbo, Michael J.; Derbyshire, Keith M. (9 July 2013).
343:
199:
281:
which guarantees successful conjugation in a variety of environments. Several proteins coded for in the
1864:
590:
and the ability to transfer relatively large amounts of genetic material (see the above discussion of
1790:
1746:
536:
55:
1785:
1715:
540:
367:
351:
346:
plasmid and the chromosome have similar sequences, allowing the F factor to insert itself into the
51:
191:
The mobile plasmid is nicked and a single strand of DNA is then transferred to the recipient cell.
1741:
1719:
131:
59:
1854:
104:
The genetic information transferred is often beneficial to the recipient. Benefits may include
1849:
1770:
440:
219:
105:
1375:
1795:
1571:
1469:
1414:
1307:
1238:
1183:
1088:
981:
599:
309:. Relaxase may work alone or in a complex of over a dozen proteins known collectively as a
1515:
Waters, Virginia L. (December 2001). "Conjugation between bacterial and mammalian cells".
8:
1291:
1143:
1077:"Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase"
936:
575:
70:
1575:
1473:
1418:
1311:
1242:
1187:
1092:
1052:
1027:
985:
1649:
1624:
1600:
1559:
1540:
1438:
1387:
1338:
1295:
1267:
1226:
1119:
1106:
1076:
1007:
945:
920:
889:
862:
637:
626:
558:) that is different and independent of the system used for inter-kingdom transfer (the
547:
of the bacteria, which are in turn endosymbionts (or parasites) of the infected plant.
339:
815:"Spontaneous zygogenesis in Escherichia coli, a form of true sexuality in prokaryotes"
435:
are formed that throw off phenotypically haploid cells, of which some show a parental
1654:
1605:
1587:
1532:
1497:
1492:
1457:
1430:
1379:
1343:
1325:
1272:
1254:
1199:
1124:
1057:
999:
950:
894:
836:
790:
751:
687:
677:
642:
587:
566:, operon). Such transfers create virulent strains from previously avirulent strains.
518:
1391:
550:
The Ti and Ri plasmids can also be transferred between bacteria using a system (the
1644:
1636:
1595:
1579:
1524:
1487:
1477:
1442:
1422:
1371:
1333:
1315:
1262:
1246:
1191:
1114:
1096:
1047:
1039:
1011:
989:
940:
932:
884:
874:
826:
603:
362:
257:
152:
125:
42:
1813:
1775:
1544:
1195:
879:
602:(TMV). While TMV is capable of infecting many plant families these are primarily
1710:
1227:"Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery"
89:
cell provides a conjugative or mobilizable genetic element that is most often a
1300:
Proceedings of the
National Academy of Sciences of the United States of America
1250:
1043:
761:
324:
215:
666:
Holmes, Randall K.; Jobling, Michael G. (1996). "Genetics". In Baron S (ed.).
1838:
1591:
1329:
1258:
555:
523:
475:
139:
conjugation in that these cases involve substantial blending of the parental
1625:"Transformation of isolated mammalian mitochondria by bacterial conjugation"
1320:
1101:
1818:
1765:
1658:
1609:
1536:
1501:
1482:
1347:
1276:
1203:
1128:
1061:
1003:
954:
898:
840:
691:
632:
583:
544:
328:
156:
135:
by distributive conjugal transfer differ from the better studied classical
1434:
1383:
831:
814:
1823:
1780:
1640:
323:) or in concert with conjugation (conjugative replication similar to the
113:
62:
although these two other mechanisms do not involve cell-to-cell contact.
1110:
36:
1728:
1583:
509:
311:
302:
207:
117:
109:
94:
78:
69:
bacterial conjugation is often regarded as the bacterial equivalent of
1167:
Michod, Richard E.; Bernstein, Harris; Nedelcu, Aurora M. (May 2008).
1757:
1426:
994:
969:
563:
436:
1289:
493:
334:
1711:
1223:
513:
423:
In addition to classical bacterial conjugation described above for
376:
294:
188:
Pilus attaches to recipient cell and brings the two cells together.
116:. Other elements can be detrimental and may be viewed as bacterial
23:
1528:
614:
481:
277:
203:
90:
81:: instead an existing organism is transformed. During classical
1290:
van
Wolferen, Marleen; Wagner, Alexander; van der Does, Chris;
579:
532:
432:
396:
347:
298:
276:
on the cell surface. The locus also includes the genes for the
140:
74:
598:-like conjugation complements other standard vehicles such as
431:. In Z-mating there is complete genetic mixing, and unstable
260:, which together are about 33 kb long and consist of about 40
167:
1805:
1560:"Designer diatom episomes delivered by bacterial conjugation"
606:
521:
conjugation. For example, the tumor-inducing (Ti) plasmid of
273:
182:
27:
1024:
261:
97:. Most conjugative plasmids have systems ensuring that the
704:
476:
Conjugation-like DNA transfer in hyperthermophilic archaea
1557:
1074:
613:-like conjugation is also primarily used for dicots, but
249:
Among other genetic information, the F-plasmid carries a
1455:
305:
in one of the strands of the conjugative plasmid at the
206:(a plasmid that can integrate itself into the bacterial
1458:"Genetic transformation of HeLa cells by Agrobacterium"
919:
Derbyshire, Keith M.; Gray, Todd A. (17 January 2014).
789:(4th ed.). New York: McGraw Hill. pp. 60–64.
1166:
860:
813:
Gratia, Jean-Pierre; Thiry, Marc (1 September 2003).
446:
569:
360:remain in contact. In common laboratory strains of
238:(denoted F). Cells that lack F plasmids are called
1360:
914:
912:
910:
908:
246:(F) and as such can function as recipient cells.
101:cell does not already contain a similar element.
1836:
1404:
968:Lederberg, Joshua; Tatum, E. L. (October 1946).
739:
737:
735:
1462:Proceedings of the National Academy of Sciences
1081:Proceedings of the National Academy of Sciences
905:
272:gene and regulatory genes, which together form
1169:"Adaptive value of sex in microbial pathogens"
967:
918:
808:
806:
676:(4th ed.). Univ of Texas Medical Branch.
292:When conjugation is initiated by a signal the
1696:
856:
854:
852:
850:
750:(7th ed.). San Francisco: W.H. Freeman.
732:
722:"F-pilus, the ultimate bacterial sex machine"
665:
594:chromosome transfer). In plant engineering,
527:and the root-tumor inducing (Ri) plasmid of
403:Since integration of the F-plasmid into the
32:parasexual mode of reproduction in bacteria.
22:is the transfer of genetic material between
1623:Yoon, Young Geol; Koob, Michael D. (2005).
812:
803:
171:Schematic drawing of bacterial conjugation.
1703:
1689:
847:
1648:
1599:
1491:
1481:
1337:
1319:
1266:
1118:
1100:
1051:
993:
944:
888:
878:
830:
780:
778:
743:
719:
488:
1622:
970:"Gene Recombination in Escherichia Coli"
492:
333:
166:
35:
1376:10.1111/j.1365-2958.1995.mmi_17020259.x
784:
726:Nature Portfolio Microbiology Community
661:
659:
657:
1837:
1514:
775:
698:
574:Conjugation is a convenient means for
1684:
1296:"The archaeal Ced system imports DNA"
1219:
1217:
1215:
1213:
937:10.1128/microbiolspec.MGM2-0022-2013
785:Ryan, K.J.; Ray, C.G., eds. (2004).
654:
112:tolerance or the ability to use new
1860:Modification of genetic information
747:An Introduction to genetic analysis
720:Patkowski, Jonasz (21 April 2023).
705:Dr.T.S.Ramarao M.sc, Ph.D. (1991).
13:
1210:
543:. The Ti and Ri plasmids are thus
129:by spontaneous zygogenesis and in
14:
1876:
1668:
1176:Infection, Genetics and Evolution
1141:
517:are an interesting case of inter-
447:Conjugal transfer in mycobacteria
16:Method of bacterial gene transfer
570:Genetic engineering applications
226:, and an origin of transfer, or
1616:
1551:
1508:
1449:
1398:
1354:
1283:
1160:
1135:
1068:
1018:
961:
713:
151:The process was discovered by
1:
648:
617:recipients are not uncommon.
576:transferring genetic material
214:) with a length of about 100
1196:10.1016/j.meegid.2008.01.002
880:10.1371/journal.pbio.1001602
787:Sherris Medical Microbiology
673:Baron's Medical Microbiology
350:of the cell. This is called
162:
79:generation of a new organism
7:
620:
416:Spontaneous zygogenesis in
10:
1881:
1251:10.1038/s41467-023-36349-8
1044:10.1038/s41467-023-37600-y
146:
1804:
1791:Sister chromatid exchange
1755:
1726:
498:Agrobacterium tumefaciens
1845:Antimicrobial resistance
1786:Horizontal gene transfer
1716:homologous recombination
1148:www.microbiologybook.org
508:Bacteria related to the
368:homologous recombination
352:homologous recombination
212:homologous recombination
52:horizontal gene transfer
40:A micrograph displaying
1720:mobile genetic elements
1321:10.1073/pnas.1513740113
1102:10.1073/pnas.0702760104
744:Griffiths, AJF (1999).
669:Genetics: Conjugation.
582:and isolated mammalian
132:Mycobacterium smegmatis
1629:Nucleic Acids Research
1483:10.1073/pnas.041327598
1364:Molecular Microbiology
505:
489:Inter-kingdom transfer
455:, like conjugation in
453:Mycobacteria smegmatis
356:
172:
47:
1771:Chromosomal crossover
1675:Bacterial conjugation
1564:Nature Communications
1231:Nature Communications
1032:Nature Communications
925:Microbiology Spectrum
832:10.1099/mic.0.26348-0
629:in algae and ciliates
496:
342:(yellow) on both the
337:
220:origin of replication
218:. It carries its own
170:
106:antibiotic resistance
50:It is a mechanism of
39:
20:Bacterial conjugation
1292:Albers, Sonja-Verena
707:B.sc Botany-Volume-1
600:tobacco mosaic virus
500:gall at the root of
467:Hfr conjugation, in
181:Donor cell produces
1677:(a Flash animation)
1576:2015NatCo...6.6925K
1474:2001PNAS...98.1871K
1419:1989Natur.340..205H
1312:2016PNAS..113.2496V
1243:2023NatCo..14..666B
1188:2008InfGE...8..267M
1093:2007PNAS..10412282L
1087:(30): 12282–12287.
986:1946Natur.158..558L
392:ecombination). The
340:insertion sequences
268:locus includes the
176:Conjugation diagram
71:sexual reproduction
1641:10.1093/nar/gni140
1584:10.1038/ncomms7925
1144:"Genetic Exchange"
638:Triparental mating
627:Sexual conjugation
506:
480:Hyperthermophilic
439:and some are true
357:
173:
48:
1865:Molecular biology
1832:
1831:
1413:(6230): 205–209.
796:978-0-8385-8529-0
757:978-0-7167-3520-5
643:Zygotic induction
588:cellular envelope
502:Carya illinoensis
1872:
1705:
1698:
1691:
1682:
1681:
1663:
1662:
1652:
1620:
1614:
1613:
1603:
1555:
1549:
1548:
1512:
1506:
1505:
1495:
1485:
1468:(4): 1871–1876.
1453:
1447:
1446:
1427:10.1038/340205a0
1402:
1396:
1395:
1358:
1352:
1351:
1341:
1323:
1306:(9): 2496–2501.
1287:
1281:
1280:
1270:
1221:
1208:
1207:
1173:
1164:
1158:
1157:
1155:
1154:
1139:
1133:
1132:
1122:
1104:
1072:
1066:
1065:
1055:
1022:
1016:
1015:
997:
995:10.1038/158558a0
965:
959:
958:
948:
916:
903:
902:
892:
882:
858:
845:
844:
834:
825:(9): 2571–2584.
810:
801:
800:
782:
773:
772:
770:
769:
760:. Archived from
741:
730:
729:
717:
711:
710:
702:
696:
695:
663:
556:transfer, operon
153:Joshua Lederberg
126:Escherichia coli
85:conjugation the
43:Escherichia coli
1880:
1879:
1875:
1874:
1873:
1871:
1870:
1869:
1835:
1834:
1833:
1828:
1814:Antigenic shift
1800:
1776:Gene conversion
1751:
1722:
1709:
1671:
1666:
1621:
1617:
1556:
1552:
1517:Nature Genetics
1513:
1509:
1454:
1450:
1403:
1399:
1359:
1355:
1288:
1284:
1222:
1211:
1171:
1165:
1161:
1152:
1150:
1140:
1136:
1073:
1069:
1023:
1019:
966:
962:
917:
906:
873:(7): e1001602.
859:
848:
811:
804:
797:
783:
776:
767:
765:
758:
742:
733:
718:
714:
703:
699:
684:
664:
655:
651:
623:
572:
510:nitrogen fixing
491:
478:
451:Conjugation in
449:
327:replication of
165:
149:
123:Conjugation in
24:bacterial cells
17:
12:
11:
5:
1878:
1868:
1867:
1862:
1857:
1852:
1847:
1830:
1829:
1827:
1826:
1821:
1816:
1810:
1808:
1802:
1801:
1799:
1798:
1793:
1788:
1783:
1778:
1773:
1768:
1762:
1760:
1753:
1752:
1750:
1749:
1747:Transformation
1744:
1739:
1733:
1731:
1724:
1723:
1708:
1707:
1700:
1693:
1685:
1679:
1678:
1670:
1669:External links
1667:
1665:
1664:
1615:
1550:
1507:
1448:
1397:
1370:(2): 259–269.
1353:
1294:(2016-03-01).
1282:
1209:
1182:(3): 267–285.
1159:
1134:
1067:
1017:
960:
904:
846:
802:
795:
774:
756:
731:
712:
697:
682:
652:
650:
647:
646:
645:
640:
635:
630:
622:
619:
571:
568:
490:
487:
477:
474:
448:
445:
325:rolling circle
196:
195:
192:
189:
186:
164:
161:
148:
145:
56:transformation
15:
9:
6:
4:
3:
2:
1877:
1866:
1863:
1861:
1858:
1856:
1855:Biotechnology
1853:
1851:
1848:
1846:
1843:
1842:
1840:
1825:
1822:
1820:
1817:
1815:
1812:
1811:
1809:
1807:
1803:
1797:
1794:
1792:
1789:
1787:
1784:
1782:
1779:
1777:
1774:
1772:
1769:
1767:
1764:
1763:
1761:
1759:
1754:
1748:
1745:
1743:
1740:
1738:
1735:
1734:
1732:
1730:
1725:
1721:
1717:
1713:
1706:
1701:
1699:
1694:
1692:
1687:
1686:
1683:
1676:
1673:
1672:
1660:
1656:
1651:
1646:
1642:
1638:
1634:
1630:
1626:
1619:
1611:
1607:
1602:
1597:
1593:
1589:
1585:
1581:
1577:
1573:
1569:
1565:
1561:
1554:
1546:
1542:
1538:
1534:
1530:
1529:10.1038/ng779
1526:
1522:
1518:
1511:
1503:
1499:
1494:
1489:
1484:
1479:
1475:
1471:
1467:
1463:
1459:
1452:
1444:
1440:
1436:
1432:
1428:
1424:
1420:
1416:
1412:
1408:
1401:
1393:
1389:
1385:
1381:
1377:
1373:
1369:
1365:
1357:
1349:
1345:
1340:
1335:
1331:
1327:
1322:
1317:
1313:
1309:
1305:
1301:
1297:
1293:
1286:
1278:
1274:
1269:
1264:
1260:
1256:
1252:
1248:
1244:
1240:
1236:
1232:
1228:
1220:
1218:
1216:
1214:
1205:
1201:
1197:
1193:
1189:
1185:
1181:
1177:
1170:
1163:
1149:
1145:
1142:Mayer, Gene.
1138:
1130:
1126:
1121:
1116:
1112:
1108:
1103:
1098:
1094:
1090:
1086:
1082:
1078:
1071:
1063:
1059:
1054:
1049:
1045:
1041:
1037:
1033:
1029:
1021:
1013:
1009:
1005:
1001:
996:
991:
987:
983:
980:(4016): 558.
979:
975:
971:
964:
956:
952:
947:
942:
938:
934:
930:
926:
922:
915:
913:
911:
909:
900:
896:
891:
886:
881:
876:
872:
868:
864:
857:
855:
853:
851:
842:
838:
833:
828:
824:
820:
816:
809:
807:
798:
792:
788:
781:
779:
764:on 2020-02-08
763:
759:
753:
749:
748:
740:
738:
736:
727:
723:
716:
708:
701:
693:
689:
685:
683:0-9631172-1-1
679:
675:
674:
670:
662:
660:
658:
653:
644:
641:
639:
636:
634:
631:
628:
625:
624:
618:
616:
612:
611:Agrobacterium
608:
605:
601:
597:
596:Agrobacterium
593:
589:
585:
581:
577:
567:
565:
561:
557:
553:
548:
546:
545:endosymbionts
542:
538:
534:
530:
529:A. rhizogenes
526:
525:
524:Agrobacterium
520:
516:
515:
511:
503:
499:
495:
486:
483:
473:
470:
466:
462:
458:
454:
444:
442:
438:
434:
430:
426:
421:
420:
419:
413:
411:
406:
401:
398:
395:
391:
387:
383:
379:
378:
371:
369:
365:
364:
353:
349:
345:
341:
336:
332:
330:
326:
322:
318:
314:
313:
308:
304:
300:
297:
296:
290:
288:
284:
279:
275:
271:
267:
263:
259:
256:
252:
247:
245:
241:
237:
233:
229:
225:
221:
217:
213:
209:
205:
201:
193:
190:
187:
184:
180:
179:
178:
177:
169:
160:
158:
154:
144:
142:
138:
134:
133:
128:
127:
121:
119:
115:
111:
107:
102:
100:
96:
92:
88:
84:
80:
76:
72:
68:
63:
61:
57:
53:
45:
44:
38:
34:
33:
29:
25:
21:
1850:Bacteriology
1819:Reassortment
1766:Transfection
1742:Transduction
1736:
1635:(16): e139.
1632:
1628:
1618:
1567:
1563:
1553:
1523:(4): 375–6.
1520:
1516:
1510:
1465:
1461:
1451:
1410:
1406:
1400:
1367:
1363:
1356:
1303:
1299:
1285:
1234:
1230:
1179:
1175:
1162:
1151:. Retrieved
1147:
1137:
1084:
1080:
1070:
1035:
1031:
1020:
977:
973:
963:
928:
924:
870:
867:PLOS Biology
866:
822:
819:Microbiology
818:
786:
766:. Retrieved
762:the original
746:
725:
715:
706:
700:
672:
668:
633:Transfection
610:
595:
591:
584:mitochondria
573:
559:
551:
549:
528:
522:
512:
507:
501:
497:
479:
469:M. smegmatis
468:
464:
460:
456:
452:
450:
441:recombinants
428:
424:
422:
417:
415:
414:
409:
404:
402:
393:
389:
388:requency of
385:
381:
375:
372:
361:
358:
329:lambda phage
320:
316:
310:
306:
293:
291:
286:
282:
269:
265:
254:
250:
248:
243:
239:
235:
231:
227:
223:
197:
175:
174:
157:Edward Tatum
150:
136:
130:
124:
122:
103:
98:
86:
82:
66:
64:
60:transduction
49:
41:
31:
19:
18:
1824:Viral shift
1781:Fusion gene
1737:Conjugation
1729:prokaryotic
1038:(1): 1879.
541:root tumors
114:metabolites
1839:Categories
1796:Transposon
1758:eukaryotes
1756:Occurs in
1727:Primarily
1237:(1): 666.
1153:2017-12-04
768:2023-08-11
649:References
604:herbaceous
537:crown gall
312:relaxosome
301:creates a
240:F-negative
232:F-positive
208:chromosome
110:xenobiotic
95:transposon
65:Classical
30:. It is a
1592:2041-1723
1330:1091-6490
1259:2041-1723
564:virulence
437:phenotype
163:Mechanism
159:in 1946.
118:parasites
99:recipient
1712:Genetics
1659:16157861
1610:25897682
1570:: 6925.
1537:11726922
1502:11172043
1392:38483513
1348:26884154
1277:36750723
1204:18295550
1129:17630285
1111:25436291
1062:37019921
1053:10076315
1004:21001945
955:25505644
899:23874149
841:12949181
692:21413277
621:See also
514:Rhizobia
433:diploids
344:F factor
317:T-strand
295:relaxase
278:proteins
200:F-factor
1650:1201378
1601:4411287
1572:Bibcode
1470:Bibcode
1443:4351266
1435:2666856
1415:Bibcode
1384:7494475
1339:4780597
1308:Bibcode
1268:9905601
1239:Bibcode
1184:Bibcode
1120:1916486
1089:Bibcode
1012:1826960
982:Bibcode
946:4259119
890:3706393
615:monocot
592:E. coli
580:diatoms
519:kingdom
482:archaea
465:E. coli
461:E. coli
457:E. coli
429:E. coli
425:E. coli
418:E. coli
410:E. coli
405:E. coli
394:E. coli
363:E. coli
244:F-minus
204:episome
147:History
141:genomes
137:E. coli
91:plasmid
83:E. coli
67:E. coli
54:as are
1657:
1647:
1608:
1598:
1590:
1543:
1535:
1500:
1490:
1441:
1433:
1407:Nature
1390:
1382:
1346:
1336:
1328:
1275:
1265:
1257:
1202:
1127:
1117:
1109:
1060:
1050:
1010:
1002:
974:Nature
953:
943:
897:
887:
839:
793:
754:
690:
680:
607:dicots
397:genome
348:genome
338:1.The
299:enzyme
264:. The
236:F-plus
222:, the
202:is an
75:mating
1806:Viral
1545:27160
1541:S2CID
1493:29349
1439:S2CID
1388:S2CID
1172:(PDF)
1107:JSTOR
1008:S2CID
931:(1).
562:, or
554:, or
533:opine
270:pilin
262:genes
258:locus
183:pilus
87:donor
28:pilus
1655:PMID
1606:PMID
1588:ISSN
1533:PMID
1498:PMID
1431:PMID
1380:PMID
1344:PMID
1326:ISSN
1273:PMID
1255:ISSN
1200:PMID
1125:PMID
1058:PMID
1000:PMID
951:PMID
895:PMID
837:PMID
791:ISBN
752:ISBN
688:PMID
678:ISBN
384:igh
321:oriV
307:oriT
303:nick
274:pili
253:and
228:oriT
224:oriV
198:The
155:and
58:and
1645:PMC
1637:doi
1596:PMC
1580:doi
1525:doi
1488:PMC
1478:doi
1423:doi
1411:340
1372:doi
1334:PMC
1316:doi
1304:113
1263:PMC
1247:doi
1192:doi
1115:PMC
1097:doi
1085:104
1048:PMC
1040:doi
990:doi
978:158
941:PMC
933:doi
885:PMC
875:doi
827:doi
823:149
671:in:
560:vir
552:tra
539:or
377:Hfr
287:trb
285:or
283:tra
266:tra
255:trb
251:tra
242:or
234:or
210:by
93:or
73:or
1841::
1718:/
1714::
1653:.
1643:.
1633:33
1631:.
1627:.
1604:.
1594:.
1586:.
1578:.
1566:.
1562:.
1539:.
1531:.
1521:29
1519:.
1496:.
1486:.
1476:.
1466:98
1464:.
1460:.
1437:.
1429:.
1421:.
1409:.
1386:.
1378:.
1368:17
1366:.
1342:.
1332:.
1324:.
1314:.
1302:.
1298:.
1271:.
1261:.
1253:.
1245:.
1235:14
1233:.
1229:.
1212:^
1198:.
1190:.
1178:.
1174:.
1146:.
1123:.
1113:.
1105:.
1095:.
1083:.
1079:.
1056:.
1046:.
1036:14
1034:.
1030:.
1006:.
998:.
988:.
976:.
972:.
949:.
939:.
927:.
923:.
907:^
893:.
883:.
871:11
869:.
865:.
849:^
835:.
821:.
817:.
805:^
777:^
734:^
724:.
686:.
656:^
609:.
443:.
370:.
216:kb
143:.
120:.
108:,
1704:e
1697:t
1690:v
1661:.
1639::
1612:.
1582::
1574::
1568:6
1547:.
1527::
1504:.
1480::
1472::
1445:.
1425::
1417::
1394:.
1374::
1350:.
1318::
1310::
1279:.
1249::
1241::
1206:.
1194::
1186::
1180:8
1156:.
1131:.
1099::
1091::
1064:.
1042::
1014:.
992::
984::
957:.
935::
929:2
901:.
877::
843:.
829::
799:.
771:.
728:.
709:.
694:.
504:.
390:r
386:f
382:h
380:(
185:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.