340:
immunocompromised patients I need of bone marrow. Gammaretroviruses carrying enhancers were then inserted into patients. The second mechanism is referred to as promoter insertion. Promoters provide our cells with the specific sequences needed to begin translation. Promoter insertion has helped researchers learn more about the HIV virus. The third mechanism is gene inactivation. An example of gene inactivation is using insertional mutagenesis to insert a retrovirus that disrupts the genome of the T cell in leukemia patients and giving them a specific antigen called CAR allowing the T cells to target cancer cells. The final mechanisms is referred to as mRNA 3' end substitution. Our genes occasionally undergo point mutations causing beta-thalassemia that interrupts red blood cell function. To fix this problem the correct gene sequence for the red blood cells are introduced and a substitution is made.
314:
mutations on protein function. Large numbers of mutants may be screened for a particular characteristic by combinatorial analysis. In this technique, multiple positions or short sequences along a DNA strand may be exhaustively modified to obtain a comprehensive library of mutant proteins. The rate of incidence of beneficial variants can be improved by different methods for constructing mutagenesis libraries. One approach to this technique is to extract and replace a portion of the DNA sequence with a library of sequences containing all possible combinations at the desired mutation site. The content of the inserted segment can include sequences of structural significance, immunogenic property, or enzymatic function. A segment may also be inserted randomly into the gene in order to assess structural or functional significance of a particular part of a protein.
336:. Engineered mutations such as these can provide important information in cancer research, such as mechanistic insights into the development of the disease. Retroviruses and transposons are the chief instrumental tools in insertional mutagenesis. Retroviruses, such as the mouse mammory tumor virus and murine leukemia virus, can be used to identify genes involved in carcinogenesis and understand the biological pathways of specific cancers. Transposons, chromosomal segments that can undergo transposition, can be designed and applied to insertional mutagenesis as an instrument for cancer gene discovery. These chromosomal segments allow insertional mutagenesis to be applied to virtually any tissue of choice while also allowing for more comprehensive, unbiased depth in DNA sequencing.
88:
238:, and N-hydroxycytidine may induce a GC to AT transition. These techniques allow specific mutations to be engineered into a protein; however, they are not flexible with respect to the kinds of mutants generated, nor are they as specific as later methods of site-directed mutagenesis and therefore have some degree of randomness. Other technologies such as cleavage of DNA at specific sites on the chromosome, addition of new nucleotides, and exchanging of base pairs it is now possible to decide where mutations can go.
277:
242:
55:. The various constituents of a gene, as well as its regulatory elements and its gene products, may be mutated so that the functioning of a genetic locus, process, or product can be examined in detail. The mutation may produce mutant proteins with interesting properties or enhanced or novel functions that may be of commercial use. Mutant strains may also be produced that have practical application or allow the molecular basis of a particular cell function to be investigated.
75:. Site-directed mutagenesis has proved useful in situations that random mutagenesis is not. Other techniques of mutagenesis include combinatorial and insertional mutagenesis. Mutagenesis that is not random can be used to clone DNA, investigate the effects of mutagens, and engineer proteins. It also has medical applications such as helping immunocompromised patients, research and treatment of diseases including HIV and cancers, and curing of diseases such as
24:
218:
random mutagenesis can produce a change in single nucleotides, it does not offer much control as to which nucleotide is being changed. Many researchers therefore seek to introduce selected changes to DNA in a precise, site-specific manner. Early attempts uses analogs of nucleotides and other chemicals were first used to generate localized
351:
can be used to produce specific mutation in an organism. Vector containing DNA sequence similar to the gene to be modified is introduced to the cell, and by a process of recombination replaces the target gene in the chromosome. This method can be used to introduce a mutation or knock out a gene, for
280:
Site saturation mutagenesis is a type of site-directed mutagenesis. This image shows the saturation mutagenesis of a single position in a theoretical 10-residue protein. The wild type version of the protein is shown at the top, with M representing the first amino acid methionine, and * representing
272:
Site directed mutagenesis allows the effect of specific mutation to be investigated. There are numerous uses; for example, it has been used to determine how susceptible certain species were to chemicals that are often used In labs. The experiment used site directed mutagenesis to mimic the expected
268:
Newer and more efficient methods of site directed mutagenesis are being constantly developed. For example, a technique called "Seamless ligation cloning extract" (or SLiCE for short) allows for the cloning of certain sequences of DNA within the genome, and more than one DNA fragment can be inserted
99:
sample sequence space. The amino acid substituted into a given position is shown. Each dot or set of connected dots is one member of the library. Error-prone PCR randomly mutates some residues to other amino acids. Alanine scanning replaces each residue of the protein with alanine, one-by-one. Site
339:
Researchers have found four mechanisms of insertional mutagenesis that can be used on humans. the first mechanism is called enhancer insertion. Enhancers boost transcription of a particular gene by interacting with a promoter of that gene. This particular mechanism was first used to help severely
313:
Combinatorial mutagenesis is a site-directed protein engineering technique whereby multiple mutants of a protein can be simultaneously engineered based on analysis of the effects of additive individual mutations. It provides a useful method to assess the combinatorial effect of a large number of
217:
Prior to the development site-directed mutagenesis techniques, all mutations made were random, and scientists had to use selection for the desired phenotype to find the desired mutation. Random mutagenesis techniques has an advantage in terms of how many mutations can be produced; however, while
374:-Cas9 technology has allowed for the efficient introduction of different types of mutations into the genome of a wide variety of organisms. The method does not require a transposon insertion site, leaves no marker, and its efficiency and simplicity has made it the preferred method for
58:
Many methods of mutagenesis exist today. Initially, the kind of mutations artificially induced in the laboratory were entirely random using mechanisms such as UV irradiation. Random mutagenesis cannot target specific regions or sequences of the genome; however, with the development of
138:, another in a minimal medium, and mutants that have specific nutritional requirements can then be identified by their inability to grow in the minimal medium. Similar procedures may be repeated with other types of cells and with different media for selection.
157:
reaction in conditions that enhance misincorporation of nucleotides (error-prone PCR), for example by reducing the fidelity of replication or using nucleotide analogues. A variation of this method for integrating non-biased mutations in a gene is
1490:
Cerchione, Derek; Loveluck, Katherine; Tillotson, Eric L.; Harbinski, Fred; DaSilva, Jen; Kelley, Chase P.; Keston-Smith, Elise; Fernandez, Cecilia A.; Myer, Vic E.; Jayaram, Hariharan; Steinberg, Barrett E.; Xu, Shuang-yong (16 April 2020).
390:
is now a viable method for introducing mutations into a gene. This method allows for extensive mutation at multiple sites, including the complete redesign of the codon usage of a gene to optimise it for a particular organism.
1362:
Müller W, Weber H, Meyer F, Weissmann C (September 1978). "Site-directed mutagenesis in DNA: generation of point mutations in cloned beta globin complementary dna at the positions corresponding to amino acids 121 to 123".
1614:
Choi GC, Zhou P, Yuen CT, Chan BK, Xu F, Bao S, Chu HY, Thean D, Tan K, Wong KH, Zheng Z, Wong AS (August 2019). "Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9".
249:
Current techniques for site-specific mutation originates from the primer extension technique developed in 1978. Such techniques commonly involve using pre-fabricated mutagenic oligonucleotides in a
544:"In Silico Site-Directed Mutagenesis Informs Species-Specific Predictions of Chemical Susceptibility Derived From the Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) Tool"
591:
Choi GC, Zhou P, Yuen CT, Chan BK, Xu F, Bao S, et al. (August 2019). "Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9".
488:"A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis"
245:
Simplified diagram of the site directed mutagenic technique using pre-fabricated oligonucleotides in a primer extension reaction with DNA polymerase
1705:
Vassiliou G, Rad R, Bradley A (2010-01-01). "The use of DNA transposons for cancer gene discovery in mice". In
Wassarman PM, Soriano PM (eds.).
265:
of small stretches of DNA at specific sites. Advances in methodology have made such mutagenesis now a relatively simple and efficient process.
104:
Early approaches to mutagenesis relied on methods which produced entirely random mutations. In such methods, cells or organisms are exposed to
273:
mutations of the specific chemical. The mutation resulted in a change in specific amino acids and the effects of this mutation were analyzed.
1447:
Reetz, M. T.; Carballeira J. D. (2007). "Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes".
1935:
1940:
1246:"Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome"
1019:
130:, mutants may be selected first by exposure to UV radiation, then plated onto an agar medium. The colonies formed are then
1657:
Uren AG, Kool J, Berns A, van
Lohuizen M (November 2005). "Retroviral insertional mutagenesis: past, present and future".
293:
in order to identify residues important to the structure or function of a protein. Another comprehensive approach is site
1305:"Site-directed mutagenesis: effect of an extracistronic mutation on the in vitro propagation of bacteriophage Qbeta RNA"
801:
641:"Retroviral Insertional Mutagenesis in Humans: Evidence for Four Genetic Mechanisms Promoting Expansion of Cell Clones"
1875:
1722:
100:
saturation substitutes each of the 20 possible amino acids (or some subset of them) at a single position, one-by-one.
1893:
159:
149:
for mutants with interesting or improved properties. These methods may involve the use of doped nucleotides in
27:
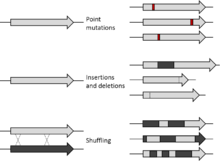
Types of mutations that can be introduced by random, site-directed, combinatorial, or insertional mutagenesis.
689:
200:
52:
910:
Hrabé de
Angelis M, Balling R (May 1998). "Large scale ENU screens in the mouse: genetics meets genomics".
323:
231:
108:
such as UV radiation or mutagenic chemicals, and mutants with desired characteristics are then selected.
1746:
250:
16:
This article is about mutagenesis as a laboratory technique. For mutagenesis as a general process, see
1493:"SMOOT libraries and phage-induced directed evolution of Cas9 to engineer reduced off-target activity"
1912:
387:
212:
154:
60:
348:
1400:"Analysis of Large-Scale Mutagenesis Data To Assess the Impact of Single Amino Acid Substitutions"
333:
117:
87:
327:
294:
281:
the termination of translation. All 19 mutants of the isoleucine at position 5 are shown below.
189:
791:
1865:
1766:"Seamless site-directed mutagenesis of the Saccharomyces cerevisiae genome using CRISPR-Cas9"
542:
Doering JA, Lee S, Kristiansen K, Evenseth L, Barron MG, Sylte I, LaLone CA (November 2018).
410:
227:
109:
1930:
1570:
1504:
1316:
1257:
919:
704:
262:
8:
953:
Flibotte S, Edgley ML, Chaudhry I, Taylor J, Neil SE, Rogula A, et al. (June 2010).
400:
365:
258:
44:
1709:. Methods in Enzymology. Vol. 477 (2nd ed.). Academic Press. pp. 91–106.
1574:
1508:
1320:
1261:
1124:
923:
708:
1841:
1816:
1792:
1765:
1707:
Guide to
Techniques in Mouse Development, Part B: Mouse Molecular Genetics, 2nd Edition
1682:
1639:
1591:
1558:
1527:
1492:
1472:
1424:
1399:
1197:"Isolation and characterization of proline peptidase mutants of Salmonella typhimurium"
1069:
1044:
979:
954:
764:
739:
665:
640:
616:
568:
543:
514:
487:
460:
435:
415:
1714:
1339:
1304:
1280:
1245:
1221:
1196:
1172:
1147:
931:
843:
818:
332:
The insertion of one or more base pairs, resulting in DNA mutations, is also known as
1871:
1846:
1797:
1728:
1718:
1674:
1643:
1631:
1596:
1532:
1464:
1429:
1380:
1376:
1344:
1285:
1226:
1177:
1128:
1074:
1025:
1015:
984:
935:
889:
848:
797:
769:
720:
670:
620:
608:
573:
519:
465:
167:
163:
92:
48:
32:
1686:
1476:
1836:
1828:
1787:
1777:
1710:
1666:
1623:
1586:
1578:
1522:
1512:
1456:
1419:
1411:
1372:
1334:
1324:
1275:
1265:
1216:
1208:
1167:
1159:
1120:
1064:
1056:
1007:
974:
966:
927:
879:
838:
830:
759:
751:
712:
660:
652:
600:
563:
555:
509:
499:
455:
447:
286:
174:
126:
76:
67:/Cas9 technology, based on a prokaryotic viral defense system, has allowed for the
1212:
755:
1517:
150:
131:
1907:
1415:
1045:"Optimizing bacteriophage engineering through an accelerated evolution platform"
1011:
970:
819:"Sequence saturation mutagenesis (SeSaM): a novel method for directed evolution"
656:
1397:
1309:
Proceedings of the
National Academy of Sciences of the United States of America
1250:
Proceedings of the
National Academy of Sciences of the United States of America
1060:
716:
451:
375:
254:
219:
196:
146:
68:
1832:
1782:
1627:
604:
504:
276:
1924:
353:
135:
884:
867:
559:
285:
The site-directed approach may be done systematically in such techniques as
241:
199:
law (as 2001/18 directive), this kind of mutagenesis may be used to produce
1850:
1801:
1732:
1678:
1670:
1635:
1600:
1536:
1468:
1433:
1270:
1078:
1029:
988:
893:
852:
724:
674:
612:
577:
523:
469:
203:
but the products are exempted from regulation: no labeling, no evaluation.
1582:
1460:
1329:
1230:
1181:
1146:
Caras IW, MacInnes MA, Persing DH, Coffino P, Martin DW (September 1982).
1132:
1002:
Bökel C (2008). "EMS Screens: From
Mutagenesis to Screening and Mapping".
939:
773:
1817:"The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans"
1763:
1398:
Vanessa E. Gray; Ronald J. Hause; Douglas M. Fowler (September 1, 2017).
1384:
1289:
1163:
834:
223:
17:
1348:
1148:"Mechanism of 2-aminopurine mutagenesis in mouse T-lymphosarcoma cells"
302:
192:(EMS) is also often used to generate animal, plant, and virus mutants.
405:
235:
436:"Development and applications of CRISPR-Cas9 for genome engineering"
63:, more specific changes can be made. Since 2013, development of the
1489:
121:
955:"Whole-genome profiling of mutagenesis in Caenorhabditis elegans"
290:
142:
105:
1006:. Methods in Molecular Biology. Vol. 420. pp. 119–38.
141:
A number of methods for generating random mutations in specific
1111:
Shortle D, DiMaio D, Nathans D (1981). "Directed mutagenesis".
1094:"GMO directive : the origins of the mutagenesis exemption"
866:
Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A (1999).
371:
120:, and went on to use the mutants he created for his studies in
64:
23:
1093:
796:(3rd ed.). Royal Society of Chemistry. pp. 191–192.
541:
298:
113:
1556:
1303:
Flavell RA, Sabo DL, Bandle EF, Weissmann C (January 1975).
1042:
952:
289:
mutagenesis, whereby residues are systematically mutated to
170:
and the mutant proteins produced can then be characterised.
1145:
1557:
Parker AS, Griswold KE, Bailey-Kellogg C (November 2011).
1302:
865:
1656:
1361:
817:
Wong TS, Tee KL, Hauer B, Schwaneberg U (February 2004).
178:
51:
of mutant genes, proteins, strains of bacteria, or other
40:
905:
903:
301:
or a set of codons may be substituted with all possible
1043:
Favor AH, Llanos CD, Youngblut MD, Bardales JA (2020).
909:
816:
1747:"Homologous Recombination Method (and Knockout Mouse)"
1446:
900:
1764:
Damien Biot-Pelletier; Vincent J. J. Martin (2016).
1110:
386:
As the cost of DNA oligonucleotide synthesis falls,
1864:Khudyakov YE, Fields HA, eds. (25 September 2002).
1704:
740:"Seventy years ago: mutation becomes experimental"
162:. PCR products which contain mutation(s) are then
1700:
1698:
1696:
1922:
1613:
785:
783:
590:
481:
479:
1863:
1693:
1552:
1550:
1548:
1546:
1243:
737:
634:
632:
630:
537:
535:
533:
433:
188:(ENU) have been used to generate mutant mice.
946:
780:
476:
39:is an important laboratory technique whereby
1607:
1194:
859:
308:
257:. This methods allows for point mutation or
206:
1559:"Optimization of combinatorial mutagenesis"
1543:
1085:
627:
530:
343:
1036:
995:
317:
1840:
1791:
1781:
1590:
1526:
1516:
1423:
1338:
1328:
1279:
1269:
1220:
1171:
1068:
978:
883:
842:
789:
763:
664:
567:
513:
503:
485:
459:
1867:Artificial DNA: Methods and Applications
584:
434:Hsu PD, Lander ES, Zhang F (June 2014).
275:
240:
86:
22:
738:Crow JF, Abrahamson S (December 1997).
638:
388:artificial synthesis of a complete gene
96:
1923:
1091:
793:Nucleic Acids in Chemistry and Biology
690:"Artificial Transmutation of the Gene"
687:
1195:McHugh GL, Miller CG (October 1974).
1001:
352:example as used in the production of
82:
1814:
1125:10.1146/annurev.ge.15.120181.001405
13:
1440:
14:
1952:
1887:
1770:Journal of Biological Engineering
1244:Shortle D, Nathans D (May 1978).
381:
1563:Journal of Computational Biology
1857:
1808:
1757:
1739:
1650:
1483:
1391:
1355:
1296:
1237:
1188:
1139:
1104:
370:Since 2013, the development of
160:sequence saturation mutagenesis
116:can cause genetic mutations in
1936:Genetically modified organisms
1152:Molecular and Cellular Biology
810:
731:
681:
427:
53:genetically modified organisms
1:
1715:10.1016/s0076-6879(10)77006-3
1213:10.1128/JB.120.1.364-371.1974
932:10.1016/s0027-5107(98)00061-x
421:
1941:Molecular biology techniques
1913:Resources in other libraries
1518:10.1371/journal.pone.0231716
1377:10.1016/0022-2836(78)90303-0
1365:Journal of Molecular Biology
639:Bushman FD (February 2020).
324:Signature tagged mutagenesis
226:, which induces an AT to GC
7:
1416:10.1534/genetics.117.300064
1012:10.1007/978-1-59745-583-1_7
971:10.1534/genetics.110.116616
756:10.1093/genetics/147.4.1491
657:10.1016/j.ymthe.2019.12.009
394:
305:at the specific positions.
153:synthesis, or conducting a
71:or mutagenesis of a genome
43:mutations are deliberately
10:
1957:
1061:10.1038/s41598-020-70841-1
790:Blackburn GM, ed. (2006).
717:10.1126/science.66.1699.84
452:10.1016/j.cell.2014.05.010
363:
321:
210:
15:
1908:Resources in your library
1870:. CRC Press. p. 13.
1833:10.1016/j.jgg.2015.06.005
1783:10.1186/s13036-016-0028-1
1628:10.1038/s41592-019-0473-0
1113:Annual Review of Genetics
605:10.1038/s41592-019-0473-0
505:10.1186/s12896-015-0162-8
486:Motohashi K (June 2015).
359:
309:Combinatorial mutagenesis
269:into the genome at once.
222:. Such chemicals include
213:Site-directed mutagenesis
207:Site-directed mutagenesis
61:site-directed mutagenesis
872:Human Molecular Genetics
349:Homologous recombination
344:Homologous recombination
253:extension reaction with
145:were later developed to
112:discovered in 1927 that
1815:Xu S (20 August 2015).
1201:Journal of Bacteriology
1092:Krinke C (March 2018).
868:"Mouse ENU mutagenesis"
688:Muller HJ (July 1927).
334:insertional mutagenesis
318:Insertional mutagenesis
1671:10.1038/sj.onc.1209043
1271:10.1073/pnas.75.5.2170
823:Nucleic Acids Research
548:Toxicological Sciences
328:Transposon mutagenesis
295:saturation mutagenesis
282:
246:
190:Ethyl methanesulfonate
101:
28:
1583:10.1089/cmb.2011.0152
1461:10.1038/nprot.2007.72
1330:10.1073/pnas.72.1.367
885:10.1093/hmg/8.10.1955
560:10.1093/toxsci/kfy186
411:Saturated mutagenesis
322:Further information:
279:
244:
90:
26:
1164:10.1128/mcb.2.9.1096
1575:2011LNCS.6577..321P
1509:2020PLoSO..1531716C
1321:1975PNAS...72..367F
1262:1978PNAS...75.2170S
924:1998MRFMM.400...25D
709:1927Sci....66...84M
401:Genetic engineering
366:CRISPR gene editing
173:In animal studies,
1049:Scientific Reports
835:10.1093/nar/gnh028
416:Directed evolution
283:
247:
102:
97:random mutagenesis
83:Random mutagenesis
29:
1894:Library resources
1021:978-1-58829-817-1
912:Mutation Research
645:Molecular Therapy
492:BMC Biotechnology
175:alkylating agents
168:expression vector
33:molecular biology
1948:
1882:
1881:
1861:
1855:
1854:
1844:
1821:J Genet Genomics
1812:
1806:
1805:
1795:
1785:
1761:
1755:
1754:
1751:Davidson College
1743:
1737:
1736:
1702:
1691:
1690:
1654:
1648:
1647:
1611:
1605:
1604:
1594:
1554:
1541:
1540:
1530:
1520:
1487:
1481:
1480:
1449:Nature Protocols
1444:
1438:
1437:
1427:
1395:
1389:
1388:
1359:
1353:
1352:
1342:
1332:
1300:
1294:
1293:
1283:
1273:
1241:
1235:
1234:
1224:
1192:
1186:
1185:
1175:
1143:
1137:
1136:
1108:
1102:
1101:
1089:
1083:
1082:
1072:
1040:
1034:
1033:
999:
993:
992:
982:
950:
944:
943:
907:
898:
897:
887:
863:
857:
856:
846:
814:
808:
807:
787:
778:
777:
767:
735:
729:
728:
694:
685:
679:
678:
668:
636:
625:
624:
588:
582:
581:
571:
539:
528:
527:
517:
507:
483:
474:
473:
463:
431:
287:alanine scanning
232:nitrosoguanidine
127:Escherichia coli
77:beta thalassemia
1956:
1955:
1951:
1950:
1949:
1947:
1946:
1945:
1921:
1920:
1919:
1918:
1917:
1902:
1901:
1897:
1890:
1885:
1878:
1862:
1858:
1813:
1809:
1762:
1758:
1745:
1744:
1740:
1725:
1703:
1694:
1665:(52): 7656–72.
1655:
1651:
1612:
1608:
1569:(11): 1743–56.
1555:
1544:
1503:(4): e0231716.
1488:
1484:
1445:
1441:
1396:
1392:
1360:
1356:
1301:
1297:
1242:
1238:
1193:
1189:
1158:(9): 1096–103.
1144:
1140:
1109:
1105:
1090:
1086:
1041:
1037:
1022:
1000:
996:
951:
947:
908:
901:
878:(10): 1955–63.
864:
860:
815:
811:
804:
788:
781:
736:
732:
692:
686:
682:
637:
628:
589:
585:
540:
531:
484:
477:
432:
428:
424:
397:
384:
368:
362:
346:
330:
320:
311:
220:point mutations
215:
209:
151:oligonucleotide
85:
21:
12:
11:
5:
1954:
1944:
1943:
1938:
1933:
1916:
1915:
1910:
1904:
1903:
1892:
1891:
1889:
1888:External links
1886:
1884:
1883:
1876:
1856:
1807:
1756:
1738:
1723:
1692:
1649:
1622:(8): 722–730.
1616:Nature Methods
1606:
1542:
1482:
1455:(4): 891–903.
1439:
1390:
1354:
1295:
1236:
1187:
1138:
1103:
1084:
1035:
1020:
994:
945:
918:(1–2): 25–32.
899:
858:
809:
803:978-0854046546
802:
779:
730:
703:(1699): 84–7.
680:
651:(2): 352–356.
626:
599:(8): 722–730.
593:Nature Methods
583:
554:(1): 131–145.
529:
475:
446:(6): 1262–78.
425:
423:
420:
419:
418:
413:
408:
403:
396:
393:
383:
382:Gene synthesis
380:
376:genome editing
364:Main article:
361:
358:
345:
342:
319:
316:
310:
307:
255:DNA polymerase
211:Main article:
208:
205:
197:European Union
132:replica-plated
110:Hermann Muller
84:
81:
9:
6:
4:
3:
2:
1953:
1942:
1939:
1937:
1934:
1932:
1929:
1928:
1926:
1914:
1911:
1909:
1906:
1905:
1900:
1895:
1879:
1877:9781420040166
1873:
1869:
1868:
1860:
1852:
1848:
1843:
1838:
1834:
1830:
1827:(8): 413–21.
1826:
1822:
1818:
1811:
1803:
1799:
1794:
1789:
1784:
1779:
1775:
1771:
1767:
1760:
1752:
1748:
1742:
1734:
1730:
1726:
1724:9780123848802
1720:
1716:
1712:
1708:
1701:
1699:
1697:
1688:
1684:
1680:
1676:
1672:
1668:
1664:
1660:
1653:
1645:
1641:
1637:
1633:
1629:
1625:
1621:
1617:
1610:
1602:
1598:
1593:
1588:
1584:
1580:
1576:
1572:
1568:
1564:
1560:
1553:
1551:
1549:
1547:
1538:
1534:
1529:
1524:
1519:
1514:
1510:
1506:
1502:
1498:
1494:
1486:
1478:
1474:
1470:
1466:
1462:
1458:
1454:
1450:
1443:
1435:
1431:
1426:
1421:
1417:
1413:
1409:
1405:
1401:
1394:
1386:
1382:
1378:
1374:
1371:(2): 343–58.
1370:
1366:
1358:
1350:
1346:
1341:
1336:
1331:
1326:
1322:
1318:
1315:(1): 367–71.
1314:
1310:
1306:
1299:
1291:
1287:
1282:
1277:
1272:
1267:
1263:
1259:
1256:(5): 2170–4.
1255:
1251:
1247:
1240:
1232:
1228:
1223:
1218:
1214:
1210:
1207:(1): 364–71.
1206:
1202:
1198:
1191:
1183:
1179:
1174:
1169:
1165:
1161:
1157:
1153:
1149:
1142:
1134:
1130:
1126:
1122:
1118:
1114:
1107:
1099:
1095:
1088:
1080:
1076:
1071:
1066:
1062:
1058:
1054:
1050:
1046:
1039:
1031:
1027:
1023:
1017:
1013:
1009:
1005:
998:
990:
986:
981:
976:
972:
968:
965:(2): 431–41.
964:
960:
956:
949:
941:
937:
933:
929:
925:
921:
917:
913:
906:
904:
895:
891:
886:
881:
877:
873:
869:
862:
854:
850:
845:
840:
836:
832:
829:(3): 26e–26.
828:
824:
820:
813:
805:
799:
795:
794:
786:
784:
775:
771:
766:
761:
757:
753:
750:(4): 1491–6.
749:
745:
741:
734:
726:
722:
718:
714:
710:
706:
702:
698:
691:
684:
676:
672:
667:
662:
658:
654:
650:
646:
642:
635:
633:
631:
622:
618:
614:
610:
606:
602:
598:
594:
587:
579:
575:
570:
565:
561:
557:
553:
549:
545:
538:
536:
534:
525:
521:
516:
511:
506:
501:
497:
493:
489:
482:
480:
471:
467:
462:
457:
453:
449:
445:
441:
437:
430:
426:
417:
414:
412:
409:
407:
404:
402:
399:
398:
392:
389:
379:
377:
373:
367:
357:
355:
354:knockout mice
350:
341:
337:
335:
329:
325:
315:
306:
304:
300:
296:
292:
288:
278:
274:
270:
266:
264:
260:
256:
252:
243:
239:
237:
233:
229:
225:
221:
214:
204:
202:
198:
193:
191:
187:
185:
181:
176:
171:
169:
165:
161:
156:
152:
148:
144:
139:
137:
133:
129:
128:
123:
119:
115:
111:
107:
98:
95:generated by
94:
93:DNA libraries
89:
80:
78:
74:
70:
66:
62:
56:
54:
50:
46:
42:
38:
34:
25:
19:
1898:
1866:
1859:
1824:
1820:
1810:
1773:
1769:
1759:
1750:
1741:
1706:
1662:
1658:
1652:
1619:
1615:
1609:
1566:
1562:
1500:
1496:
1485:
1452:
1448:
1442:
1410:(1): 53–61.
1407:
1403:
1393:
1368:
1364:
1357:
1312:
1308:
1298:
1253:
1249:
1239:
1204:
1200:
1190:
1155:
1151:
1141:
1116:
1112:
1106:
1097:
1087:
1055:(1): 13981.
1052:
1048:
1038:
1003:
997:
962:
958:
948:
915:
911:
875:
871:
861:
826:
822:
812:
792:
747:
743:
733:
700:
696:
683:
648:
644:
596:
592:
586:
551:
547:
495:
491:
443:
439:
429:
385:
369:
347:
338:
331:
312:
284:
271:
267:
248:
216:
194:
186:-nitrosourea
183:
179:
172:
140:
125:
103:
72:
57:
36:
30:
1931:Mutagenesis
1899:Mutagenesis
303:amino acids
224:aminopurine
136:rich medium
134:, one in a
118:fruit flies
47:to produce
37:mutagenesis
18:mutagenesis
1925:Categories
1119:: 265–94.
1004:Drosophila
422:References
297:where one
228:transition
45:engineered
1644:196811756
621:196811756
406:Oncomouse
263:insertion
236:bisulfite
49:libraries
1851:26336798
1802:27134651
1733:20699138
1687:14441244
1679:16299527
1659:Oncogene
1636:31308554
1601:21923411
1537:32298334
1497:PLOS ONE
1477:37361631
1469:17446890
1434:28751422
1404:Genetics
1079:32814789
1030:18641944
989:20439774
959:Genetics
894:10469849
853:14872057
744:Genetics
725:17802387
675:31951833
613:31308554
578:30060110
524:26037246
470:24906146
395:See also
259:deletion
230:, while
177:such as
166:into an
143:proteins
122:genetics
106:mutagens
1842:4560834
1793:4850645
1592:5220575
1571:Bibcode
1528:7161989
1505:Bibcode
1425:5586385
1317:Bibcode
1258:Bibcode
1231:4607625
1182:6983647
1133:6279018
1098:Inf'OGM
1070:7438504
980:2881127
940:9685575
920:Bibcode
774:9409815
765:1208325
705:Bibcode
697:Science
666:7001082
569:6390969
515:4453199
461:4343198
291:alanine
182:-ethyl-
73:in vivo
69:editing
1896:about
1874:
1849:
1839:
1800:
1790:
1731:
1721:
1685:
1677:
1642:
1634:
1599:
1589:
1535:
1525:
1475:
1467:
1432:
1422:
1385:712841
1383:
1347:
1340:432306
1337:
1290:209457
1288:
1281:392513
1278:
1229:
1222:245771
1219:
1180:
1173:369902
1170:
1131:
1077:
1067:
1028:
1018:
987:
977:
938:
892:
851:
844:373423
841:
800:
772:
762:
723:
673:
663:
619:
611:
576:
566:
522:
512:
498:: 47.
468:
458:
372:CRISPR
360:CRISPR
251:primer
164:cloned
147:screen
124:. For
114:X-rays
65:CRISPR
1776:: 6.
1683:S2CID
1640:S2CID
1473:S2CID
1349:47176
693:(PDF)
617:S2CID
299:codon
195:In a
1872:ISBN
1847:PMID
1798:PMID
1729:PMID
1719:ISBN
1675:PMID
1632:PMID
1597:PMID
1533:PMID
1465:PMID
1430:PMID
1381:PMID
1345:PMID
1286:PMID
1227:PMID
1178:PMID
1129:PMID
1075:PMID
1026:PMID
1016:ISBN
985:PMID
936:PMID
890:PMID
849:PMID
798:ISBN
770:PMID
721:PMID
671:PMID
609:PMID
574:PMID
520:PMID
466:PMID
440:Cell
326:and
201:GMOs
91:How
1837:PMC
1829:doi
1788:PMC
1778:doi
1711:doi
1667:doi
1624:doi
1587:PMC
1579:doi
1523:PMC
1513:doi
1457:doi
1420:PMC
1412:doi
1408:207
1373:doi
1369:124
1335:PMC
1325:doi
1276:PMC
1266:doi
1217:PMC
1209:doi
1205:120
1168:PMC
1160:doi
1121:doi
1065:PMC
1057:doi
1008:doi
975:PMC
967:doi
963:185
928:doi
916:400
880:doi
839:PMC
831:doi
760:PMC
752:doi
748:147
713:doi
661:PMC
653:doi
601:doi
564:PMC
556:doi
552:166
510:PMC
500:doi
456:PMC
448:doi
444:157
261:or
155:PCR
41:DNA
31:In
1927::
1845:.
1835:.
1825:42
1823:.
1819:.
1796:.
1786:.
1774:10
1772:.
1768:.
1749:.
1727:.
1717:.
1695:^
1681:.
1673:.
1663:24
1661:.
1638:.
1630:.
1620:16
1618:.
1595:.
1585:.
1577:.
1567:18
1565:.
1561:.
1545:^
1531:.
1521:.
1511:.
1501:15
1499:.
1495:.
1471:.
1463:.
1451:.
1428:.
1418:.
1406:.
1402:.
1379:.
1367:.
1343:.
1333:.
1323:.
1313:72
1311:.
1307:.
1284:.
1274:.
1264:.
1254:75
1252:.
1248:.
1225:.
1215:.
1203:.
1199:.
1176:.
1166:.
1154:.
1150:.
1127:.
1117:15
1115:.
1096:.
1073:.
1063:.
1053:10
1051:.
1047:.
1024:.
1014:.
983:.
973:.
961:.
957:.
934:.
926:.
914:.
902:^
888:.
874:.
870:.
847:.
837:.
827:32
825:.
821:.
782:^
768:.
758:.
746:.
742:.
719:.
711:.
701:66
699:.
695:.
669:.
659:.
649:28
647:.
643:.
629:^
615:.
607:.
597:16
595:.
572:.
562:.
550:.
546:.
532:^
518:.
508:.
496:15
494:.
490:.
478:^
464:.
454:.
442:.
438:.
378:.
356:.
234:,
79:.
35:,
1880:.
1853:.
1831::
1804:.
1780::
1753:.
1735:.
1713::
1689:.
1669::
1646:.
1626::
1603:.
1581::
1573::
1539:.
1515::
1507::
1479:.
1459::
1453:2
1436:.
1414::
1387:.
1375::
1351:.
1327::
1319::
1292:.
1268::
1260::
1233:.
1211::
1184:.
1162::
1156:2
1135:.
1123::
1100:.
1081:.
1059::
1032:.
1010::
991:.
969::
942:.
930::
922::
896:.
882::
876:8
855:.
833::
806:.
776:.
754::
727:.
715::
707::
677:.
655::
623:.
603::
580:.
558::
526:.
502::
472:.
450::
184:N
180:N
20:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.