1478:(EMP) is an initiative to collect natural samples and analyze the microbial community around the globe. Microbes are highly abundant, diverse and have an important role in the ecological system. Yet as of 2010, it was estimated that the total global environmental DNA sequencing effort had produced less than 1 percent of the total DNA found in a liter of seawater or a gram of soil, and the specific interactions between microbes are largely unknown. The EMP aims to process as many as 200,000 samples in different biomes, generating a complete database of microbes on earth to characterize environments and ecosystems by microbial composition and interaction. Using these data, new ecological and evolutionary theories can be proposed and tested.
414:
susceptibility to allergic airway disease. The frequency of certain subsets of microbes has been linked to disease severity. The presence of specific microbes early in postnatal life, instruct future immune responses. In gnotobiotic mice certain gut bacteria were found to transmit a particular phenotype to recipient germ-free mice, that promoted accumulation of colonic regulatory T cells, and strains that modulated mouse adiposity and cecal metabolite concentrations. This combinatorial approach enables a systems-level understanding of microbial contributions to human biology. But also other mucoide tissues as lung and vagina have been studied in relation to diseases such as asthma, allergy and vaginosis.
1336:(HVRs); the former can be used to design broad primers while the latter allow for finer taxonomic distinction. However, species-level resolution is not typically possible using the 16S rDNA. Primer selection is an important step, as anything that cannot be targeted by the primer will not be amplified and thus will not be detected, moreover different sets of primers can be selected to amplify different HVRs in the gene, or pairs of them. The appropriate choice of which HVRs to amplify has to be made according to the taxonomic groups of interest, as different target regions has been shown to influence taxonomical classification.
459:
313:
6284:
505:
189:
7026:
6851:
1532:
amoxicillin and macrolides cause significant shifts in the gut microbiota characterized by a change in the bacterial classes
Bifidobacteria, Enterobacteria and Clostridia. A single course of antibiotics in adults causes changes in both the bacterial and fungal microbiota, with even more persistent changes in the fungal communities. The bacteria and fungi live together in the gut and there is most likely a competition for nutrient sources present. Seelbinder
1275:
7052:
724:
605:
6162:
74:
32:
3580:
1660:
259:, when disadvantageous to the host. Other authors define a situation as mutualistic where both benefit, and commensal, where the unaffected host benefits the symbiont. A nutrient exchange may be bidirectional or unidirectional, may be context dependent and may occur in diverse ways. Microbiota that are expected to be present, and that under normal circumstances do not cause disease, are deemed
7064:
570:. As the infant microbiome is established, commensal bacteria quickly populate the gut, prompting a range of immune responses and "programming" the immune system with long-lasting effects. The bacteria are able to stimulate lymphoid tissue associated with the gut mucosa, which enables the tissue to produce antibodies for pathogens that may enter the gut.
1409:. One drawback of this approach is that many members of microbial communities do not have a representative sequenced genome, but this applies to 16S rRNA amplicon sequencing as well and is a fundamental problem. With shotgun sequencing, it can be resolved by having a high coverage (50-100x) of the unknown genome, effectively doing a de novo
1332:"16S rDNA", the sequence of DNA which encodes the ribosomal RNA molecule). Since ribosomes are present in all living organisms, using 16S rDNA allows for DNA to be amplified from many more organisms than if another marker were used. The 16S rRNA gene contains both slowly evolving regions and 9 fast evolving regions, also known as
474:
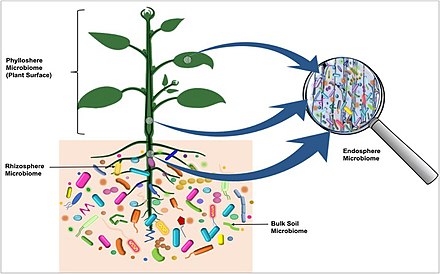
The plant microbiome was recently discovered to originate from the seed. Microorganism which are transmitted via seed migrate into the developing seedling in a specific route in which certain community move to the leaves and others to the roots. In the diagram on the right, microbiota colonizing the
406:
microbiome to convert cellulose into proteins, short chain fatty acids, and gases. Culture methods cannot provide information on all microorganisms present. Comparative metagenomic studies yielded the surprising result that individual cattle possess markedly different community structures, predicted
1323:
sequencing relies on having some expectations about the composition of the community that is being studied. In target amplicon sequencing a phylogenetically informative marker is targeted for sequencing. Such a marker should be present in ideally all the expected organisms. It should also evolve in
5910:
Korpela K, Helve O, Kolho KL, Saisto T, Skogberg K, Dikareva E, Stefanovic V, Salonen A, Andersson S, de Vos WM. Maternal Fecal
Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell. 2020 Oct 15;183(2):324-334.e5. doi:
1366:
distances are usually defined between microbiome samples, and downstream multivariate methods are carried out on the distance matrices. An important point is that the scale of data is extensive, and further approaches must be taken to identify patterns from the available information. Tools used to
421:
form huge underground colonies harvesting hundreds of kilograms of leaves each year and are unable to digest the cellulose in the leaves directly. They maintain fungus gardens as the colony's primary food source. While the fungus itself does not digest cellulose, a microbial community containing a
1428:
Despite the fact that metagenomics is limited by the availability of reference sequences, one significant advantage of metagenomics over targeted amplicon sequencing is that metagenomics data can elucidate the functional potential of the community DNA. Targeted gene surveys cannot do this as they
1544:
has previously been associated with IBD and further it has been observed to be increased in non-responders to a biological drug, infliximab, given to IBD patients with severe IBD. Propionate and acetic acid are both short-chain fatty acids (SCFAs) that have been observed to be beneficial to gut
413:
have become the most studied mammalian regarding their microbiomes. The gut microbiota have been studied in relation to allergic airway disease, obesity, gastrointestinal diseases and diabetes. Perinatal shifting of microbiota through low dose antibiotics can have long-lasting effects on future
299:
zone or anthosphere. The stability of the rhizosphere microbiota over generations depends upon the plant type but even more on the soil composition, i.e. living and non living environment. Clinically, new microbiota can be acquired through fecal microbiota transplant to treat infections such as
6004:
Rebecka Ventin-Holmberg, Anja Eberl, Schahzad Saqib, Katri
Korpela, Seppo Virtanen, Taina Sipponen, Anne Salonen, Päivi Saavalainen, Eija Nissilä, Bacterial and Fungal Profiles as Markers of Infliximab Drug Response in Inflammatory Bowel Disease, Journal of Crohn's and Colitis, 2020;, jjaa252,
1486:
The gut microbiota are very important for the host health because they play role in degradation of non-digestible polysaccharides (fermentation of resistant starch, oligosaccharides, inulin) strengthening gut integrity or shaping the intestinal epithelium, harvesting energy, protecting against
4938:
Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K. S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; Mende, D. R.; Li, J.; Xu, J.; Li, S.; Li, D.; Cao, J.; Wang, B.; Liang, H.; Zheng, H.; Xie, Y.; Tap, J.; Lepage, P.; Bertalan, M.; Batto, J. M.; Hansen, T.; Le
Paslier, D.;
4663:
Caporaso, J. G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F. D.; Costello, E. K.; Fierer, N.; Peña, A. G.; Goodrich, J. K.; Gordon, J. I.; Huttley, G. A.; Kelley, S. T.; Knights, D.; Koenig, J. E.; Ley, R. E.; Lozupone, C. A.; McDonald, D.; Muegge, B. D.; Pirrung, M.; Reeder, J.;
685:
was simply that of opportunistic colonization. If this is true, the basic observation leading to the theory would be invalid. The theory has gained significant popularity as a way of explaining rapid changes in adaptation that cannot otherwise be explained by traditional mechanisms of natural
1531:
Another factor that has been observed to cause huge changes in the gut microbiota, particularly in children, is the use of antibiotics, associating with health issues such as higher BMI, and further an increased risk towards metabolic diseases such as obesity. In infants it was observed that
398:
Newborn marsupials are born with histologically immature immune tissues and unable to mount their own specific immune defence. They are therefore heavily reliant on their mother's immune system and the milk for their protection. Most marsupials have pouches, and their own microbiota changes
1470:
initiative to identify and characterize microorganisms found in both healthy and diseased humans. The five-year project, best characterized as a feasibility study with a budget of $ 115 million, tested how changes in the human microbiome are associated with human health or disease.
1392:
is also used extensively for studying microbial communities. In metagenomic sequencing, DNA is recovered directly from environmental samples in an untargeted manner with the goal of obtaining an unbiased sample from all genes of all members of the community. Recent studies use shotgun
565:
of mammals. In many animals, the immune system and microbiota may engage in "cross-talk" by exchanging chemical signals, which may enable the microbiota to influence immune reactivity and targeting. Bacteria can be transferred from mother to child through direct contact and after
6017:
El Hage, R., Hernandez-Sanabria, E., Calatayud Arroyo, M., Props, R., & Van de Wiele, T. (2019). Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Frontiers in microbiology, 10,
626:. The hologenome theory originated in studies on coral reefs. Coral reefs are the largest structures created by living organisms, and contain abundant and highly complex microbial communities. Over the past several decades, major declines in coral populations have occurred.
5965:
Seelbinder, B., Chen, J., Brunke, S., Vazquez-Uribe, R., Santhaman, R., Meyer, A. C., ... & Panagiotou, G. (2020). Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome, 8(1),
630:, water pollution and over-fishing are three stress factors that have been described as leading to disease susceptibility. Over twenty different coral diseases have been described, but of these, only a handful have had their causative agents isolated and characterized.
6027:
Tian, X., Hellman, J., Horswill, A. R., Crosby, H. A., Francis, K. P., & Prakash, A. (2019). Elevated gut microbiome-derived propionate levels are associated with reduced sterile lung inflammation and bacterial immunity in mice. Frontiers in microbiology, 10,
673:
can adapt to changing environmental conditions far more rapidly than by genetic mutation and selection alone. Extrapolating this hypothesis to other organisms, including higher plants and animals, led to the proposal of the hologenome theory of evolution.
1324:
such a way that it is conserved enough that primers can target genes from a wide range of organisms while evolving quickly enough to allow for finer resolution at the taxonomic level. A common marker for human microbiome studies is the gene for bacterial
172:. The presence of microbiota in human and other metazoan guts has been critical for understanding the co-evolution between metazoans and bacteria. Microbiota play key roles in the intestinal immune and metabolic responses via their fermentation product (
422:
diversity of bacteria is doing so. Analysis of the microbial population's genome revealed many genes with a role in cellulose digestion. This microbiome's predicted carbohydrate-degrading enzyme profile is similar to that of the bovine rumen, but the
5950:
Korpela, K., Salonen, A., Saxen, H., Nikkonen, A., Peltola, V., Jaakkola, T., ... & Kolho, K. L. (2020). Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatric
Research,
5920:
Korpela, K., Salonen, A., Saxen, H., Nikkonen, A., Peltola, V., Jaakkola, T., ... & Kolho, K. L. (2020). Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatric
Research,
668:
The puzzle of how corals managed to acquire resistance to a specific pathogen led to a 2007 proposal, that a dynamic relationship exists between corals and their symbiotic microbial communities. It is thought that by altering its composition, the
5994:
Sokol H, Leducq V, Aschard H, Pham H P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039–1048. doi:
577:
in the intestines, a type of pattern recognition receptor host cells use to recognize dangers and repair damage. Pathogens can influence this coexistence leading to immune dysregulation including and susceptibility to diseases, mechanisms of
4996:
Tringe, S. G.; Von Mering, C.; Kobayashi, A.; Salamov, A. A.; Chen, K.; Chang, H. W.; Podar, M.; Short, J. M.; Mathur, E. J.; Detter, J. C.; Bork, P.; Hugenholtz, P.; Rubin, E. M. (2005). "Comparative
Metagenomics of Microbial Communities".
365:
Humans are colonized by many microorganisms; the traditional estimate was that humans live with ten times more non-human cells than human cells; more recent estimates have lowered this to 3:1 and even to about 1:1 by number (1:350 by mass).
4713:
Schloss, P. D.; Westcott, S. L.; Ryabin, T.; Hall, J. R.; Hartmann, M.; Hollister, E. B.; Lesniewski, R. A.; Oakley, B. B.; Parks, D. H.; Robinson, C. J.; Sahl, J. W.; Stres, B.; Thallinger, G. G.; Van Horn, D. J.; Weber, C. F. (2009).
5940:
Korpela, K., Salonen, A., Virta, L. J., Kekkonen, R. A., Forslund, K., Bork, P., & De Vos, W. M. (2016). Intestinal microbiome is related to lifetime antibiotic use in
Finnish pre-school children. Nature communications, 7,
1553:
Microbial DNA inhabiting a person's human body can uniquely identify the person. A person's privacy may be compromised if the person anonymously donated microbe DNA data. Their medical condition and identity could be revealed.
377:
sequenced the genome of the human microbiota, focusing particularly on the microbiota that normally inhabit the skin, mouth, nose, digestive tract, and vagina. It reached a milestone in 2012 when it published initial results.
4483:
Soriano-Lerma, Ana; Pérez-Carrasco, Virginia; Sánchez-Marañón, Manuel; Ortiz-González, Matilde; Sánchez-Martín, Victoria; Gijón, Juan; Navarro-Mari, José María; García-Salcedo, José Antonio; Soriano, Miguel (December 2020).
5975:
Cabral, D. J., Penumutchu, S., Norris, C., Morones-Ramirez, J. R., & Belenky, P. (2018). Microbial competition between
Escherichia coli and Candida albicans reveals a soluble fungicidal factor. Microbial cell, 5(5),
4880:
Turnbaugh, P. J.; Hamady, M.; Yatsunenko, T.; Cantarel, B. L.; Duncan, A.; Ley, R. E.; Sogin, M. L.; Jones, W. J.; Roe, B. A.; Affourtit, J. P.; Egholm, M.; Henrissat, B.; Heath, A. C.; Knight, R.; Gordon, J. I. (2008).
1527:
The colonization of the human gut microbiota may start already before birth. There are multiple factors in the environment that affects the development of the microbiota with birthmode being one of the most impactful.
399:
throughout the reproductive stages: oestrus, birth/oestrus, and post-oestrus. Some pouch and skin secretions have had antimicrobial peptides identified, that presumably support the young at this vulnerable time.
369:
In fact, these are so small that there are around 100 trillion microbiota on the human body, around 39 trillion by revised estimates, with only 0.2 kg of total mass in a "reference" 70 kg human body.
5380:
Markowitz, V. M.; Ivanova, N. N.; Szeto, E.; Palaniappan, K.; Chu, K.; Dalevi, D.; Chen, I. M. A.; Grechkin, Y.; Dubchak, I.; Anderson, I.; Lykidis, A.; Mavromatis, K.; Hugenholtz, P.; Kyrpides, N. C. (2007).
1445:
Metatranscriptomics studies have been performed to study the gene expression of microbial communities through methods such as the pyrosequencing of extracted RNA. Structure based studies have also identified
5930:
Schei, K., Simpson, M. R., Avershina, E., Rudi, K., Øien, T., Júlíusson, P. B., ... & Ødegård, R. A. (2020). Early Gut Fungal and
Bacterial Microbiota and Childhood Growth. Frontiers in pediatrics, 8,
1339:
Targeted studies of eukaryotic and viral communities are limited and subject to the challenge of excluding host DNA from amplification and the reduced eukaryotic and viral biomass in the human microbiome.
5900:
Vandenplas, Y., Carnielli, V. P., Ksiazyk, J., Luna, M. S., Migacheva, N., Mosselmans, J. M., ... & Wabitsch, M. (2020), Factors affecting early-life intestinal microbiota development. Nutrition, 78,
536:
situations the plant often exchanges hexose sugars for inorganic phosphate from the fungal symbiont. It is speculated that such very ancient associations have aided plants when they first colonized land.
1429:
only reveal the phylogenetic relationship between the same gene from different organisms. Functional analysis is done by comparing the recovered sequences to databases of metagenomic annotations such as
549:, direct enhancement of mineral uptake, and protection from pathogens. PGPBs may protect plants from pathogens by competing with the pathogen for an ecological niche or a substrate, producing inhibitory
686:
selection. Within the hologenome theory, the holobiont has not only become the principal unit of natural selection but also the result of other step of integration that it is also observed at the cell (
2066:"Evolution of the human gut flora". Andrew H. Moeller, Yingying Li, Eitel Mpoudi Ngole, Steve Ahuka-Mundeke, Elizabeth V. Lonsdorf, Anne E. Pusey, Martine Peeters, Beatrice H. Hahn, Howard Ochman.
407:
phenotype, and metabolic potentials, even though they were fed identical diets, were housed together, and were apparently functionally identical in their utilization of plant cell wall resources.
1516:, which is the increased level of circulating Lipopolysaccharides from gram negative bacterial cells wall. It is found that endotoxemia has association with development of insulin resistance.
442:
cells sense the gut microbiota-derived metabolites and coordinate antibacterial, mechanical, and metabolic branches of the host intestinal innate immune response to the commensal microbiota.
6132:
500:
The bacterial microbiota of potato tubers consists of bacteria transmitted from one tuber generation to the next and bacteria recruited from the soil colonize potato plants via the root.
1589:
1519:
In addition that butyrate production affects serotonin level. Elevated serotonin level has contribution in obesity, which is known to be a risk factor for development of diabetes.
657:
infection, although other diseases still cause bleaching. The surprise stems from the knowledge that corals are long lived, with lifespans on the order of decades, and do not have
3561:
Buchholz, F., Antonielli, L., Kostić, T., Sessitsch, A. and Mitter, B. (2019) "The bacterial community in potato is recruited from soil and partly inherited across generations".
2767:
Bataille, A; Lee-Cruz, L; Tripathi, B; Kim, H; Waldman, B (Jan 2016). "Microbiome Variation Across Amphibian Skin Regions: Implications for Chytridiomycosis Mitigation Efforts".
5117:
Watson, Mick; Roehe, Rainer; Walker, Alan W.; Dewhurst, Richard J.; Snelling, Timothy J.; Ivan Liachko; Langford, Kyle W.; Press, Maximilian O.; Wiser, Andrew H. (2018-02-28).
1639:
Dastogeer, K.M., Tumpa, F.H., Sultana, A., Akter, M.A. and Chakraborty, A. (2020) "Plant microbiome–an account of the factors that shape community composition and diversity".
1545:
microbiota health. When antibiotics affect the growth of bacteria in the gut, there might be an overgrowth of certain fungi, which might be pathogenic when not regulated.
620:
proposes that an object of natural selection is not the individual organism, but the organism together with its associated organisms, including its microbial communities.
1299:
430:
can affect the way its gut looks, by impacting epithelial renewal rate, cellular spacing, and the composition of different cell types in the epithelium. When the moth
449:(turquoise killifish). Transferring the gut microbiota from young killfish into middle-aged killifish significantly extends the lifespans of the middle-aged killfish.
251:
during the nineteenth century is central to the microbiome, where microbiota colonize a host in a non-harmful coexistence. The relationship with their host is called
6037:
Li, Y., Faden, H. S., & Zhu, L. (2020). The response of the gut microbiota to dietary changes in the first two years of life. Frontiers in pharmacology, 11, 334.
5280:
Meyer, F.; Paarmann, D.; d'Souza, M.; Olson, R.; Glass, E. M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; Wilkening, J.; Edwards, R. A. (2008).
3100:
Russell SL, Gold MJ, et al. (Aug 2014). "Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases".
53:
2432:
6492:
1413:. As soon as there is a complete genome of an unknown organism available it can be compared phylogenetically and the organism put into its place in the
2743:"On and in You." Micropia, www.micropia.nl/en/discover/stories/on-and-in-you/#:~:text=They're%20on%20you%2C%20in,re%20known%20as%20human%20microbiota.
5985:
Peleg, A. Y., Hogan, D. A., & Mylonakis, E. (2010). Medically important bacterial–fungal interactions. Nature Reviews Microbiology, 8(5), 340-349
3543:
2953:
Old JM, Deane EM (1998). "The effect of oestrus and the presence of pouch young on aerobic bacteria isolated from the pouch of the tammar wallaby,
5182:
Muller, J.; Szklarczyk, D.; Julien, P.; Letunic, I.; Roth, A.; Kuhn, M.; Powell, S.; Von Mering, C.; Doerks, T.; Jensen, L. J.; Bork, P. (2009).
2068:
642:
6140:
5184:"EggNOG v2.0: Extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations"
4592:"Blocking primers to enhance PCR amplification of rare sequences in mixed samples – a case study on prey DNA in Antarctic krill stomachs"
2629:
2600:
248:
6048:
4185:
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007). "The role of microorganisms in coral health, disease and evolution".
4716:"Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities"
5532:
Maron, PA; Ranjard, L.; Mougel, C.; Lemanceau, P. (2007). "Metaproteomics: A New Approach for Studying Functional Microbial Ecology".
5331:
Sun, S.; Chen, J.; Li, W.; Altintas, I.; Lin, A.; Peltier, S.; Stocks, K.; Allen, E. E.; Ellisman, M.; Grethe, J.; Wooley, J. (2010).
211:
the ability to perform genomic and gene expression analyses of single cells and of entire microbial communities in the disciplines of
207:
to humans, live in close association with microbial organisms. Several advances have driven the perception of microbiomes, including:
40:
6831:
6821:
5858:
Blandino, G.; Inturri, R.; Lazzara, F.; Di Rosa, M.; Malaguarnera, L. (2016-11-01). "Impact of gut microbiota on diabetes mellitus".
5823:
Muñoz-Garach, Araceli; Diaz-Perdigones, Cristina; Tinahones, Francisco J. (December 2016). "Microbiota y diabetes mellitus tipo 2".
5430:
Shi, Y.; Tyson, G. W.; Delong, E. F. (2009). "Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column".
665:
do not produce antibodies, and they should seemingly not be able to respond to new challenges except over evolutionary time scales.
6208:
283:
is at birth, and may even occur through the germ cell line. In plants, the colonizing process can be initiated below ground in the
6607:
6582:
2466:"Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects"
1362:
Phylogenetic relationships are then inferred between the sequences. Due to the complexity of the data, distance measures such as
1405:. To determine the phylogenetic identity of a sequence, it is compared to available full genome sequences using methods such as
6942:
4771:
Callahan, Benjamin J.; McMurdie, Paul J.; Rosen, Michael J.; Han, Andrew W.; Johnson, Amy Jo A.; Holmes, Susan P. (July 2016).
1579:
5282:"The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes"
3818:
1306:
3834:
Bloemberg, G. V.; Lugtenberg, B. J. J. (2001). "Molecular basis of plant growth promotion and biocontrol by rhizobacteria".
487:, passing through the stolons and migrating into the plant as well as into the next generation of tubers are shown in blue.
6887:
6628:
906:
538:
395:
depending on their microbiome, resisting pathogen colonization or inhibiting their growth with antimicrobial skin peptides.
3665:
Berlec, Aleš (2012-09-01). "Novel techniques and findings in the study of plant microbiota: Search for plant probiotics".
512:
Plants are attractive hosts for microorganisms since they provide a variety of nutrients. Microorganisms on plants can be
81:, and are found on the outside surfaces and in the internal tissues of the host plant, as well as in the surrounding soil.
3135:
Turnbaugh PJ, et al. (Dec 2006). "An obesity-associated gut microbiome with increased capacity for energy harvest".
1434:
1012:
3809:
Kloepper, J. W (1993). "Plant growth-promoting rhizobacteria as biological control agents". In Metting, F. B. Jr (ed.).
508:
Light micrograph of a cross section of a coralloid root of a cycad, showing the layer that hosts symbiotic cyanobacteria
6816:
1490:
Several studies showed that the gut bacterial composition in diabetic patients became altered with increased levels of
1433:. The metabolic pathways that these genes are involved in can then be predicted with tools such as MG-RAST, CAMERA and
2994:"Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases"
4566:
3602:"Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root"
2302:
1007:
276:
528:
have, through convergent evolution, developed similar morphology and occupy similar ecological niches. They develop
6643:
6623:
5663:"The importance of metagenomic surveys to microbial ecology: Or why Darwin would have been a metagenomic scientist"
5586:
3705:
2867:
Old JM, Deane EM (2000). "Development of the immune system and immunological protection in marsupial pouch young".
1951:
911:
561:
The symbiotic relationship between a host and its microbiota is under laboratory research for how it may shape the
2440:
438:
immune-related genes are downregulated and the amount of its gut microbiota increases. In the dipteran intestine,
6937:
4664:
Sevinsky, J. R.; Turnbaugh, P. J.; Walters, W. A.; Widmann, J.; Yatsunenko, T.; Zaneveld, J.; Knight, R. (2010).
3496:"The influence of the microbiota on immune development, chronic inflammation, and cancer in the context of aging"
2091:"Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex"
1619:
1422:
6177:
5589:. US National Institutes of Health, Department of Health and Human Services, US Government. 2016. Archived from
1454:
is an approach that studies the proteins expressed by microbiota, giving insight into its functional potential.
1512:
The decrease in butyrate production is associated with defects in intestinal permeability, which could lead to
1092:
617:
599:
4383:
Kuczynski, J.; Lauber, C. L.; Walters, W. A.; Parfrey, L. W.; Clemente, J. C.; Gevers, D.; Knight, R. (2011).
6727:
6592:
4278:
Leggat W, Ainsworth T, Bythell J, Dove S, Gates R, Hoegh-Guldberg O, Iglesias-Prieto R, Yellowlees D (2007).
1467:
4144:
4086:
Blander, J Magarian; Longman, Randy S; Iliev, Iliyan D; Sonnenberg, Gregory F; Artis, David (19 July 2017).
6325:
6283:
6201:
2617:
1191:
3188:"Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice"
6668:
5333:"Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: The CAMERA resource"
845:
20:
616:
Organisms evolve within ecosystems so that the change of one organism affects the change of others. The
7042:
6698:
6587:
6413:
6385:
2579:
2010:
1352:
818:
4486:"Influence of 16S rRNA target region on the outcome of microbiome studies in soil and saliva samples"
4145:"Reciprocity in microbiome and immune system interactions and its implications in disease and health"
3235:
Barfod, KK; Roggenbuck, M; Hansen, LH; Schjørring, S; Larsen, ST; Sørensen, SJ; Krogfelt, KA (2013).
2523:"Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition"
1356:
677:
As of 2007 the hologenome theory was still being debated. A major criticism has been the claim that
244:
5019:
4866:
7006:
6880:
6717:
6428:
3764:"Common themes in nutrient acquisition by plant symbiotic microbes, described by the Gene Ontology"
2597:
1836:
Turnbaugh, P. J.; Ley, R. E.; Hamady, M.; Fraser-Liggett, C. M.; Knight, R.; Gordon, J. I. (2007).
1513:
1475:
1463:
1258:
1253:
961:
374:
5614:"Meeting Report: The Terabase Metagenomics Workshop and the Vision of an Earth Microbiome Project"
6854:
6768:
6763:
6658:
6572:
6320:
6194:
4436:"A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria"
4434:
Chakravorty, Soumitesh; Helb, Danica; Burday, Michele; Connell, Nancy; Alland, David (May 2007).
4389:
2521:
Tkacz, Andrzej; Cheema, Jitender; Chandra, Govind; Grant, Alastair; Poole, Philip S. (Nov 2015).
1641:
427:
45:
1780:
6809:
6753:
6310:
6300:
5014:
3339:"Microbiota-Induced Changes in Drosophila melanogaster Host Gene Expression and Gut Morphology"
1683:
1347:
methods are used to infer the composition of the microbial community. This can be done through
1344:
1186:
681:
was misidentified as the causative agent of coral bleaching, and that its presence in bleached
658:
608:
Bleached branching coral (foreground) and normal branching coral (background). Keppel Islands,
446:
173:
105:
3439:"Microbial Control of Intestinal Homeostasis via Enteroendocrine Cell Innate Immune Signaling"
3053:"Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma"
1679:"The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract"
7089:
7084:
6990:
6799:
6743:
6648:
6547:
6423:
6356:
4853:
3537:
2643:"Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans"
1406:
1196:
997:
840:
3237:"The murine lung microbiome in relation to the intestinal and vaginal bacterial communities"
458:
228:
Biologists have come to appreciate that microbes make up an important part of an organism's
7099:
7029:
6949:
6705:
6418:
6315:
6087:
5541:
5439:
5130:
5071:
5006:
4952:
4894:
4727:
4497:
4336:
4244:
3843:
3670:
3613:
3144:
3005:
2831:
2776:
2534:
2477:
2385:
2225:
2161:
1851:
1794:
1333:
1292:
1279:
1157:
1017:
1002:
859:
662:
439:
8:
6959:
6904:
6873:
6521:
6053:
5233:"KEGG for representation and analysis of molecular networks involving diseases and drugs"
1213:
1076:
864:
809:
799:
587:
574:
533:
497:
Bacteria might colonize the tubers predominantly from the inside of plants via the stolon
423:
252:
216:
97:
6167:
6091:
5545:
5443:
5134:
5075:
5010:
4956:
4898:
4731:
4501:
4482:
4340:
4248:
4229:
3847:
3674:
3617:
3148:
3009:
2835:
2780:
2538:
2481:
2389:
2229:
2165:
1855:
1798:
387:
Amphibians have microbiota on their skin. Some species are able to carry a fungus named
6954:
6758:
6722:
6710:
6557:
6443:
6373:
6361:
6351:
6305:
6113:
5795:
5762:
5743:
5689:
5662:
5638:
5613:
5565:
5509:
5482:
5463:
5407:
5382:
5357:
5332:
5308:
5281:
5257:
5232:
5208:
5183:
5159:
5118:
5094:
5059:
5040:
4973:
4940:
4915:
4882:
4841:
4805:
4772:
4748:
4715:
4690:
4665:
4618:
4591:
4526:
4485:
4460:
4435:
4411:
4384:
4362:
4301:
4210:
4120:
4087:
4063:
4038:
4014:
3989:
3960:
3935:
3911:
3878:
3786:
3763:
3744:
3647:
3520:
3495:
3471:
3438:
3414:
3387:
3363:
3338:
3314:
3287:
3263:
3236:
3212:
3187:
3168:
3077:
3052:
3028:
2993:
2930:
2903:
2849:
2800:
2721:
2694:
2672:
2555:
2522:
2498:
2465:
2344:
2327:
2246:
2213:
2182:
2149:
2125:
2095:
2090:
1986:
1928:
1897:"Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine"
1872:
1837:
1818:
1756:
1729:
1710:
1206:
1066:
870:
854:
649:
occurred every summer in the eastern Mediterranean. Surprisingly, however, after 2003,
609:
4558:
4160:
3855:
2970:
2880:
2753:
1421:. An emerging approach is to combine shotgun sequencing with proximity-ligation data (
7094:
6969:
6922:
6683:
6633:
6526:
6465:
6433:
6395:
6378:
6105:
5883:
5875:
5840:
5800:
5782:
5735:
5694:
5643:
5557:
5514:
5455:
5412:
5362:
5313:
5262:
5213:
5164:
5146:
5099:
5032:
4978:
4920:
4810:
4792:
4753:
4695:
4623:
4572:
4562:
4531:
4513:
4465:
4416:
4354:
4260:
4202:
4164:
4125:
4107:
4068:
4019:
4005:
3965:
3916:
3898:
3859:
3814:
3791:
3736:
3728:
3724:
3686:
3651:
3639:
3631:
3525:
3476:
3458:
3419:
3368:
3319:
3268:
3217:
3160:
3117:
3082:
3033:
2974:
2935:
2884:
2844:
2819:
2792:
2726:
2664:
2560:
2503:
2413:
2408:
2373:
2349:
2308:
2298:
2251:
2187:
2130:
2112:
2027:
1978:
1920:
1877:
1810:
1785:
1761:
1702:
1584:
1394:
1325:
976:
791:
770:
542:
431:
5569:
3748:
3600:
Abdelfattah, Ahmed; Wisniewski, Michael; Schena, Leonardo; Tack, Ayco J. M. (2021).
3388:"Increase in Gut Microbiota after Immune Suppression in Baculovirus-infected Larvae"
2853:
2804:
2489:
1990:
1932:
1714:
1536:. found that commensal bacteria in the gut regulate the growth and pathogenicity of
312:
6896:
6653:
6577:
6511:
6366:
6117:
6095:
5867:
5832:
5790:
5774:
5747:
5725:
5684:
5674:
5633:
5625:
5549:
5504:
5494:
5467:
5447:
5402:
5394:
5352:
5344:
5303:
5293:
5252:
5244:
5203:
5195:
5154:
5138:
5089:
5079:
5024:
4968:
4960:
4910:
4902:
4833:
4829:
4800:
4784:
4743:
4735:
4685:
4677:
4613:
4603:
4554:
4521:
4505:
4455:
4447:
4406:
4398:
4366:
4344:
4305:
4291:
4252:
4214:
4194:
4156:
4115:
4099:
4058:
4050:
4009:
4001:
3955:
3947:
3906:
3890:
3851:
3781:
3771:
3720:
3682:
3678:
3621:
3570:
3515:
3507:
3466:
3450:
3409:
3399:
3358:
3350:
3309:
3299:
3258:
3248:
3207:
3199:
3172:
3152:
3109:
3072:
3064:
3023:
3013:
2966:
2925:
2915:
2876:
2839:
2784:
2716:
2706:
2676:
2654:
2550:
2542:
2493:
2485:
2403:
2393:
2339:
2241:
2233:
2177:
2169:
2120:
2104:
2073:
2051:
2019:
1968:
1960:
1910:
1867:
1859:
1842:
1822:
1802:
1779:
Backhed, F.; Ley, R. E.; Sonnenburg, J. L.; Peterson, D. A.; Gordon, J. I. (2005).
1751:
1743:
1692:
1650:
1348:
1126:
1071:
1045:
936:
832:
804:
733:
583:
494:
Bacteria are recruited from the soil more or less independent of the potato variety
469:
418:
392:
355:
333:
150:
78:
5044:
4845:
3706:"Phyllosphere microbiology with special reference to diversity and plant genotype"
634:
is the most serious of these diseases. In the Mediterranean Sea, the bleaching of
7068:
6917:
6678:
6562:
6516:
6408:
5871:
5836:
5084:
4830:"UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing"
4549:
Marchesi, J. R. (2010). "Prokaryotic and Eukaryotic Diversity of the Human Gut".
3811:
Soil microbial ecology: applications in agricultural and environmental management
3574:
3404:
3304:
3203:
2711:
2604:
2292:
2108:
1964:
1734:
1540:
by their metabolites, particularly by propionate, acetic acid and 5-dodecenoate.
1447:
1410:
1056:
1040:
971:
765:
631:
550:
5713:
5119:"Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen"
4451:
2904:"Marsupial and monotreme milk – a review of its nutrients and immune properties"
1498:
and Clostridiales members, with decrease in butyrate-producing bacteria such as
7056:
6794:
6789:
6748:
6673:
6552:
6487:
6448:
6438:
5142:
4509:
4256:
3951:
3894:
3584:
3113:
2659:
2642:
2237:
1915:
1901:
1896:
1697:
1678:
1664:
1451:
1398:
1181:
1061:
966:
627:
546:
504:
5553:
3776:
3454:
2788:
2055:
2008:; Berlanga, M. (2013). "Symbiogenesis: The holobiont as a unit of evolution".
1654:
1506:
This alteration is due to many factors such as antibiotic abuse, diet, and age
7078:
6932:
6693:
6638:
6602:
6460:
6253:
6243:
6225:
6217:
5879:
5786:
5739:
5483:"Structure-based Search Reveals Hammerhead Ribozymes in the Human Microbiome"
5150:
4796:
4517:
4111:
3902:
3732:
3635:
3462:
3253:
2398:
2312:
2116:
2005:
1614:
1233:
1228:
986:
951:
691:
687:
641:
was first described in 1994 and shortly determined to be due to infection by
562:
132:, and have been found to be crucial for immunologic, hormonal, and metabolic
89:
5730:
5714:"Gut Microbiota and Type 2 Diabetes Mellitus : What is The Link ?"
5499:
5298:
5028:
4941:"A human gut microbial gene catalogue established by metagenomic sequencing"
4321:"Bacteria are not the primary cause of bleaching in the Mediterranean coral
3626:
3601:
3511:
3018:
2077:
1806:
1401:
to recover the sequences of the reads. The reads can then be assembled into
267:; normal flora can not only be harmless, but can be protective of the host.
6964:
6804:
6531:
6292:
6109:
6006:
5887:
5844:
5804:
5698:
5647:
5561:
5518:
5459:
5416:
5366:
5317:
5266:
5217:
5168:
5103:
5036:
4982:
4924:
4814:
4757:
4699:
4627:
4608:
4576:
4535:
4469:
4420:
4358:
4296:
4279:
4264:
4206:
4168:
4129:
4072:
4023:
3969:
3920:
3863:
3795:
3740:
3690:
3643:
3529:
3480:
3423:
3372:
3323:
3288:"An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity"
3272:
3221:
3164:
3121:
3086:
3037:
2939:
2888:
2820:"Innate immune defenses of amphibian skin: antimicrobial peptides and more"
2796:
2730:
2668:
2564:
2507:
2417:
2353:
2255:
2191:
2134:
2031:
2023:
1982:
1924:
1881:
1814:
1765:
1706:
1604:
1574:
1414:
1389:
1384:
1218:
1201:
1121:
1087:
775:
746:
579:
351:
320:
292:
288:
240:
212:
188:
93:
5679:
5348:
5058:
Wooley, J. C.; Godzik, A.; Friedberg, I. (2010). Bourne, Philip E. (ed.).
4642:"VAMPS: The Visualization and Analysis of Microbial Population Structures"
4349:
4320:
4054:
3936:"Coordination of tolerogenic immune responses by the commensal microbiota"
3354:
3286:
Suen; Scott JJ; Aylward FO; et al. (2010). Sonnenburg, Justin (ed.).
3068:
2978:
2754:"NIH Human Microbiome Project defines normal bacterial makeup of the body"
2695:"Revised Estimates for the Number of Human and Bacteria Cells in the Body"
2546:
1747:
6836:
6597:
6453:
5629:
5398:
5248:
5199:
4739:
2173:
2046:
1171:
900:
760:
755:
476:
435:
284:
165:
133:
5778:
5590:
5451:
5231:
Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. (2009).
4964:
4906:
4681:
4198:
4039:"Microbes drive evolution of animals and plants: the hologenome concept"
3877:
Palm, Noah W.; de Zoete, Marcel R.; Flavell, Richard A. (30 June 2015).
3156:
2630:
Scientists bust myth that our bodies have more bacteria than human cells
1863:
6927:
6688:
6567:
6470:
6258:
4788:
2920:
1973:
1609:
1594:
1564:
1223:
1176:
1137:
1116:
981:
875:
741:
715:
491:
The soil is the main reservoir for bacteria that colonize potato tubers
256:
255:
when organisms perform tasks that are known to be useful for the host,
197:
141:
101:
77:
Diverse microbial communities of characteristic microbiota are part of
73:
2818:
Woodhams DC, Rollins-Smith LA, Alford RA, Simon MA, Harris RN (2007).
479:, entering the roots and colonizing the next tuber generation via the
7011:
6985:
6912:
6663:
6480:
4939:
Linneberg, A.; Nielsen, H. B. R.; Pelletier, E.; Renault, P. (2010).
4773:"DADA2: High-resolution sample inference from Illumina amplicon data"
4385:"Experimental and analytical tools for studying the human microbiome"
4088:"Regulation of inflammation by microbiota interactions with the host"
1599:
1162:
1032:
941:
895:
670:
521:
517:
339:
229:
169:
168:
and genetic characteristics, sometimes collectively referred to as a
157:
6100:
6075:
5822:
4666:"QIIME allows analysis of high-throughput community sequencing data"
4402:
4103:
2270:
7051:
6865:
6773:
6335:
6330:
6268:
6238:
4837:
4662:
1355:(OTUs); or alternatively with denoising methodologies, identifying
1320:
946:
723:
513:
445:
Fish have their own microbiomes, including the short-lived species
316:
193:
113:
3386:
Jakubowska, Agata K.; Vogel, Heiko; Herrero, Salvador (May 2013).
2271:
Biologie et complexité : histoire et modèles du commensalisme
2089:
Jugder, Bat-Erdene; Kamareddine, Layla; Watnick, Paula I. (2021).
1949:
Salvucci, E. (2016). "Microbiome, holobiont and the net of life".
604:
483:, are visualized with a red color. Bacteria present in the mother
6403:
6263:
6233:
6186:
6049:"Microbial DNA in Human Body Can Be Used to Identify Individuals"
5612:
Gilbert, J. A.; Meyer, F.; Antonopoulos, D.; et al. (2010).
4879:
3990:"Microbial-immune cross-talk and regulation of the immune system"
3934:
Round, June L.; O'Connell, Ryan M.; Mazmanian, Sarkis K. (2010).
3583:
Material was copied from this source, which is available under a
2991:
2291:
Sherwood, Linda; Willey, Joanne; Woolverton, Christopher (2013).
1663:
Material was copied from this source, which is available under a
1363:
1132:
636:
553:, or inducing systemic resistance in host plants to the pathogen
347:
204:
177:
161:
121:
117:
5379:
4995:
3337:
Broderick, Nichole A.; Buchon, Nicolas; Lemaitre, Bruno (2014).
3234:
2817:
2374:"Four hundred-million-year-old vesicular arbuscular mycorrhizae"
1835:
31:
6248:
4318:
3599:
2044:
Davenport, Emily R et al. "The human microbiome in evolution".
1402:
1372:
1108:
529:
480:
359:
343:
296:
280:
224:
methods of mathematical analysis suitable for complex data sets
221:
databases accessible to researchers across multiple disciplines
146:
125:
5383:"IMG/M: A data management and analysis system for metagenomes"
3704:
Whipps, J.m.; Hand, P.; Pink, D.; Bending, G.d. (2008-12-01).
1677:
De Sordi, Luisa; Lourenço, Marta; Debarbieux, Laurent (2019).
545:, solubilization of minerals such as phosphorus, synthesis of
6826:
6273:
5857:
5279:
5181:
4382:
4085:
3579:
1659:
1569:
1522:
1418:
1368:
567:
525:
484:
403:
129:
109:
6133:"Can The Microbes You Leave Behind Be Used to Identify You?"
4433:
1778:
1425:) to assemble complete microbial genomes without culturing.
5911:
10.1016/j.cell.2020.08.047. Epub 2020 Oct 1. PMID 33007265.
5531:
5230:
4227:
3988:
Cahenzli, Julia; Balmer, Maria L.; McCoy, Kathy D. (2012).
3933:
2992:
Brulc JM; Antonopoulos DA; Miller MEB; et al. (2009).
2959:
Comparative Immunology Microbiology and Infectious Diseases
2766:
1430:
410:
5661:
Gilbert, J. A.; O'Dor, R.; King, N.; Vogel, T. M. (2011).
4277:
4184:
1676:
573:
The human microbiome may play a role in the activation of
5660:
5116:
4712:
4036:
2618:
Ten Times More Microbial Cells than Body Cells in Humans?
2464:
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005).
2297:(9th ed.). New York: McGraw Hill. pp. 713–721.
541:(PGPB) provide the plant with essential services such as
532:, threadlike structures that penetrate the host cell. In
5718:
Afro-Egyptian Journal of Infectious and Endemic Diseases
5611:
4770:
4641:
3186:
Faith JJ, Ahern PP, Ridaura VK, et al. (Jan 2014).
2463:
2290:
2088:
2003:
4319:
Ainsworth TD, Fine M, Roff G, Hoegh-Guldberg O (2008).
3336:
2520:
5961:
5959:
5957:
4280:"The hologenome theory disregards the coral holobiont"
3879:"Immune–microbiota interactions in health and disease"
3585:
Creative Commons Attribution 4.0 International License
3385:
3285:
1665:
Creative Commons Attribution 4.0 International License
402:
In mammals, herbivores such as cattle depend on their
235:
7040:
5057:
3703:
2274:. PhD Dissertation, University of Lyon, France, 2014.
1481:
5480:
4228:
Baird AH, Bhagooli R, Ralph PJ, Takahashi S (2009).
3437:
Watnick, Paula I.; Jugder, Bat-Erdene (2020-02-01).
2150:"Cross-kingdom similarities in microbiome functions"
426:
is almost entirely different. Gut microbiota of the
5954:
4037:Rosenberg, Eugene; Zilber-Rosenberg, Ilana (2016).
3987:
3876:
3833:
3050:
1895:Ley, R. E.; Peterson, D. A.; Gordon, J. I. (2006).
1590:
Microbiota of the lower reproductive tract of women
1440:
653:in the eastern Mediterranean has been resistant to
463:
Routes of colonization of potato tubers by bacteria
2693:Sender, Ron; Fuchs, Shai; Milo, Ron (2016-08-19).
2371:
232:, far beyond the occasional symbiotic case study.
5761:Thursby, Elizabeth; Juge, Nathalie (2017-06-01).
5373:
5330:
4937:
3827:
3813:. New York: Marcel Dekker Inc. pp. 255–274.
3185:
2901:
2147:
1894:
1781:"Host-Bacterial Mutualism in the Human Intestine"
417:Insects have their own microbiomes. For example,
7076:
5224:
4989:
3493:
2616:Judah L. Rosner for Microbe Magazine, Feb 2014.
2211:
1351:methodologies, by clustering the amplicons into
291:, or originate from the above ground parts, the
5481:Jimenez, R. M.; Delwart, E.; Luptak, A (2011).
4883:"A core gut microbiome in obese and lean twins"
4706:
3557:
3555:
3553:
3179:
3128:
3093:
3044:
2692:
2640:
2069:Proceedings of the National Academy of Sciences
702:
391:, which in others can cause a deadly infection
354:which live on the human body are excluded. The
5654:
5474:
5429:
5175:
4873:
4589:
4142:
2319:
1944:
1942:
1727:
6881:
6202:
5273:
3761:
3542:: CS1 maint: DOI inactive as of April 2024 (
3436:
3051:Russell SL, Gold MJ; et al. (May 2012).
1300:
593:
5525:
5423:
4656:
4542:
3550:
3487:
2286:
2284:
2282:
2280:
2212:Bosch, T. C. G.; McFall-Ngai, M. J. (2011).
1728:Peterson, J; Garges, S; et al. (2009).
1450:(ncRNAs) such as ribozymes from microbiota.
645:. From 1994 to 2002, bacterial bleaching of
5760:
5324:
4378:
4376:
3983:
3981:
3979:
3099:
2207:
2205:
2203:
2201:
1939:
6888:
6874:
6209:
6195:
5763:"Introduction to the human gut microbiota"
5051:
4551:Advances in Applied Microbiology Volume 72
2628:Alison Abbott for Nature News. Jan 8 2016
2372:Remy W, Taylor TN, Hass H, Kerp H (1994).
2141:
1523:Gut microbiota development and antibiotics
1307:
1293:
6832:Physical factors affecting microbial life
6822:Microbially induced sedimentary structure
6099:
5794:
5729:
5688:
5678:
5667:Microbial Informatics and Experimentation
5637:
5508:
5498:
5406:
5356:
5307:
5297:
5256:
5207:
5158:
5093:
5083:
5018:
4972:
4914:
4804:
4747:
4689:
4617:
4607:
4525:
4459:
4410:
4348:
4295:
4119:
4062:
4030:
4013:
3959:
3910:
3785:
3775:
3625:
3519:
3470:
3413:
3403:
3362:
3313:
3303:
3262:
3252:
3211:
3134:
3076:
3027:
3017:
2929:
2919:
2843:
2720:
2710:
2658:
2641:Sender, R; Fuchs, S; Milo, R (Jan 2016).
2554:
2497:
2407:
2397:
2343:
2277:
2245:
2181:
2124:
1972:
1914:
1871:
1755:
1696:
1487:pathogens, and regulating host immunity.
1378:
270:
6073:
6007:https://doi.org/10.1093/ecco-jcc/jjaa252
4583:
4548:
4373:
4221:
3976:
3808:
2952:
2869:Developmental and Comparative Immunology
2866:
2577:
2430:
2198:
1948:
1742:(12). NIH HMP Working Group: 2317–2323.
603:
503:
457:
311:
247:(1809–1894), a Belgian professor at the
187:
72:
56:of all important aspects of the article.
6608:International Census of Marine Microbes
6583:Hydrothermal vent microbial communities
5711:
4931:
4230:"Coral bleaching: the role of the host"
4079:
2902:Stannard HJ, Miller RD, Old JM (2020).
2746:
2688:
2686:
2590:
2459:
2457:
2367:
2365:
2363:
2325:
156:The microbiome and host emerged during
7077:
5581:
5579:
4720:Applied and Environmental Microbiology
3664:
3494:Tibbs TN, Lopez LR, Arthur JC (2019).
3379:
2634:
2622:
2610:
2148:Mendes, R.; Raaijmakers, J.M. (2015).
1580:List of bacterial vaginosis microbiota
52:Please consider expanding the lead to
6869:
6190:
5818:
5816:
5814:
4827:
4180:
4178:
3927:
3595:
3593:
1466:launched in 2008 was a United States
307:
279:in animals from mammalians to marine
6895:
6629:Microbiomes of the built environment
6130:
6076:"Microbiomes raise privacy concerns"
6046:
4590:Vestheim, H.; Jarman, S. N. (2008).
3870:
2683:
2454:
2360:
2328:"Gut bacteria in health and disease"
1375:and DADA2 or UNOISE3 for denoising.
381:
25:
5576:
2214:"Metaorganisms as the new frontier"
1343:After the amplicons are sequenced,
697:
287:, around the germinating seed, the
236:Types of microbe-host relationships
13:
6817:Lines on the Antiquity of Microbes
6216:
5811:
4644:. Bay Paul Center, MBL, Woods Hole
4440:Journal of Microbiological Methods
4175:
3590:
2072:. Nov 2014, 111 (46) 16431–16435;
1730:"The NIH Human Microbiome Project"
1721:
1482:Gut microbiota and type 2 diabetes
149:of the microbes that reside in an
14:
7111:
4161:10.2174/1871528113666140330201056
2596:American Academy of Microbiology
2437:Healthcare Journal of New Orleans
1548:
277:initial acquisition of microbiota
153:or else the microbes themselves.
7062:
7050:
7025:
7024:
6850:
6849:
6644:Microbial symbiosis and immunity
6282:
6160:
6124:
6067:
6040:
6031:
6021:
6011:
5998:
5988:
5979:
4553:. Vol. 72. pp. 43–62.
4006:10.1111/j.1365-2567.2012.03624.x
3836:Current Opinion in Plant Biology
3725:10.1111/j.1365-2672.2008.03906.x
3578:
2845:10.1111/j.1469-1795.2007.00150.x
2580:"What is Clostridium difficile?"
1952:Critical Reviews in Microbiology
1658:
1441:RNA and protein-based approaches
1367:analyze the data include VAMPS,
1274:
1273:
722:
145:describes either the collective
30:
5969:
5944:
5934:
5924:
5914:
5904:
5894:
5851:
5754:
5705:
5605:
5487:Journal of Biological Chemistry
5110:
4828:Edgar, Robert C. (2016-10-15).
4821:
4764:
4634:
4476:
4427:
4312:
4271:
4237:Trends in Ecology and Evolution
4143:Nikoopour, E; Singh, B (2014).
4136:
3802:
3755:
3713:Journal of Applied Microbiology
3697:
3658:
3430:
3330:
3279:
3228:
2985:
2946:
2895:
2860:
2811:
2760:
2737:
2571:
2514:
2490:10.1128/AEM.71.9.4951-4959.2005
2424:
2262:
2082:
2060:
2050:. vol. 15,1 127. 27 Dec. 2017,
1620:Vaginal microbiota in pregnancy
539:Plant-growth promoting bacteria
183:
44:may be too short to adequately
5587:"NIH Human Microbiome Project"
3762:Chibucos MC, Tyler BM (2009).
3683:10.1016/j.plantsci.2012.05.010
2578:Copeland, CS (19 April 2019).
2038:
1997:
1888:
1838:"The Human Microbiome Project"
1829:
1772:
1670:
1633:
618:hologenome theory of evolution
600:Hologenome theory of evolution
389:Batrachochytrium dendrobatidis
338:The human microbiota includes
54:provide an accessible overview
1:
6728:Synthetic microbial consortia
6593:Microbial oxidation of sulfur
6493:Host microbe interactions in
5712:Ibrahim, Nesma (2018-07-01).
5618:Standards in Genomic Sciences
5393:(Database issue): D534–D538.
5343:(Database issue): D546–D551.
5243:(Database issue): D355–D360.
5194:(Database issue): D190–D195.
4559:10.1016/S0065-2164(10)72002-5
4290:(10): Online Correspondence.
3856:10.1016/S1369-5266(00)00183-7
2971:10.1016/s0147-9571(98)00022-8
2881:10.1016/S0145-305X(00)00008-2
2431:Copeland, CS (Sep–Oct 2017).
1626:
1504:Faecalibacterium prausnitzii.
1468:National Institutes of Health
520:(found inside plant tissue).
203:All plants and animals, from
6326:Microbial population biology
5872:10.1016/j.diabet.2016.04.004
5837:10.1016/j.endonu.2016.07.008
5085:10.1371/journal.pcbi.1000667
4149:Inflamm Allergy Drug Targets
3770:. 9(Suppl 1) (Suppl 1): S6.
3575:10.1371/journal.pone.0223691
3405:10.1371/journal.ppat.1003379
3305:10.1371/journal.pgen.1001129
3204:10.1126/scitranslmed.3008051
2712:10.1371/journal.pbio.1002533
2109:10.1016/j.immuni.2021.05.017
1965:10.3109/1040841X.2014.962478
1192:Microbial population biology
703:Targeted amplicon sequencing
7:
4452:10.1016/j.mimet.2007.02.005
4284:Nature Reviews Microbiology
4187:Nature Reviews Microbiology
2332:Gastroenterol Hepatol (N Y)
2326:Quigley, E. M. (Sep 2013).
1557:
1457:
1353:operational taxonomic units
556:
358:refers to their collective
192:The predominant species of
21:Microbiota (disambiguation)
16:Community of microorganisms
10:
7116:
6588:Marine microbial symbiosis
6414:Kill the Winner hypothesis
6386:Bacteria collective motion
5995:10.1136/gutjnl-2015-310746
5825:Endocrinología y Nutrición
5143:10.1038/s41467-018-03317-6
5064:PLOS Computational Biology
5060:"A Primer on Metagenomics"
4510:10.1038/s41598-020-70141-8
4257:10.1016/j.tree.2008.09.005
3952:10.1016/j.jaut.2009.11.007
3895:10.1016/j.clim.2015.05.014
3606:Environmental Microbiology
3114:10.1016/j.jaci.2014.06.027
2998:Proc. Natl. Acad. Sci. USA
2660:10.1016/j.cell.2016.01.013
2378:Proc. Natl. Acad. Sci. USA
2238:10.1016/j.zool.2011.04.001
2011:International Microbiology
1916:10.1016/j.cell.2006.02.017
1698:10.1016/j.chom.2019.01.018
1382:
1357:amplicon sequence variants
819:Marine microbial symbiosis
597:
594:Co-evolution of microbiota
467:
331:
18:
7020:
6999:
6978:
6903:
6845:
6782:
6736:
6624:Microbes in human culture
6616:
6540:
6504:
6394:
6344:
6291:
6280:
6224:
5860:Diabetes & Metabolism
5554:10.1007/s00248-006-9196-8
3777:10.1186/1471-2180-9-S1-S6
3455:10.1016/j.tim.2019.09.005
2789:10.1007/s00248-015-0653-0
2756:. NIH News. 13 June 2012.
2056:10.1186/s12915-017-0454-7
1655:10.1016/j.cpb.2020.100161
516:(found on the plants) or
453:
327:
245:Pierre-Joseph van Beneden
243:, a concept developed by
7007:Human Microbiome Project
6718:Human Microbiome Project
6429:Microbial biodegradation
3254:10.1186/1471-2180-13-303
2399:10.1073/pnas.91.25.11841
1476:Earth Microbiome Project
1464:Human Microbiome Project
1259:Earth Microbiome Project
1254:Human Microbiome Project
1013:Accessible carbohydrates
375:Human Microbiome Project
6979:Disorders and therapies
6769:Microbiological culture
6764:Microbial DNA barcoding
6573:Antarctic microorganism
6321:Microbial phylogenetics
6131:Yong, Ed (2015-05-11).
6074:Callaway, Ewen (2015).
5731:10.21608/aeji.2018.9950
5500:10.1074/jbc.C110.209288
5299:10.1186/1471-2105-9-386
5029:10.1126/science.1107851
4390:Nature Reviews Genetics
3940:Journal of Autoimmunity
3627:10.1111/1462-2920.15392
3514:(inactive 2024-04-15).
3512:10.15698/mic2019.08.685
3019:10.1073/pnas.0806191105
2294:Prescott's Microbiology
2078:10.1073/pnas.1419136111
1807:10.1126/science.1104816
1684:Cell Host & Microbe
659:adaptive immune systems
106:multicellular organisms
6943:In bacterial vaginosis
6754:Impedance microbiology
6495:Caenorhabditis elegans
6311:Microbial intelligence
6301:Microbial biogeography
5387:Nucleic Acids Research
5337:Nucleic Acids Research
5237:Nucleic Acids Research
5188:Nucleic Acids Research
4861:Cite journal requires
4609:10.1186/1742-9994-5-12
4297:10.1038/nrmicro1635-c1
3443:Trends in Microbiology
3102:J Allergy Clin Immunol
2470:Appl Environ Microbiol
2024:10.2436/20.1501.01.188
1500:Roseburia intestinalis
1379:Metagenomic sequencing
1345:molecular phylogenetic
1187:Biological dark matter
694:) and genomic levels.
613:
509:
465:
447:Nothobranchius furzeri
324:
271:Acquisition and change
200:
174:short-chain fatty acid
82:
6800:Microbial dark matter
6744:Dark-field microscopy
6548:Marine microorganisms
6424:Microbial cooperation
5680:10.1186/2042-5783-1-5
5123:Nature Communications
4350:10.1038/ismej.2007.88
4055:10.1128/mbio.01395-15
3355:10.1128/mBio.01117-14
3069:10.1038/embor.2012.32
2598:FAQ: Human Microbiome
2547:10.1038/ismej.2015.41
2433:"The World Within Us"
1748:10.1101/gr.096651.109
1642:Current Plant Biology
1492:Lactobacillus gasseri
1334:hypervariable regions
1197:Microbial cooperation
663:innate immune systems
607:
507:
461:
315:
249:University of Louvain
191:
112:. Microbiota include
76:
6419:Microbial consortium
6316:Microbial metabolism
5630:10.4056/sigs.1433550
4740:10.1128/AEM.01541-09
4596:Frontiers in Zoology
2174:10.1038/ismej.2015.7
1496:Streptococcus mutans
1158:Biomass partitioning
1093:hologenome evolution
1018:Flora (microbiology)
104:found in and on all
19:For other uses, see
6522:Seagrass microbiome
6137:National Geographic
6092:2015Natur.521..136C
6054:Scientific American
5779:10.1042/BCJ20160510
5767:Biochemical Journal
5546:2007MicEc..53..486M
5452:10.1038/nature08055
5444:2009Natur.459..266S
5349:10.1093/nar/gkq1102
5135:2018NatCo...9..870S
5076:2010PLSCB...6E0667W
5011:2005Sci...308..554T
4965:10.1038/nature08821
4957:2010Natur.464...59.
4907:10.1038/nature07540
4899:2009Natur.457..480T
4732:2009ApEnM..75.7537S
4682:10.1038/nmeth.f.303
4502:2020NatSR..1013637S
4341:2008ISMEJ...2...67A
4249:2009TEcoE..24...16B
4199:10.1038/nrmicro1635
3883:Clinical Immunology
3848:2001COPB....4..343B
3675:2012PlnSc.193...96B
3669:. 193–194: 96–102.
3618:2021EnvMi..23.2199A
3157:10.1038/nature05414
3149:2006Natur.444.1027T
3143:(7122): 1027–1031.
3010:2009PNAS..106.1948B
2836:2007AnCon..10..425W
2824:Animal Conservation
2781:2016MicEc..71..221B
2539:2015ISMEJ...9.2349T
2482:2005ApEnM..71.4951C
2390:1994PNAS...9111841R
2384:(25): 11841–11843.
2230:2011Zool..114..185B
2166:2015ISMEJ...9.1905M
2103:(8): 1683–1697.e3.
1864:10.1038/nature06244
1856:2007Natur.449..804T
1799:2005Sci...307.1915B
1793:(5717): 1915–1920.
1214:Metatranscriptomics
1008:Initial acquisition
1003:Microbial community
710:Part of a series on
588:autoimmune diseases
575:toll-like receptors
424:species composition
319:microbiota causing
217:metatranscriptomics
6759:Microbial cytology
6723:Protein production
6558:Marine prokaryotes
6444:Microbial food web
6374:Protist locomotion
6352:Bacterial motility
6306:Microbial genetics
5399:10.1093/nar/gkm869
5286:BMC Bioinformatics
5249:10.1093/nar/gkp896
5200:10.1093/nar/gkp951
4789:10.1038/nmeth.3869
4490:Scientific Reports
4323:Oculina patagonica
2921:10.7717/peerj.9335
2603:2016-12-31 at the
1417:, by creating new
792:Marine microbiomes
614:
610:Great Barrier Reef
510:
466:
325:
308:Microbiota by host
201:
83:
7038:
7037:
6863:
6862:
6634:Food microbiology
6527:Soil microbiology
6466:Microbial synergy
6434:Microbial ecology
5773:(11): 1823–1836.
5534:Microbial Ecology
5493:(10): 7737–7743.
5438:(7244): 266–269.
5005:(5721): 554–557.
4893:(7228): 480–484.
4726:(23): 7537–7541.
4092:Nature Immunology
3820:978-0-8247-8737-0
2533:(11): 2349–2359.
1850:(7164): 804–810.
1585:Marine microbiota
1395:Sanger sequencing
1317:
1316:
907:Built environment
889:Other microbiomes
833:Human microbiomes
734:Plant microbiomes
543:nitrogen fixation
434:is infected with
432:Spodoptera exigua
382:Non-human animals
265:normal microbiota
205:simple life forms
88:are the range of
79:plant microbiomes
71:
70:
7107:
7067:
7066:
7065:
7055:
7054:
7046:
7028:
7027:
6897:Human microbiota
6890:
6883:
6876:
6867:
6866:
6853:
6852:
6654:Human microbiome
6578:Coral microbiome
6512:Plant microbiome
6286:
6211:
6204:
6197:
6188:
6187:
6164:
6163:
6152:
6151:
6149:
6148:
6139:. Archived from
6128:
6122:
6121:
6103:
6071:
6065:
6064:
6062:
6061:
6047:magazine, Ewen.
6044:
6038:
6035:
6029:
6025:
6019:
6015:
6009:
6002:
5996:
5992:
5986:
5983:
5977:
5973:
5967:
5963:
5952:
5948:
5942:
5938:
5932:
5928:
5922:
5918:
5912:
5908:
5902:
5898:
5892:
5891:
5855:
5849:
5848:
5820:
5809:
5808:
5798:
5758:
5752:
5751:
5733:
5709:
5703:
5702:
5692:
5682:
5658:
5652:
5651:
5641:
5609:
5603:
5602:
5600:
5598:
5583:
5574:
5573:
5529:
5523:
5522:
5512:
5502:
5478:
5472:
5471:
5427:
5421:
5420:
5410:
5377:
5371:
5370:
5360:
5328:
5322:
5321:
5311:
5301:
5277:
5271:
5270:
5260:
5228:
5222:
5221:
5211:
5179:
5173:
5172:
5162:
5114:
5108:
5107:
5097:
5087:
5055:
5049:
5048:
5022:
4993:
4987:
4986:
4976:
4935:
4929:
4928:
4918:
4877:
4871:
4870:
4864:
4859:
4857:
4849:
4825:
4819:
4818:
4808:
4768:
4762:
4761:
4751:
4710:
4704:
4703:
4693:
4660:
4654:
4653:
4651:
4649:
4638:
4632:
4631:
4621:
4611:
4587:
4581:
4580:
4546:
4540:
4539:
4529:
4480:
4474:
4473:
4463:
4431:
4425:
4424:
4414:
4380:
4371:
4370:
4352:
4329:The ISME Journal
4316:
4310:
4309:
4299:
4275:
4269:
4268:
4234:
4225:
4219:
4218:
4182:
4173:
4172:
4140:
4134:
4133:
4123:
4083:
4077:
4076:
4066:
4049:(2): e01395–15.
4034:
4028:
4027:
4017:
3985:
3974:
3973:
3963:
3946:(3): J220–J225.
3931:
3925:
3924:
3914:
3874:
3868:
3867:
3831:
3825:
3824:
3806:
3800:
3799:
3789:
3779:
3768:BMC Microbiology
3759:
3753:
3752:
3719:(6): 1744–1755.
3710:
3701:
3695:
3694:
3662:
3656:
3655:
3629:
3612:(4): 2199–2214.
3597:
3588:
3582:
3569:(11): e0223691.
3559:
3548:
3547:
3541:
3533:
3523:
3491:
3485:
3484:
3474:
3434:
3428:
3427:
3417:
3407:
3383:
3377:
3376:
3366:
3349:(3): e01117–14.
3334:
3328:
3327:
3317:
3307:
3283:
3277:
3276:
3266:
3256:
3232:
3226:
3225:
3215:
3192:Sci. Transl. Med
3183:
3177:
3176:
3132:
3126:
3125:
3097:
3091:
3090:
3080:
3048:
3042:
3041:
3031:
3021:
3004:(6): 1948–1953.
2989:
2983:
2982:
2955:Macropus eugenii
2950:
2944:
2943:
2933:
2923:
2899:
2893:
2892:
2864:
2858:
2857:
2847:
2815:
2809:
2808:
2764:
2758:
2757:
2750:
2744:
2741:
2735:
2734:
2724:
2714:
2690:
2681:
2680:
2662:
2638:
2632:
2626:
2620:
2614:
2608:
2594:
2588:
2587:
2575:
2569:
2568:
2558:
2518:
2512:
2511:
2501:
2476:(9): 4951–4959.
2461:
2452:
2451:
2449:
2448:
2439:. Archived from
2428:
2422:
2421:
2411:
2401:
2369:
2358:
2357:
2347:
2323:
2317:
2316:
2288:
2275:
2266:
2260:
2259:
2249:
2209:
2196:
2195:
2185:
2160:(9): 1905–1907.
2154:The ISME Journal
2145:
2139:
2138:
2128:
2086:
2080:
2064:
2058:
2042:
2036:
2035:
2001:
1995:
1994:
1976:
1946:
1937:
1936:
1918:
1892:
1886:
1885:
1875:
1833:
1827:
1826:
1776:
1770:
1769:
1759:
1725:
1719:
1718:
1700:
1674:
1668:
1662:
1637:
1538:Candida albicans
1309:
1302:
1295:
1282:
1277:
1276:
1046:Marine holobiont
846:Fecal transplant
726:
707:
706:
698:Research methods
584:immune tolerance
470:Plant microbiome
419:leaf-cutter ants
393:Chytridiomycosis
356:human microbiome
334:Human microbiota
151:ecological niche
66:
63:
57:
34:
26:
7115:
7114:
7110:
7109:
7108:
7106:
7105:
7104:
7075:
7074:
7073:
7063:
7061:
7049:
7041:
7039:
7034:
7016:
6995:
6991:Faecal transfer
6974:
6899:
6894:
6864:
6859:
6841:
6778:
6732:
6612:
6563:Marine protists
6536:
6517:Root microbiome
6500:
6409:Biological pump
6390:
6340:
6287:
6278:
6220:
6215:
6185:
6184:
6183:
6165:
6161:
6156:
6155:
6146:
6144:
6143:on May 30, 2015
6129:
6125:
6101:10.1038/521136a
6072:
6068:
6059:
6057:
6045:
6041:
6036:
6032:
6026:
6022:
6016:
6012:
6003:
5999:
5993:
5989:
5984:
5980:
5974:
5970:
5964:
5955:
5949:
5945:
5939:
5935:
5929:
5925:
5919:
5915:
5909:
5905:
5899:
5895:
5856:
5852:
5831:(10): 560–568.
5821:
5812:
5759:
5755:
5710:
5706:
5659:
5655:
5610:
5606:
5596:
5594:
5593:on 11 June 2016
5585:
5584:
5577:
5530:
5526:
5479:
5475:
5428:
5424:
5378:
5374:
5329:
5325:
5278:
5274:
5229:
5225:
5180:
5176:
5115:
5111:
5070:(2): e1000667.
5056:
5052:
5020:10.1.1.377.2288
4994:
4990:
4951:(7285): 59–65.
4936:
4932:
4878:
4874:
4862:
4860:
4851:
4850:
4826:
4822:
4769:
4765:
4711:
4707:
4661:
4657:
4647:
4645:
4640:
4639:
4635:
4588:
4584:
4569:
4547:
4543:
4481:
4477:
4432:
4428:
4403:10.1038/nrg3129
4381:
4374:
4317:
4313:
4276:
4272:
4232:
4226:
4222:
4183:
4176:
4141:
4137:
4104:10.1038/ni.3780
4084:
4080:
4035:
4031:
3986:
3977:
3932:
3928:
3875:
3871:
3832:
3828:
3821:
3807:
3803:
3760:
3756:
3708:
3702:
3698:
3663:
3659:
3598:
3591:
3560:
3551:
3535:
3534:
3492:
3488:
3435:
3431:
3398:(5): e1003379.
3384:
3380:
3335:
3331:
3298:(9): e1001129.
3284:
3280:
3233:
3229:
3184:
3180:
3133:
3129:
3098:
3094:
3049:
3045:
2990:
2986:
2951:
2947:
2900:
2896:
2865:
2861:
2816:
2812:
2765:
2761:
2752:
2751:
2747:
2742:
2738:
2705:(8): e1002533.
2691:
2684:
2639:
2635:
2627:
2623:
2615:
2611:
2605:Wayback Machine
2595:
2591:
2576:
2572:
2519:
2515:
2462:
2455:
2446:
2444:
2429:
2425:
2370:
2361:
2324:
2320:
2305:
2289:
2278:
2267:
2263:
2210:
2199:
2146:
2142:
2087:
2083:
2065:
2061:
2043:
2039:
2002:
1998:
1947:
1940:
1893:
1889:
1834:
1830:
1777:
1773:
1735:Genome Research
1726:
1722:
1675:
1671:
1638:
1634:
1629:
1624:
1560:
1551:
1525:
1484:
1460:
1448:non-coding RNAs
1443:
1411:genome assembly
1387:
1381:
1313:
1272:
1265:
1264:
1263:
1248:
1240:
1239:
1238:
1167:
1152:
1144:
1143:
1142:
1129:
1111:
1101:
1100:
1099:
1083:
1050:
1041:Plant holobiont
1035:
1025:
1024:
1023:
1022:
993:
931:
921:
920:
919:
903:
890:
882:
881:
880:
867:
850:
835:
825:
824:
823:
814:
794:
784:
783:
782:
771:soil microbiome
766:root microbiome
751:
736:
705:
700:
632:Coral bleaching
602:
596:
559:
551:allelochemicals
472:
464:
456:
440:enteroendocrine
384:
336:
330:
310:
273:
238:
186:
136:of their host.
67:
61:
58:
51:
39:This article's
35:
24:
17:
12:
11:
5:
7113:
7103:
7102:
7097:
7092:
7087:
7072:
7071:
7059:
7036:
7035:
7033:
7032:
7021:
7018:
7017:
7015:
7014:
7009:
7003:
7001:
6997:
6996:
6994:
6993:
6988:
6982:
6980:
6976:
6975:
6973:
6972:
6967:
6962:
6957:
6952:
6947:
6946:
6945:
6940:
6930:
6925:
6920:
6915:
6909:
6907:
6901:
6900:
6893:
6892:
6885:
6878:
6870:
6861:
6860:
6858:
6857:
6846:
6843:
6842:
6840:
6839:
6834:
6829:
6824:
6819:
6814:
6813:
6812:
6802:
6797:
6795:Deep biosphere
6792:
6790:Bioremediation
6786:
6784:
6780:
6779:
6777:
6776:
6771:
6766:
6761:
6756:
6751:
6749:DNA sequencing
6746:
6740:
6738:
6734:
6733:
6731:
6730:
6725:
6720:
6715:
6714:
6713:
6708:
6703:
6702:
6701:
6691:
6686:
6681:
6676:
6671:
6666:
6661:
6651:
6646:
6641:
6636:
6631:
6626:
6620:
6618:
6614:
6613:
6611:
6610:
6605:
6600:
6595:
6590:
6585:
6580:
6575:
6570:
6565:
6560:
6555:
6553:Marine viruses
6550:
6544:
6542:
6538:
6537:
6535:
6534:
6529:
6524:
6519:
6514:
6508:
6506:
6502:
6501:
6499:
6498:
6490:
6488:Quorum sensing
6485:
6484:
6483:
6478:
6468:
6463:
6458:
6457:
6456:
6451:
6449:microbial loop
6441:
6439:Microbial cyst
6436:
6431:
6426:
6421:
6416:
6411:
6406:
6400:
6398:
6392:
6391:
6389:
6388:
6383:
6382:
6381:
6371:
6370:
6369:
6364:
6359:
6357:run-and-tumble
6348:
6346:
6342:
6341:
6339:
6338:
6333:
6328:
6323:
6318:
6313:
6308:
6303:
6297:
6295:
6289:
6288:
6281:
6279:
6277:
6276:
6271:
6266:
6261:
6256:
6251:
6246:
6241:
6236:
6230:
6228:
6222:
6221:
6218:Microorganisms
6214:
6213:
6206:
6199:
6191:
6166:
6159:
6158:
6157:
6154:
6153:
6123:
6066:
6039:
6030:
6020:
6010:
5997:
5987:
5978:
5968:
5953:
5943:
5933:
5923:
5913:
5903:
5893:
5866:(5): 303–315.
5850:
5827:(in Spanish).
5810:
5753:
5724:(2): 112–119.
5704:
5653:
5624:(3): 243–248.
5604:
5575:
5540:(3): 486–493.
5524:
5473:
5422:
5372:
5323:
5272:
5223:
5174:
5109:
5050:
4988:
4930:
4872:
4863:|journal=
4838:10.1101/081257
4820:
4783:(7): 581–583.
4777:Nature Methods
4763:
4705:
4676:(5): 335–336.
4670:Nature Methods
4655:
4633:
4582:
4567:
4541:
4475:
4446:(2): 330–339.
4426:
4372:
4311:
4270:
4220:
4193:(5): 355–362.
4174:
4135:
4098:(8): 851–860.
4078:
4029:
3975:
3926:
3889:(2): 122–127.
3869:
3842:(4): 343–350.
3826:
3819:
3801:
3754:
3696:
3657:
3589:
3549:
3506:(8): 324–334.
3500:Microbial Cell
3486:
3449:(2): 141–149.
3429:
3378:
3329:
3278:
3227:
3178:
3127:
3108:(1): 100–109.
3092:
3063:(5): 440–447.
3043:
2984:
2965:(4): 237–245.
2945:
2894:
2875:(5): 445–454.
2859:
2830:(4): 425–428.
2810:
2775:(1): 221–232.
2759:
2745:
2736:
2682:
2653:(3): 337–340.
2633:
2621:
2609:
2589:
2570:
2513:
2453:
2423:
2359:
2338:(9): 560–569.
2318:
2303:
2276:
2261:
2224:(4): 185–190.
2197:
2140:
2081:
2059:
2037:
2018:(3): 133–143.
2006:Margulis, Lynn
2004:Guerrero, R.;
1996:
1959:(3): 485–494.
1938:
1909:(4): 837–848.
1887:
1828:
1771:
1720:
1691:(2): 210–218.
1669:
1631:
1630:
1628:
1625:
1623:
1622:
1617:
1612:
1607:
1602:
1597:
1592:
1587:
1582:
1577:
1572:
1567:
1561:
1559:
1556:
1550:
1549:Privacy issues
1547:
1524:
1521:
1483:
1480:
1459:
1456:
1452:Metaproteomics
1442:
1439:
1399:pyrosequencing
1383:Main article:
1380:
1377:
1315:
1314:
1312:
1311:
1304:
1297:
1289:
1286:
1285:
1284:
1283:
1267:
1266:
1262:
1261:
1256:
1250:
1249:
1246:
1245:
1242:
1241:
1237:
1236:
1231:
1226:
1221:
1216:
1211:
1210:
1209:
1199:
1194:
1189:
1184:
1182:Quorum sensing
1179:
1174:
1168:
1166:
1165:
1160:
1154:
1153:
1150:
1149:
1146:
1145:
1141:
1140:
1135:
1130:
1124:
1119:
1113:
1112:
1107:
1106:
1103:
1102:
1098:
1097:
1096:
1095:
1084:
1082:
1081:
1080:
1079:
1074:
1069:
1064:
1059:
1051:
1049:
1048:
1043:
1037:
1036:
1031:
1030:
1027:
1026:
1021:
1020:
1015:
1010:
1005:
1000:
994:
992:
991:
990:
989:
984:
979:
974:
969:
958:
957:
956:
955:
954:
949:
944:
933:
932:
927:
926:
923:
922:
918:
917:
909:
904:
898:
892:
891:
888:
887:
884:
883:
879:
878:
873:
868:
862:
857:
855:Gut–brain axis
851:
849:
848:
843:
837:
836:
831:
830:
827:
826:
822:
821:
815:
813:
812:
807:
802:
796:
795:
790:
789:
786:
785:
781:
780:
779:
778:
773:
768:
763:
752:
750:
749:
744:
738:
737:
732:
731:
728:
727:
719:
718:
712:
711:
704:
701:
699:
696:
628:Climate change
598:Main article:
595:
592:
558:
555:
547:plant hormones
502:
501:
498:
495:
492:
462:
455:
452:
451:
450:
443:
415:
408:
400:
396:
383:
380:
332:Main article:
329:
326:
309:
306:
272:
269:
237:
234:
226:
225:
222:
219:
185:
182:
90:microorganisms
69:
68:
48:the key points
38:
36:
29:
15:
9:
6:
4:
3:
2:
7112:
7101:
7098:
7096:
7093:
7091:
7088:
7086:
7083:
7082:
7080:
7070:
7060:
7058:
7053:
7048:
7047:
7044:
7031:
7023:
7022:
7019:
7013:
7010:
7008:
7005:
7004:
7002:
6998:
6992:
6989:
6987:
6984:
6983:
6981:
6977:
6971:
6968:
6966:
6963:
6961:
6958:
6956:
6953:
6951:
6948:
6944:
6941:
6939:
6936:
6935:
6934:
6931:
6929:
6926:
6924:
6921:
6919:
6916:
6914:
6911:
6910:
6908:
6906:
6902:
6898:
6891:
6886:
6884:
6879:
6877:
6872:
6871:
6868:
6856:
6848:
6847:
6844:
6838:
6835:
6833:
6830:
6828:
6825:
6823:
6820:
6818:
6815:
6811:
6808:
6807:
6806:
6803:
6801:
6798:
6796:
6793:
6791:
6788:
6787:
6785:
6781:
6775:
6772:
6770:
6767:
6765:
6762:
6760:
6757:
6755:
6752:
6750:
6747:
6745:
6742:
6741:
6739:
6735:
6729:
6726:
6724:
6721:
6719:
6716:
6712:
6709:
6707:
6704:
6700:
6697:
6696:
6695:
6692:
6690:
6687:
6685:
6682:
6680:
6677:
6675:
6672:
6670:
6667:
6665:
6662:
6660:
6657:
6656:
6655:
6652:
6650:
6647:
6645:
6642:
6640:
6639:Microbial oil
6637:
6635:
6632:
6630:
6627:
6625:
6622:
6621:
6619:
6617:Human related
6615:
6609:
6606:
6604:
6603:Picoeukaryote
6601:
6599:
6596:
6594:
6591:
6589:
6586:
6584:
6581:
6579:
6576:
6574:
6571:
6569:
6566:
6564:
6561:
6559:
6556:
6554:
6551:
6549:
6546:
6545:
6543:
6539:
6533:
6530:
6528:
6525:
6523:
6520:
6518:
6515:
6513:
6510:
6509:
6507:
6503:
6497:
6496:
6491:
6489:
6486:
6482:
6479:
6477:
6474:
6473:
6472:
6469:
6467:
6464:
6462:
6461:Microbial mat
6459:
6455:
6452:
6450:
6447:
6446:
6445:
6442:
6440:
6437:
6435:
6432:
6430:
6427:
6425:
6422:
6420:
6417:
6415:
6412:
6410:
6407:
6405:
6402:
6401:
6399:
6397:
6393:
6387:
6384:
6380:
6377:
6376:
6375:
6372:
6368:
6365:
6363:
6360:
6358:
6355:
6354:
6353:
6350:
6349:
6347:
6343:
6337:
6334:
6332:
6329:
6327:
6324:
6322:
6319:
6317:
6314:
6312:
6309:
6307:
6304:
6302:
6299:
6298:
6296:
6294:
6290:
6285:
6275:
6272:
6270:
6267:
6265:
6262:
6260:
6257:
6255:
6254:Nanobacterium
6252:
6250:
6247:
6245:
6244:Cyanobacteria
6242:
6240:
6237:
6235:
6232:
6231:
6229:
6227:
6223:
6219:
6212:
6207:
6205:
6200:
6198:
6193:
6192:
6189:
6181:
6180:
6179:
6173:
6169:
6142:
6138:
6134:
6127:
6119:
6115:
6111:
6107:
6102:
6097:
6093:
6089:
6086:(7551): 136.
6085:
6081:
6077:
6070:
6056:
6055:
6050:
6043:
6034:
6024:
6014:
6008:
6001:
5991:
5982:
5972:
5962:
5960:
5958:
5947:
5937:
5927:
5917:
5907:
5897:
5889:
5885:
5881:
5877:
5873:
5869:
5865:
5861:
5854:
5846:
5842:
5838:
5834:
5830:
5826:
5819:
5817:
5815:
5806:
5802:
5797:
5792:
5788:
5784:
5780:
5776:
5772:
5768:
5764:
5757:
5749:
5745:
5741:
5737:
5732:
5727:
5723:
5719:
5715:
5708:
5700:
5696:
5691:
5686:
5681:
5676:
5672:
5668:
5664:
5657:
5649:
5645:
5640:
5635:
5631:
5627:
5623:
5619:
5615:
5608:
5592:
5588:
5582:
5580:
5571:
5567:
5563:
5559:
5555:
5551:
5547:
5543:
5539:
5535:
5528:
5520:
5516:
5511:
5506:
5501:
5496:
5492:
5488:
5484:
5477:
5469:
5465:
5461:
5457:
5453:
5449:
5445:
5441:
5437:
5433:
5426:
5418:
5414:
5409:
5404:
5400:
5396:
5392:
5388:
5384:
5376:
5368:
5364:
5359:
5354:
5350:
5346:
5342:
5338:
5334:
5327:
5319:
5315:
5310:
5305:
5300:
5295:
5291:
5287:
5283:
5276:
5268:
5264:
5259:
5254:
5250:
5246:
5242:
5238:
5234:
5227:
5219:
5215:
5210:
5205:
5201:
5197:
5193:
5189:
5185:
5178:
5170:
5166:
5161:
5156:
5152:
5148:
5144:
5140:
5136:
5132:
5128:
5124:
5120:
5113:
5105:
5101:
5096:
5091:
5086:
5081:
5077:
5073:
5069:
5065:
5061:
5054:
5046:
5042:
5038:
5034:
5030:
5026:
5021:
5016:
5012:
5008:
5004:
5000:
4992:
4984:
4980:
4975:
4970:
4966:
4962:
4958:
4954:
4950:
4946:
4942:
4934:
4926:
4922:
4917:
4912:
4908:
4904:
4900:
4896:
4892:
4888:
4884:
4876:
4868:
4855:
4847:
4843:
4839:
4835:
4831:
4824:
4816:
4812:
4807:
4802:
4798:
4794:
4790:
4786:
4782:
4778:
4774:
4767:
4759:
4755:
4750:
4745:
4741:
4737:
4733:
4729:
4725:
4721:
4717:
4709:
4701:
4697:
4692:
4687:
4683:
4679:
4675:
4671:
4667:
4659:
4643:
4637:
4629:
4625:
4620:
4615:
4610:
4605:
4601:
4597:
4593:
4586:
4578:
4574:
4570:
4568:9780123809896
4564:
4560:
4556:
4552:
4545:
4537:
4533:
4528:
4523:
4519:
4515:
4511:
4507:
4503:
4499:
4495:
4491:
4487:
4479:
4471:
4467:
4462:
4457:
4453:
4449:
4445:
4441:
4437:
4430:
4422:
4418:
4413:
4408:
4404:
4400:
4396:
4392:
4391:
4386:
4379:
4377:
4368:
4364:
4360:
4356:
4351:
4346:
4342:
4338:
4334:
4330:
4326:
4324:
4315:
4307:
4303:
4298:
4293:
4289:
4285:
4281:
4274:
4266:
4262:
4258:
4254:
4250:
4246:
4242:
4238:
4231:
4224:
4216:
4212:
4208:
4204:
4200:
4196:
4192:
4188:
4181:
4179:
4170:
4166:
4162:
4158:
4155:(2): 94–104.
4154:
4150:
4146:
4139:
4131:
4127:
4122:
4117:
4113:
4109:
4105:
4101:
4097:
4093:
4089:
4082:
4074:
4070:
4065:
4060:
4056:
4052:
4048:
4044:
4040:
4033:
4025:
4021:
4016:
4011:
4007:
4003:
3999:
3995:
3991:
3984:
3982:
3980:
3971:
3967:
3962:
3957:
3953:
3949:
3945:
3941:
3937:
3930:
3922:
3918:
3913:
3908:
3904:
3900:
3896:
3892:
3888:
3884:
3880:
3873:
3865:
3861:
3857:
3853:
3849:
3845:
3841:
3837:
3830:
3822:
3816:
3812:
3805:
3797:
3793:
3788:
3783:
3778:
3773:
3769:
3765:
3758:
3750:
3746:
3742:
3738:
3734:
3730:
3726:
3722:
3718:
3714:
3707:
3700:
3692:
3688:
3684:
3680:
3676:
3672:
3668:
3667:Plant Science
3661:
3653:
3649:
3645:
3641:
3637:
3633:
3628:
3623:
3619:
3615:
3611:
3607:
3603:
3596:
3594:
3586:
3581:
3576:
3572:
3568:
3564:
3558:
3556:
3554:
3545:
3539:
3531:
3527:
3522:
3517:
3513:
3509:
3505:
3501:
3497:
3490:
3482:
3478:
3473:
3468:
3464:
3460:
3456:
3452:
3448:
3444:
3440:
3433:
3425:
3421:
3416:
3411:
3406:
3401:
3397:
3393:
3389:
3382:
3374:
3370:
3365:
3360:
3356:
3352:
3348:
3344:
3340:
3333:
3325:
3321:
3316:
3311:
3306:
3301:
3297:
3293:
3289:
3282:
3274:
3270:
3265:
3260:
3255:
3250:
3246:
3242:
3241:BMC Microbiol
3238:
3231:
3223:
3219:
3214:
3209:
3205:
3201:
3197:
3193:
3189:
3182:
3174:
3170:
3166:
3162:
3158:
3154:
3150:
3146:
3142:
3138:
3131:
3123:
3119:
3115:
3111:
3107:
3103:
3096:
3088:
3084:
3079:
3074:
3070:
3066:
3062:
3058:
3054:
3047:
3039:
3035:
3030:
3025:
3020:
3015:
3011:
3007:
3003:
2999:
2995:
2988:
2980:
2976:
2972:
2968:
2964:
2960:
2956:
2949:
2941:
2937:
2932:
2927:
2922:
2917:
2913:
2909:
2905:
2898:
2890:
2886:
2882:
2878:
2874:
2870:
2863:
2855:
2851:
2846:
2841:
2837:
2833:
2829:
2825:
2821:
2814:
2806:
2802:
2798:
2794:
2790:
2786:
2782:
2778:
2774:
2770:
2763:
2755:
2749:
2740:
2732:
2728:
2723:
2718:
2713:
2708:
2704:
2700:
2696:
2689:
2687:
2678:
2674:
2670:
2666:
2661:
2656:
2652:
2648:
2644:
2637:
2631:
2625:
2619:
2613:
2606:
2602:
2599:
2593:
2585:
2581:
2574:
2566:
2562:
2557:
2552:
2548:
2544:
2540:
2536:
2532:
2528:
2524:
2517:
2509:
2505:
2500:
2495:
2491:
2487:
2483:
2479:
2475:
2471:
2467:
2460:
2458:
2443:on 2019-12-07
2442:
2438:
2434:
2427:
2419:
2415:
2410:
2405:
2400:
2395:
2391:
2387:
2383:
2379:
2375:
2368:
2366:
2364:
2355:
2351:
2346:
2341:
2337:
2333:
2329:
2322:
2314:
2310:
2306:
2304:9780073402406
2300:
2296:
2295:
2287:
2285:
2283:
2281:
2273:
2272:
2265:
2257:
2253:
2248:
2243:
2239:
2235:
2231:
2227:
2223:
2219:
2215:
2208:
2206:
2204:
2202:
2193:
2189:
2184:
2179:
2175:
2171:
2167:
2163:
2159:
2155:
2151:
2144:
2136:
2132:
2127:
2122:
2118:
2114:
2110:
2106:
2102:
2098:
2097:
2092:
2085:
2079:
2075:
2071:
2070:
2063:
2057:
2053:
2049:
2048:
2041:
2033:
2029:
2025:
2021:
2017:
2013:
2012:
2007:
2000:
1992:
1988:
1984:
1980:
1975:
1970:
1966:
1962:
1958:
1954:
1953:
1945:
1943:
1934:
1930:
1926:
1922:
1917:
1912:
1908:
1904:
1903:
1898:
1891:
1883:
1879:
1874:
1869:
1865:
1861:
1857:
1853:
1849:
1845:
1844:
1839:
1832:
1824:
1820:
1816:
1812:
1808:
1804:
1800:
1796:
1792:
1788:
1787:
1782:
1775:
1767:
1763:
1758:
1753:
1749:
1745:
1741:
1737:
1736:
1731:
1724:
1716:
1712:
1708:
1704:
1699:
1694:
1690:
1686:
1685:
1680:
1673:
1666:
1661:
1656:
1652:
1648:
1644:
1643:
1636:
1632:
1621:
1618:
1616:
1615:Vaginal flora
1613:
1611:
1608:
1606:
1603:
1601:
1598:
1596:
1593:
1591:
1588:
1586:
1583:
1581:
1578:
1576:
1573:
1571:
1568:
1566:
1563:
1562:
1555:
1546:
1543:
1539:
1535:
1529:
1520:
1517:
1515:
1510:
1509:
1505:
1501:
1497:
1493:
1488:
1479:
1477:
1472:
1469:
1465:
1455:
1453:
1449:
1438:
1436:
1432:
1426:
1424:
1420:
1416:
1412:
1408:
1404:
1400:
1396:
1391:
1386:
1376:
1374:
1370:
1365:
1360:
1358:
1354:
1350:
1346:
1341:
1337:
1335:
1331:
1327:
1322:
1310:
1305:
1303:
1298:
1296:
1291:
1290:
1288:
1287:
1281:
1271:
1270:
1269:
1268:
1260:
1257:
1255:
1252:
1251:
1244:
1243:
1235:
1234:Symbiogenesis
1232:
1230:
1229:Superorganism
1227:
1225:
1222:
1220:
1217:
1215:
1212:
1208:
1205:
1204:
1203:
1200:
1198:
1195:
1193:
1190:
1188:
1185:
1183:
1180:
1178:
1175:
1173:
1170:
1169:
1164:
1161:
1159:
1156:
1155:
1148:
1147:
1139:
1136:
1134:
1131:
1128:
1125:
1123:
1120:
1118:
1115:
1114:
1110:
1105:
1104:
1094:
1091:
1090:
1089:
1086:
1085:
1078:
1075:
1073:
1070:
1068:
1065:
1063:
1060:
1058:
1055:
1054:
1053:
1052:
1047:
1044:
1042:
1039:
1038:
1034:
1029:
1028:
1019:
1016:
1014:
1011:
1009:
1006:
1004:
1001:
999:
996:
995:
988:
985:
983:
980:
978:
975:
973:
970:
968:
965:
964:
963:
960:
959:
953:
952:rhizobacteria
950:
948:
945:
943:
940:
939:
938:
935:
934:
930:
925:
924:
916:
914:
910:
908:
905:
902:
899:
897:
894:
893:
886:
885:
877:
874:
872:
869:
866:
863:
861:
858:
856:
853:
852:
847:
844:
842:
839:
838:
834:
829:
828:
820:
817:
816:
811:
808:
806:
803:
801:
798:
797:
793:
788:
787:
777:
774:
772:
769:
767:
764:
762:
759:
758:
757:
754:
753:
748:
745:
743:
740:
739:
735:
730:
729:
725:
721:
720:
717:
714:
713:
709:
708:
695:
693:
692:endosymbiosis
689:
688:symbiogenesis
684:
683:O. patagonica
680:
675:
672:
666:
664:
660:
656:
652:
651:O. patagonica
648:
647:O. patagonica
644:
643:Vibrio shiloi
640:
638:
633:
629:
625:
621:
619:
611:
606:
601:
591:
589:
585:
581:
576:
571:
569:
564:
563:immune system
554:
552:
548:
544:
540:
535:
531:
527:
523:
519:
515:
506:
499:
496:
493:
490:
489:
488:
486:
482:
478:
471:
460:
448:
444:
441:
437:
433:
429:
425:
420:
416:
412:
409:
405:
401:
397:
394:
390:
386:
385:
379:
376:
371:
367:
363:
361:
357:
353:
352:Micro-animals
350:and viruses.
349:
345:
341:
335:
322:
318:
314:
305:
303:
298:
294:
290:
286:
282:
278:
268:
266:
262:
258:
254:
250:
246:
242:
233:
231:
223:
220:
218:
214:
210:
209:
208:
206:
199:
195:
190:
181:
179:
175:
171:
167:
163:
159:
154:
152:
148:
144:
143:
137:
135:
131:
127:
123:
119:
115:
111:
107:
103:
99:
95:
91:
87:
80:
75:
65:
55:
49:
47:
42:
37:
33:
28:
27:
22:
7090:Bacteriology
7085:Microbiology
6965:Human virome
6938:In pregnancy
6805:Microswimmer
6699:in pregnancy
6649:Nylon-eating
6532:Spermosphere
6494:
6475:
6293:Microbiology
6176:
6175:
6174:profile for
6171:
6145:. Retrieved
6141:the original
6136:
6126:
6083:
6079:
6069:
6058:. Retrieved
6052:
6042:
6033:
6023:
6013:
6000:
5990:
5981:
5971:
5946:
5936:
5926:
5916:
5906:
5896:
5863:
5859:
5853:
5828:
5824:
5770:
5766:
5756:
5721:
5717:
5707:
5670:
5666:
5656:
5621:
5617:
5607:
5595:. Retrieved
5591:the original
5537:
5533:
5527:
5490:
5486:
5476:
5435:
5431:
5425:
5390:
5386:
5375:
5340:
5336:
5326:
5289:
5285:
5275:
5240:
5236:
5226:
5191:
5187:
5177:
5126:
5122:
5112:
5067:
5063:
5053:
5002:
4998:
4991:
4948:
4944:
4933:
4890:
4886:
4875:
4854:cite journal
4823:
4780:
4776:
4766:
4723:
4719:
4708:
4673:
4669:
4658:
4646:. Retrieved
4636:
4599:
4595:
4585:
4550:
4544:
4496:(1): 13637.
4493:
4489:
4478:
4443:
4439:
4429:
4397:(1): 47–58.
4394:
4388:
4335:(1): 67–73.
4332:
4328:
4322:
4314:
4287:
4283:
4273:
4243:(1): 16–20.
4240:
4236:
4223:
4190:
4186:
4152:
4148:
4138:
4095:
4091:
4081:
4046:
4042:
4032:
4000:(1): 12–22.
3997:
3993:
3943:
3939:
3929:
3886:
3882:
3872:
3839:
3835:
3829:
3810:
3804:
3767:
3757:
3716:
3712:
3699:
3666:
3660:
3609:
3605:
3566:
3562:
3538:cite journal
3503:
3499:
3489:
3446:
3442:
3432:
3395:
3391:
3381:
3346:
3342:
3332:
3295:
3291:
3281:
3244:
3240:
3230:
3198:(220): 220.
3195:
3191:
3181:
3140:
3136:
3130:
3105:
3101:
3095:
3060:
3056:
3046:
3001:
2997:
2987:
2962:
2958:
2954:
2948:
2911:
2907:
2897:
2872:
2868:
2862:
2827:
2823:
2813:
2772:
2769:Microb. Ecol
2768:
2762:
2748:
2739:
2702:
2699:PLOS Biology
2698:
2650:
2646:
2636:
2624:
2612:
2607:January 2014
2592:
2583:
2573:
2530:
2526:
2516:
2473:
2469:
2445:. Retrieved
2441:the original
2436:
2426:
2381:
2377:
2335:
2331:
2321:
2293:
2269:
2264:
2221:
2217:
2157:
2153:
2143:
2100:
2094:
2084:
2067:
2062:
2045:
2040:
2015:
2009:
1999:
1956:
1950:
1906:
1900:
1890:
1847:
1841:
1831:
1790:
1784:
1774:
1739:
1733:
1723:
1688:
1682:
1672:
1646:
1640:
1635:
1605:Psychobiotic
1575:Human virome
1552:
1541:
1537:
1533:
1530:
1526:
1518:
1511:
1507:
1503:
1499:
1495:
1491:
1489:
1485:
1473:
1461:
1444:
1427:
1415:tree of life
1390:Metagenomics
1388:
1385:Metagenomics
1361:
1342:
1338:
1329:
1318:
1219:Metabolomics
1202:Metagenomics
1088:Hologenomics
928:
912:
776:spermosphere
747:Phyllosphere
682:
678:
676:
667:
654:
650:
646:
635:
623:
622:
615:
580:inflammation
572:
560:
511:
473:
388:
372:
368:
364:
337:
321:inflammation
302:C. difficile
301:
293:phyllosphere
289:spermosphere
274:
264:
261:normal flora
260:
241:Commensalism
239:
227:
213:metagenomics
202:
184:Introduction
155:
140:
138:
108:, including
92:that may be
85:
84:
62:October 2018
59:
43:
41:lead section
7100:Microbiomes
6905:Human flora
6837:Siderophore
6598:Phycosphere
6454:viral shunt
3392:PLOS Pathog
2268:Poreau B.,
2047:BMC Biology
1974:11336/33456
1514:endotoxemia
1172:Gnotobiosis
901:Phycosphere
761:laimosphere
756:Rhizosphere
716:Microbiomes
624:Coral reefs
534:mutualistic
477:rhizosphere
436:baculovirus
323:in the lung
304:infection.
253:mutualistic
166:epigenetics
162:synergistic
134:homeostasis
98:mutualistic
7079:Categories
6737:Techniques
6568:Microalgae
6476:microbiota
6471:Microbiome
6259:Prokaryote
6178:microbiome
6147:2015-05-17
6060:2015-05-17
5129:(1): 870.
4832:: 081257.
3994:Immunology
3292:PLOS Genet
2447:2019-12-07
1649:: 100161.
1627:References
1610:Skin flora
1595:Phytobiome
1565:Anagenesis
1349:clustering
1224:Pan-genome
1177:Phytobiome
1138:Virosphere
1033:Holobionts
929:Microbiota
913:Drosophila
876:Necrobiome
841:Human milk
742:Endosphere
639:patagonica
518:endophytes
468:See also:
317:Pathogenic
198:human skin
164:unit from
142:microbiome
102:pathogenic
86:Microbiota
7012:OpenBiome
6986:Dysbiosis
6970:Mycobiota
6810:biohybrid
6664:dysbiosis
6481:holobiont
6379:amoeboids
6362:twitching
5880:1262-3636
5787:0264-6021
5740:2090-7184
5151:2041-1723
5015:CiteSeerX
4797:1548-7105
4518:2045-2322
4112:1529-2908
3903:1521-6616
3733:1365-2672
3652:231576517
3636:1462-2920
3463:0966-842X
2914:: e9335.
2313:886600661
2117:1074-7613
1600:Probiotic
1319:Targeted
1163:Dysbiosis
1077:rhodolith
942:endophyte
896:Mycobiome
860:Placental
679:V. shiloi
671:holobiont
655:V. shiloi
522:Oomycetes
514:epiphytes
428:fruit fly
285:root zone
257:parasitic
230:phenotype
170:holobiont
158:evolution
139:The term
94:commensal
46:summarize
7095:Bacteria
7069:Medicine
7030:Category
6960:Salivary
6950:Placenta
6855:Category
6774:Staining
6706:placenta
6336:Virology
6331:Mycology
6269:Protozoa
6239:Bacteria
6110:25971486
5888:27179626
5845:27633134
5805:28512250
5699:22587826
5673:(1): 5.
5648:21304727
5570:26953155
5562:17431707
5519:21257745
5460:19444216
5417:17932063
5367:21045053
5318:18803844
5267:19880382
5218:19900971
5169:29491419
5104:20195499
5037:15845853
4983:20203603
4925:19043404
4815:27214047
4758:19801464
4700:20383131
4648:11 March
4628:18638418
4577:20602987
4536:32788589
4470:17391789
4421:22179717
4359:18059488
4265:19022522
4207:17384666
4169:24678760
4130:28722709
4073:27034283
4024:22804726
3970:19963349
3921:26141651
3864:11418345
3796:19278554
3749:35055151
3741:19120625
3691:22794922
3644:33427409
3563:PLOS One
3530:31403049
3481:31699645
3424:23717206
3373:24865556
3324:20885794
3273:24373613
3222:24452263
3165:17183312
3122:25145536
3087:22422004
3057:EMBO Rep
3038:19181843
2940:32612884
2889:10785270
2854:84293044
2805:12951957
2797:26271741
2731:27541692
2669:26824647
2601:Archived
2584:Vitalacy
2565:25909975
2508:16151072
2418:11607500
2354:24729765
2256:21737250
2192:25647346
2135:34107298
2096:Immunity
2032:24568029
1991:30677140
1983:25430522
1933:17203181
1925:16497592
1882:17943116
1815:15790844
1766:19819907
1715:73455329
1707:30763535
1558:See also
1458:Projects
1359:(ASVs).
1326:16S rRNA
1321:amplicon
1280:Category
1247:Projects
1127:Mangrove
1067:seagrass
947:epiphyte
865:Salivary
810:Cetacean
800:Seagrass
661:. Their
557:Research
340:bacteria
300:chronic
295:and the
194:bacteria
122:protists
114:bacteria
7057:Biology
7043:Portals
7000:Related
6404:Biofilm
6396:Ecology
6367:gliding
6264:Protist
6234:Archaea
6168:Scholia
6118:4393347
6088:Bibcode
5901:110812.
5796:5433529
5748:3900880
5690:3348666
5639:3035311
5597:14 June
5542:Bibcode
5510:3048661
5468:4340144
5440:Bibcode
5408:2238950
5358:3013694
5309:2563014
5292:: 386.
5258:2808910
5209:2808932
5160:5830445
5131:Bibcode
5095:2829047
5072:Bibcode
5007:Bibcode
4999:Science
4974:3779803
4953:Bibcode
4916:2677729
4895:Bibcode
4806:4927377
4749:2786419
4728:Bibcode
4691:3156573
4619:2517594
4527:7423937
4498:Bibcode
4461:2562909
4412:5119550
4367:1032896
4337:Bibcode
4306:9031305
4245:Bibcode
4215:2967190
4121:5800875
4064:4817260
4015:3533697
3961:3155383
3912:4943041
3844:Bibcode
3787:2654666
3671:Bibcode
3614:Bibcode
3521:6685047
3472:6980660
3415:3662647
3364:4045073
3315:2944797
3264:3878784
3247:: 303.
3213:3973144
3173:4400297
3145:Bibcode
3078:3343350
3029:2633212
3006:Bibcode
2979:9775355
2931:7319036
2832:Bibcode
2777:Bibcode
2722:4991899
2677:1790146
2556:4611498
2535:Bibcode
2499:1214602
2478:Bibcode
2386:Bibcode
2345:3983973
2247:3992624
2226:Bibcode
2218:Zoology
2183:4542044
2162:Bibcode
2126:8363570
1873:3709439
1852:Bibcode
1823:6332272
1795:Bibcode
1786:Science
1757:2792171
1542:Candida
1403:contigs
1364:UniFrac
1151:Related
1133:Viriome
1109:Viromes
987:vaginal
871:Uterine
637:Oculina
481:stolons
360:genomes
348:archaea
281:sponges
178:acetate
147:genomes
130:viruses
118:archaea
6955:Uterus
6933:Vagina
6711:uterus
6694:vagina
6659:asthma
6541:Marine
6505:Plants
6345:Motion
6226:Groups
6170:has a
6116:
6108:
6080:Nature
5886:
5878:
5843:
5803:
5793:
5785:
5746:
5738:
5697:
5687:
5646:
5636:
5568:
5560:
5517:
5507:
5466:
5458:
5432:Nature
5415:
5405:
5365:
5355:
5316:
5306:
5265:
5255:
5216:
5206:
5167:
5157:
5149:
5102:
5092:
5045:161283
5043:
5035:
5017:
4981:
4971:
4945:Nature
4923:
4913:
4887:Nature
4846:784388
4844:
4813:
4803:
4795:
4756:
4746:
4698:
4688:
4626:
4616:
4602:: 12.
4575:
4565:
4534:
4524:
4516:
4468:
4458:
4419:
4409:
4365:
4357:
4304:
4263:
4213:
4205:
4167:
4128:
4118:
4110:
4071:
4061:
4022:
4012:
3968:
3958:
3919:
3909:
3901:
3862:
3817:
3794:
3784:
3747:
3739:
3731:
3689:
3650:
3642:
3634:
3528:
3518:
3479:
3469:
3461:
3422:
3412:
3371:
3361:
3322:
3312:
3271:
3261:
3220:
3210:
3171:
3163:
3137:Nature
3120:
3085:
3075:
3036:
3026:
2977:
2938:
2928:
2887:
2852:
2803:
2795:
2729:
2719:
2675:
2667:
2563:
2553:
2527:ISME J
2506:
2496:
2416:
2406:
2352:
2342:
2311:
2301:
2254:
2244:
2190:
2180:
2133:
2123:
2115:
2030:
1989:
1981:
1931:
1923:
1880:
1870:
1843:Nature
1821:
1813:
1764:
1754:
1713:
1705:
1373:mothur
1278:
1072:sponge
998:Marine
586:, and
530:hyphae
454:Plants
328:Humans
297:flower
128:, and
110:plants
6923:Mouth
6827:Omics
6783:Other
6684:mouth
6669:fecal
6274:Virus
6249:Fungi
6172:topic
6114:S2CID
6018:1206.
5941:10410
5744:S2CID
5566:S2CID
5464:S2CID
5041:S2CID
4842:S2CID
4363:S2CID
4302:S2CID
4233:(PDF)
4211:S2CID
3745:S2CID
3709:(PDF)
3648:S2CID
3169:S2CID
2908:PeerJ
2850:S2CID
2801:S2CID
2673:S2CID
2409:45331
1987:S2CID
1929:S2CID
1819:S2CID
1711:S2CID
1570:Biome
1534:et al
1435:IMG/M
1407:BLAST
1369:QIIME
1207:viral
1122:Human
1057:coral
962:Human
937:Plant
805:Coral
568:birth
526:fungi
485:tuber
404:rumen
344:fungi
160:as a
126:fungi
100:, or
6928:Skin
6918:Lung
6689:skin
6679:lung
6106:PMID
6028:159.
5966:1-20
5951:1-6.
5884:PMID
5876:ISSN
5841:PMID
5801:PMID
5783:ISSN
5736:ISSN
5695:PMID
5644:PMID
5599:2016
5558:PMID
5515:PMID
5456:PMID
5413:PMID
5363:PMID
5314:PMID
5263:PMID
5214:PMID
5165:PMID
5147:ISSN
5100:PMID
5033:PMID
4979:PMID
4921:PMID
4867:help
4811:PMID
4793:ISSN
4754:PMID
4696:PMID
4650:2012
4624:PMID
4573:PMID
4563:ISBN
4532:PMID
4514:ISSN
4466:PMID
4417:PMID
4355:PMID
4261:PMID
4203:PMID
4165:PMID
4126:PMID
4108:ISSN
4069:PMID
4043:mBio
4020:PMID
3966:PMID
3917:PMID
3899:ISSN
3860:PMID
3815:ISBN
3792:PMID
3737:PMID
3729:ISSN
3687:PMID
3640:PMID
3632:ISSN
3544:link
3526:PMID
3477:PMID
3459:ISSN
3420:PMID
3369:PMID
3343:mBio
3320:PMID
3269:PMID
3218:PMID
3161:PMID
3118:PMID
3083:PMID
3034:PMID
2975:PMID
2936:PMID
2885:PMID
2793:PMID
2727:PMID
2665:PMID
2647:Cell
2561:PMID
2504:PMID
2414:PMID
2350:PMID
2309:OCLC
2299:ISBN
2252:PMID
2188:PMID
2131:PMID
2113:ISSN
2028:PMID
1979:PMID
1921:PMID
1902:Cell
1878:PMID
1811:PMID
1762:PMID
1703:PMID
1502:and
1474:The
1462:The
1431:KEGG
1423:Hi-C
1419:taxa
1330:i.e.
1062:crab
982:skin
977:oral
972:lung
524:and
411:Mice
373:The
275:The
215:and
6913:Gut
6674:gut
6096:doi
6084:521
5976:249
5931:658
5921:1-6
5868:doi
5833:doi
5791:PMC
5775:doi
5771:474
5726:doi
5685:PMC
5675:doi
5634:PMC
5626:doi
5550:doi
5505:PMC
5495:doi
5491:286
5448:doi
5436:459
5403:PMC
5395:doi
5353:PMC
5345:doi
5304:PMC
5294:doi
5253:PMC
5245:doi
5204:PMC
5196:doi
5155:PMC
5139:doi
5090:PMC
5080:doi
5025:doi
5003:308
4969:PMC
4961:doi
4949:464
4911:PMC
4903:doi
4891:457
4834:doi
4801:PMC
4785:doi
4744:PMC
4736:doi
4686:PMC
4678:doi
4614:PMC
4604:doi
4555:doi
4522:PMC
4506:doi
4456:PMC
4448:doi
4407:PMC
4399:doi
4345:doi
4292:doi
4253:doi
4195:doi
4157:doi
4116:PMC
4100:doi
4059:PMC
4051:doi
4010:PMC
4002:doi
3998:138
3956:PMC
3948:doi
3907:PMC
3891:doi
3887:159
3852:doi
3782:PMC
3772:doi
3721:doi
3717:105
3679:doi
3622:doi
3571:doi
3516:PMC
3508:doi
3467:PMC
3451:doi
3410:PMC
3400:doi
3359:PMC
3351:doi
3310:PMC
3300:doi
3259:PMC
3249:doi
3208:PMC
3200:doi
3153:doi
3141:444
3110:doi
3106:135
3073:PMC
3065:doi
3024:PMC
3014:doi
3002:106
2967:doi
2957:".
2926:PMC
2916:doi
2877:doi
2840:doi
2785:doi
2717:PMC
2707:doi
2655:doi
2651:164
2551:PMC
2543:doi
2494:PMC
2486:doi
2404:PMC
2394:doi
2340:PMC
2242:PMC
2234:doi
2222:114
2178:PMC
2170:doi
2121:PMC
2105:doi
2074:doi
2052:doi
2020:doi
1969:hdl
1961:doi
1911:doi
1907:124
1868:PMC
1860:doi
1848:449
1803:doi
1791:307
1752:PMC
1744:doi
1693:doi
1651:doi
1397:or
1117:Bat
967:gut
915:gut
263:or
196:on
176:),
7081::
6135:.
6112:.
6104:.
6094:.
6082:.
6078:.
6051:.
5956:^
5882:.
5874:.
5864:42
5862:.
5839:.
5829:63
5813:^
5799:.
5789:.
5781:.
5769:.
5765:.
5742:.
5734:.
5720:.
5716:.
5693:.
5683:.
5669:.
5665:.
5642:.
5632:.
5620:.
5616:.
5578:^
5564:.
5556:.
5548:.
5538:53
5536:.
5513:.
5503:.
5489:.
5485:.
5462:.
5454:.
5446:.
5434:.
5411:.
5401:.
5391:36
5389:.
5385:.
5361:.
5351:.
5341:39
5339:.
5335:.
5312:.
5302:.
5288:.
5284:.
5261:.
5251:.
5241:38
5239:.
5235:.
5212:.
5202:.
5192:38
5190:.
5186:.
5163:.
5153:.
5145:.
5137:.
5125:.
5121:.
5098:.
5088:.
5078:.
5066:.
5062:.
5039:.
5031:.
5023:.
5013:.
5001:.
4977:.
4967:.
4959:.
4947:.
4943:.
4919:.
4909:.
4901:.
4889:.
4885:.
4858::
4856:}}
4852:{{
4840:.
4809:.
4799:.
4791:.
4781:13
4779:.
4775:.
4752:.
4742:.
4734:.
4724:75
4722:.
4718:.
4694:.
4684:.
4672:.
4668:.
4622:.
4612:.
4598:.
4594:.
4571:.
4561:.
4530:.
4520:.
4512:.
4504:.
4494:10
4492:.
4488:.
4464:.
4454:.
4444:69
4442:.
4438:.
4415:.
4405:.
4395:13
4393:.
4387:.
4375:^
4361:.
4353:.
4343:.
4331:.
4327:.
4300:.
4286:.
4282:.
4259:.
4251:.
4241:24
4239:.
4235:.
4209:.
4201:.
4189:.
4177:^
4163:.
4153:13
4151:.
4147:.
4124:.
4114:.
4106:.
4096:18
4094:.
4090:.
4067:.
4057:.
4045:.
4041:.
4018:.
4008:.
3996:.
3992:.
3978:^
3964:.
3954:.
3944:34
3942:.
3938:.
3915:.
3905:.
3897:.
3885:.
3881:.
3858:.
3850:.
3838:.
3790:.
3780:.
3766:.
3743:.
3735:.
3727:.
3715:.
3711:.
3685:.
3677:.
3646:.
3638:.
3630:.
3620:.
3610:23
3608:.
3604:.
3592:^
3577:.
3567:14
3565:,
3552:^
3540:}}
3536:{{
3524:.
3502:.
3498:.
3475:.
3465:.
3457:.
3447:28
3445:.
3441:.
3418:.
3408:.
3394:.
3390:.
3367:.
3357:.
3345:.
3341:.
3318:.
3308:.
3294:.
3290:.
3267:.
3257:.
3245:13
3243:.
3239:.
3216:.
3206:.
3194:.
3190:.
3167:.
3159:.
3151:.
3139:.
3116:.
3104:.
3081:.
3071:.
3061:13
3059:.
3055:.
3032:.
3022:.
3012:.
3000:.
2996:.
2973:.
2963:21
2961:.
2934:.
2924:.
2910:.
2906:.
2883:.
2873:24
2871:.
2848:.
2838:.
2828:10
2826:.
2822:.
2799:.
2791:.
2783:.
2773:71
2771:.
2725:.
2715:.
2703:14
2701:.
2697:.
2685:^
2671:.
2663:.
2649:.
2645:.
2582:.
2559:.
2549:.
2541:.
2529:.
2525:.
2502:.
2492:.
2484:.
2474:71
2472:.
2468:.
2456:^
2435:.
2412:.
2402:.
2392:.
2382:91
2380:.
2376:.
2362:^
2348:.
2334:.
2330:.
2307:.
2279:^
2250:.
2240:.
2232:.
2220:.
2216:.
2200:^
2186:.
2176:.
2168:.
2156:.
2152:.
2129:.
2119:.
2111:.
2101:54
2099:.
2093:.
2026:.
2016:16
2014:.
1985:.
1977:.
1967:.
1957:42
1955:.
1941:^
1927:.
1919:.
1905:.
1899:.
1876:.
1866:.
1858:.
1846:.
1840:.
1817:.
1809:.
1801:.
1789:.
1783:.
1760:.
1750:.
1740:19
1738:.
1732:.
1709:.
1701:.
1689:25
1687:.
1681:.
1657:.
1647:23
1645:,
1494:,
1437:.
1371:,
690:,
590:.
582:,
362:.
346:,
342:,
180:.
124:,
120:,
116:,
96:,
7045::
6889:e
6882:t
6875:v
6210:e
6203:t
6196:v
6182:.
6150:.
6120:.
6098::
6090::
6063:.
5890:.
5870::
5847:.
5835::
5807:.
5777::
5750:.
5728::
5722:6
5701:.
5677::
5671:1
5650:.
5628::
5622:3
5601:.
5572:.
5552::
5544::
5521:.
5497::
5470:.
5450::
5442::
5419:.
5397::
5369:.
5347::
5320:.
5296::
5290:9
5269:.
5247::
5220:.
5198::
5171:.
5141::
5133::
5127:9
5106:.
5082::
5074::
5068:6
5047:.
5027::
5009::
4985:.
4963::
4955::
4927:.
4905::
4897::
4869:)
4865:(
4848:.
4836::
4817:.
4787::
4760:.
4738::
4730::
4702:.
4680::
4674:7
4652:.
4630:.
4606::
4600:5
4579:.
4557::
4538:.
4508::
4500::
4472:.
4450::
4423:.
4401::
4369:.
4347::
4339::
4333:2
4325:"
4308:.
4294::
4288:5
4267:.
4255::
4247::
4217:.
4197::
4191:5
4171:.
4159::
4132:.
4102::
4075:.
4053::
4047:7
4026:.
4004::
3972:.
3950::
3923:.
3893::
3866:.
3854::
3846::
3840:4
3823:.
3798:.
3774::
3751:.
3723::
3693:.
3681::
3673::
3654:.
3624::
3616::
3587:.
3573::
3546:)
3532:.
3510::
3504:6
3483:.
3453::
3426:.
3402::
3396:9
3375:.
3353::
3347:5
3326:.
3302::
3296:6
3275:.
3251::
3224:.
3202::
3196:6
3175:.
3155::
3147::
3124:.
3112::
3089:.
3067::
3040:.
3016::
3008::
2981:.
2969::
2942:.
2918::
2912:8
2891:.
2879::
2856:.
2842::
2834::
2807:.
2787::
2779::
2733:.
2709::
2679:.
2657::
2586:.
2567:.
2545::
2537::
2531:9
2510:.
2488::
2480::
2450:.
2420:.
2396::
2388::
2356:.
2336:9
2315:.
2258:.
2236::
2228::
2194:.
2172::
2164::
2158:9
2137:.
2107::
2076::
2054::
2034:.
2022::
1993:.
1971::
1963::
1935:.
1913::
1884:.
1862::
1854::
1825:.
1805::
1797::
1768:.
1746::
1717:.
1695::
1667:.
1653::
1508:.
1328:(
1308:e
1301:t
1294:v
612:.
64:)
60:(
50:.
23:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.