1425:, has led to significant financial losses within the Chinese aquaculture industry of this prawn. Generally, mature male and female prawns are 10 cm and 8 cm in body length respectively, however when impacted by GRD, the prawns exhibit early maturity, wherein males demonstrate mating behaviours at 6 cm and females possess eggs at 5 cm. The impacts of GRD have led to decreases in prawn production of more than 50% in some cases. Through metatranscriptomic sequencing of the viromes of healthy prawns and prawns with GRD, and phylogenetic analysis of the resulting RNA sequences, four marna-like viruses were placed in the
217:
1395:(GPD), a highly pathogenic disease that spreads quickly within aquaculture sites, the industry has suffered significant losses in recent years, resulting in economic instabilities. Shrimp infected with GPD show signs of slower movements and responses, along with color change in liver pancreatic tissues (full brown to light brown). In the 24 to 48 hours after infection, shrimps also exhibit swollen gill filaments, as well as the entire body becoming more transparent and brittle (hence the name of the disease). Discovered through metagenomic sequencing of a
33:
976:
formation of vacuoles, appearance of fibrous matter in vacuolated parts of the cell, and the deterioration of the cytoplasm. Virus particles accumulate in the cytoplasm and may form crystalline arrays or random aggregates. The virus then exits the host cell by tubule-guided viral movement and virions are released by cell lysis. Burst sizes differ greatly between species and range from 66 to 10. As infected cells are lysed shortly after showing signs of infection, proportions of infected cells in the environment remain low.
206:
1004:, a toxic bloom-forming alga, it replicates inside the cytoplasm of its host, and is distributed either singly or in groups as crystalline arrays. Within those that are aligned in crystalline arrays, sectioned particles show that they are either packaged completely (electron dense), or incompletely (ring-shaped). Once infected, the host cells exhibit symptoms such as swelling of the endoplasmic reticulum, disintegration and vacuolation of the cytoplasm, as well as fibrous material appearing in those vacuolated areas.
823:
55:
913:) have been determined. In both species, VP1 surrounds the five-fold axes while VP2 and VP3 form the two- and three-fold axes. The jelly roll folds on the MCPs are non-canonical with VP1 having nine β-strands and VP2 and VP3 having seven. VP1 also contains a unique EF-loop connected to the extra β-strand in the jelly roll fold which protrudes from the surface of the virus particle. A CD-loop is also present on VP1 which obscures the canyon structure that binds host receptors in the related
835:
851:
1263:(HaNIV), it is demonstrated that although both viruses infect host strains from the Northeast Pacific (with HaRNAV also able to infect host strains from Japan), they each targeted different subsets of host strains. The difference in host range between the two viruses suggests that they likely coexisted within the local environment, each infecting their specific hosts, therefore it is possible to infer the strain composition of
863:
1129:, the transcriptomes and viral metagenomes of 38 echinoderm species, with representation of species from all five classes of echinoderms, were analyzed and categorized into their respective families and genera. From this analysis, it was found that of the viral contigs recovered, more than half were classified within the viral order
1051:
temperatures above 20 °C. Strain 2–10, however, is more susceptible below 20 °C which is not optimal for host growth. Iron-limitation has also been shown to reduce viral infection rates. Stationary phase infection and virus-induced lysis of host cells is also impaired by bacteria of the genera
1074:
is the natural host of CsfrRNAV01. Infection with the virus induces spore formation in host cells, and the spores formed take longer to germinate than spores induced by host nutrient limitation. Although viral RNA is present within these spores, those that germinate are not able to produce infectious
1184:
In the structural analysis of the capsid's cryo-EM maps and atomic models, PyMOL 1.4 was used to obtain a Root-Mean-Square-Deviated (RMSD) per residue, which provided a value for structural diversity. Analysis for the structural proteins VP1, VP2, and VP3, suggested that VP1 was responsible for the
1050:
Meunier. It was first isolated from strain 2-10 but is able to infect other strains of the same species. Host cells in stationary phase are more susceptible to the virus than logarithmic phase cells. Host susceptibility is also affected by temperature, with various strains being more susceptible at
930:
genomes consist of linear, non-segmented, positive-sense single-stranded RNA 8.6-9.6 kb in length. The genomic RNA generally contains either one or two open-reading frames (ORFs) that may overlap. For species whose genomes have two ORFs, ORF1 encodes proteins involved in replication (RNA helicase,
975:
Replication occurs in the cytoplasm and relies on the viral RdRp. The positive-sense RNA genome is directly translated to produce viral proteins. After a latent period between 8–48 hours, host cells are observed to experience cytopathic symptoms such as swelling of the endoplasmic reticulum,
1271:'s susceptibility to viruses tends to shift over the course of the bloom, with the cells being much more resistant to viral infection after the bloom has collapsed. This suggests that viruses may have played an important role in regulating the
287:
has been suggested to be the possible pathogen of glass post-larvae disease, which is prevalent in shrimp aquaculture. Viruses detected in a cultured prawn species that had been affected by growth retardation disease have also been placed in
249:
that infect various photosynthetic marine protists. Members of the family have non-enveloped, icosahedral capsids. Replication occurs in the cytoplasm and causes lysis of the host cell. The first species of this family that was isolated is
1275:
bloom. Similarly, a study done on the viral assemblage composition of HaRNAV has shown that the diversity of HaRNAV is correlated with the composition of its host, therefore also suggesting that HaRNAV influences the composition of
1158:
socialis forma radians RNA virus (CsfrRNAV), full and empty capsids were analyzed by their atomic structure to which identified common and diverse structural features of the VP1 protein surface between different virus species under
1372:
bloom), while SmDNAV did not demonstrate this pattern. This suggests that AuRNAV likely targets hosts that can effectively utilize dead phytoplankton cells, therefore the virus is also a vital part of the bloom dynamic.
1433:
viruses (MRV) MRV3, MRV4, MRV5, and MRV6, have sequences which are 9242, 8887, 8548, and 9153 base pairs in length respectively. MRV3, MRV5, and MRV6 were found to be novel viruses, with MRV3 being placed in the genus
530:
527:
523:
946:
single-strand RNA virus 01 (AuRNAV01) which has a third ORF that encodes a protein with unknown function. In AuRNAV01, ORFs 2 and 3 are thought to be translated from subgenomic RNA rather than using an IRES.
1260:
1256:
885:
are non-enveloped, with icosahedral geometries, and T=pseudo3 symmetry. Virus particles are 22-35 nm in diameter. The capsid consists of three major capsid proteins (MCPs: VP1, VP2, VP3), each having a
359:
355:
604:
600:
1011:
in the
Pacific Ocean, namely 3 strains (NEPCC 102, NEPCC 522, NEPCC 764) from the Northeast Pacific region, and 2 strains (H93616, H94608) from Japan. It is noted that HaRNAV infect different strains of
1303:
in natural environments, with different species targeting specific strains of the diatom. Studies have suggested that these viruses play important roles in controlling and regulating the population of
270:. As of 2021, there are twenty species across seven genera in this family, as well as many other related virus sequences discovered through metagenomic sequencing that are currently unclassified.
1364:(SmDNAV). Studies have revealed that these two viruses maintain different fluctuation patterns from each other, with AuRNAV showing a significant increase in abundance following an increase of
543:
539:
1358:
553:
549:
3070:
Ying N, Wang Y, Song X, Qin B, Wu Y, Yang L, Fang W (May 2022). "Transcriptome analysis of
Macrobrachium rosenbergii: Identification of precocious puberty and slow-growing information".
890:. Most species also encode a minor capsid protein (mCP: VP4). All VPs are composed of β-sheets and α-helices. that is located around the five-fold axes on the inside of the capsid.
1152:
use a narrow range of hosts, as they lyse their host species, which is thought to connect to a receptor binding mechanism unique to the family. In tests involving the species
2919:"Isolation and characterization of a single-stranded RNA virus that infects the marine planktonic diatom Chaetoceros sp. (SS08-C03): New ssRNA virus infecting Chaetoceros"
1117:, as well as by comparing genetic codes in other virus groups in the Yangshan assemblage, it has been inferenced that these viruses likely infect unicellular eukaryotes.
1392:
3253:
780:
1055:
that coexist with the host diatom in the marine environment. This is thought to be a mechanism allowing for susceptible diatom species to survive viral infections.
672:
shares 22.4-30.3% of amino acid sequence identity of RdRp and 18.5-26.4% of capsid sequences. It is the most divergent and deeply branched taxon within the family.
1540:"Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo"
1488:
1141:
or that members of this viral family are capable of infecting a wider range of hosts than the single-celled eukaryotes they were initially known to infect.
1411:
is theorized to be the primary pathogen of the disease, providing a possible entryway into finding preventive measures against GPD in the shrimp industry.
5550:
794:
730:
723:
6024:
787:
773:
632:
6601:
6123:
5318:
3597:
766:
759:
716:
709:
696:
653:
646:
639:
459:
191:
2613:"Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom"
1113:
Given the nature of metaviromic analysis, it is difficult to determine the exact host range of these viruses. However, since the species fall under
6420:
6088:
6031:
6003:
5975:
5645:
5329:
5260:
4520:
4513:
3604:
3562:
3555:
277:
family and their hosts have notable significance in marine ecology, and are also relevant within the aquaculture industry. HaRNAV and viruses from
7135:
6615:
6469:
5996:
5989:
4809:
3625:
3611:
3569:
1877:"Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order
6707:
6658:
6608:
6587:
6038:
3548:
3246:
746:
619:
576:
567:
466:
445:
431:
198:
177:
163:
1173:
have an extra EF-Loop, implying the usage of a unique receptor-binding mechanism. A possible binding site for algal hosts were also found in E
7148:
6427:
5500:
5493:
1650:
Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, Suttle CA (2003). "Characterization of HaRNAV, a single-stranded RNA virus causing lysis of
1353:
2329:"Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp"
3013:"Ecological Dynamics of Two Distinct Viruses Infecting Marine Eukaryotic Decomposer Thraustochytrids (Labyrinthulomycetes, Stramenopiles)"
1333:
This suggests that the virus may play a role in decreasing host populations at the end of the summer bloom, although other parasites of
1489:"Marnaviridae - Positive Sense RNA Viruses - Positive Sense RNA Viruses (2011) - International Committee on Taxonomy of Viruses (ICTV)"
2272:"Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier"
3239:
7122:
1419:
Prevalence of growth retardation disease (GRD), which causes decreased growth rates and precocious puberty in the freshwater prawn
1329:
present in temperate coastal waters. Strains of this virus have only been successfully isolated shortly after the summer bloom of
1233:
family, thereby further expanding the family's potential distribution southwards to a cold, freshwater environment in
Antarctica.
1295:
is considered to be a major taxonomic group, playing an essential role as primary producers during bloom periods. Viruses under
1192:
based on VP1s, local structural diversity that is reflected can be used to better predict the targeted algal hosts of different
3216:
1225:
being widespread across the
Pacific Ocean from North America to Asia. Furthermore, sequencing of RNA viromes originating from
1209:
While HaRNAV was initially isolated from
British Columbian waters, it has been found that HaRNAV is also capable of infecting
411:-like sequences, prompting the proposal for a taxonomic reorganization to incorporate the new data into existing frameworks.
1229:
in the
Antarctic revealed four Antarctia picorna-like viruses (APLV), one of which, APLV-3, was found to cluster with the
931:
RdRp) and a cysteine protease which cleaves the polyprotein, while ORF2 encodes structural proteins. Genomes also contain
2805:"Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo"
919:
family. Unlike other species, VP4 is not present in the CsfrRNAV01 capsid, although the corresponding gene still exists.
1391:
is an important shrimp aquaculture species that is cultured globally, and most notably in China. However, due to the
623:
species share 52.4-59.1% identities of RdRp and 32.2-38.3% of capsid sequences, when compared with each other within
514:
species share 57.8-64.4% of RdRp and 29.1-33.4% of capsid amino acid sequences, when compared with each other within
1137:
family. This finding suggests the protists, such as symbionts, that are affiliated with echinoderms are infected by
754:. When compared with other genera of the family, they share 24.3-47.2% of RdRp and 17.1-47.9% for capsid sequences.
7161:
7153:
704:. When compared with other genera of the family, they share 24.3-47.2% of RdRp and 18.2-31.5% of capsid sequences.
627:. When compared with other genera of the family, they share 24.0-48.2% of RdRp and 17.2-27.4% of capsid sequences.
518:
When compared with other genera of the family, they share 23.8-47.8% of RdRp and 18.3-29.4% of capsid sequences.
283:
are known to have roles in regulating dynamics and composition of their hosts’ blooms. An unclassified sequence,
1751:"A novel virus in the family Marnaviridae as a potential pathogen of Penaeus vannamei glass post-larvae disease"
1221:
family have been identified from shrimp and prawn aquaculture species in
Chinese waters, this further supports
681:
677:
2216:"Structural Insights into Common and Host-Specific Receptor-Binding Mechanisms in Algal Picorna-like Viruses"
1928:
Lawrence JE, Chan AM, Suttle CA (2001). "A novel virus (HaNIV) causes lysis of the toxic bloom-forming alga
1442:
In contrast, MRV4 was characterized as being a previously identified Beihai picorna-like virus in the genus
1125:
Through a study aiming to investigate the diversity of the RNA viruses infecting species within the phylum
932:
495:
381:
750:
species share 48.8-77.0% of RdRp and 27.6-59.0% of capsid sequences, when compared with each other within
700:
species share 35.6-63.6% of RdRp and 24.9-34.4% of capsid sequences, when compared with each other within
5039:
3113:
An Z, Sun L, Chen J. "Investigation of the causes of the "iron shell" phenomenon of Roche marsh shrimp".
54:
6783:
6288:
5362:
2918:
595:
shares 22.4-33.6% identities of RdRp and 17.1-22.5% of capsid sequences. It is a deep-branching taxon.
7166:
1421:
1307:, thus influencing the spring bloom and subsequently affecting the local marine ecosystems as well.
3262:
935:(IRES) in intergenic regions and on the 5' end. The 3' end of the genome contains a poly(A) tail.
7194:
7189:
3223:
2427:"Temperature alters algicidal activity of DNA and RNA viruses infecting Chaetoceros tenuissimus"
1024:
cells also demonstrate more viral resistance after its bloom has collapsed, suggesting that the
7100:
7062:
2089:
571:
shares 25.4-48.2% of RdRp and 21.7-47.9% of capsid sequences, with other genera in the family.
3126:
2523:"Coculture with marine bacteria confers resistance to complete viral lysis of diatom cultures"
392:
was discovered to have a larger variety of viruses classified under it. Previously unassigned
7140:
2956:
Arsenieff L, Simon N, Rigaut-Jalabert F, Le Gall F, Chaffron S, Corre E, et al. (2019).
1817:"Virome Analysis of Normal and Growth Retardation Disease-Affected Macrobrachium rosenbergii"
1689:
Arsenieff L, Simon N, Rigaut-Jalabert F, Le Gall F, Chaffron S, Corre E, et al. (2019).
343:(NEPCC 522). It must not be confused with two other unrelated viruses that infect this host,
1181:
and/or CD loops, which could play a critical role in its unique receptor-binding mechanism.
3266:
3024:
2873:
2816:
2668:
Jackson EW, Wilhelm RC, Buckley DH, Hewson I (June 2022). "The RNA virome of echinoderms".
2624:
2562:
Pelusi A, De Luca P, Manfellotto F, Thamatrakoln K, Bidle KD, Montresor M (February 2021).
2479:
2468:"Impaired viral infection and reduced mortality of diatoms in iron-limited oceanic regions"
2466:
Kranzler CF, Brzezinski MA, Cohen NR, Lampe RH, Maniscalco M, Till CP, et al. (2021).
2381:
2283:
1941:
1246:
1000:
316:
266:
49:
2917:
Tomaru Y, Toyoda K, Kimura K, Takao Y, Sakurada K, Nakayama N, Nagasaki K (January 2013).
1357:
RNA virus (AuRNAV) has been shown to infect thraustochytrids together with another virus,
1028:
strains may change over the course of the algal bloom, which is regulated by the viruses.
1020:
viruses, suggesting co-existence, with each virus having a slightly different host range.
8:
6248:
4007:
3482:
1953:
221:
216:
210:
3028:
2877:
2820:
2628:
2483:
2426:
2385:
2287:
1945:
1777:
1750:
4566:
4201:
4187:
3779:
3174:
3141:
3095:
3047:
3012:
2988:
2957:
2938:
2894:
2861:
2780:
2753:
2734:
2645:
2612:
2588:
2563:
2503:
2402:
2369:
2304:
2271:
2242:
2215:
2178:
2147:
2057:
2030:
2006:
1982:"Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome"
1981:
1957:
1905:
1876:
1841:
1816:
1790:
1721:
1690:
1671:
1616:
1587:
498:(RdRp) sequences and capsid amino acid sequences, species are listed under each genus.
3200:
2121:
482:
has several characteristics that are similar to those of other viruses in the broader
314:
The family was proposed following the discovery of an RNA virus (HaRNAV) that infects
7109:
6723:
5682:
5557:
5536:
5429:
5085:
4743:
4385:
3948:
3541:
3468:
3179:
3099:
3087:
3052:
2993:
2934:
2899:
2842:
2837:
2828:
2804:
2785:
2738:
2726:
2685:
2650:
2593:
2544:
2507:
2495:
2448:
2407:
2350:
2309:
2247:
2183:
2062:
2011:
1910:
1846:
1815:
Zhou D, Liu S, Guo G, He X, Xing C, Miao Q, et al. (December 2022). He B (ed.).
1794:
1782:
1726:
1667:
1621:
1561:
490:
are placed through analysis of the RdRp sequence of amino acids. The seven genera of
332:
237:
2942:
1961:
1675:
822:
7114:
6513:
6441:
6334:
5909:
5851:
5814:
5543:
5274:
5232:
5180:
5143:
5113:
5074:
4928:
4854:
4711:
4033:
3996:
3970:
3928:
3729:
3719:
3534:
3386:
3357:
3231:
3169:
3161:
3122:
3079:
3042:
3032:
2983:
2973:
2930:
2889:
2881:
2832:
2824:
2775:
2765:
2716:
2677:
2640:
2632:
2583:
2575:
2534:
2487:
2438:
2397:
2389:
2340:
2299:
2291:
2237:
2227:
2173:
2163:
2052:
2042:
2001:
1993:
1980:
Wolf YI, Silas S, Wang Y, Wu S, Bocek M, Kazlauskas D, et al. (October 2020).
1949:
1900:
1892:
1836:
1828:
1772:
1767:
1762:
1716:
1706:
1663:
1611:
1603:
1551:
1387:
1348:
336:
321:
1185:
host-specificity of the virus, and was based on the diversity in the VP1 protein.
205:
7085:
6973:
6844:
6679:
6627:
6540:
6496:
6485:
6323:
6175:
6116:
5940:
5661:
5591:
5479:
5462:
5397:
4994:
4949:
4670:
4177:
3959:
3870:
3790:
3686:
3037:
1226:
1217:
may have a wide distribution throughout the
Pacific Ocean. As viruses within the
964:
Entry of the virus is achieved by penetration into the host cell. EF-, CD-, and E
887:
510:
424:
377:
279:
156:
115:
3140:
Dong X, Wang G, Hu T, Li J, Li C, Cao Z, et al. (June 2021). Heck M (ed.).
1556:
1539:
972:-loops are hypothesized to interact with host receptors to trigger viral entry.
7016:
6994:
6962:
6927:
6833:
6798:
6734:
6577:
6566:
6529:
6371:
6255:
6145:
6074:
6064:
5825:
5617:
5580:
5486:
5281:
4865:
4838:
4485:
4474:
4378:
4343:
4336:
4322:
4315:
4264:
4236:
4166:
4155:
4120:
3904:
3707:
3576:
3513:
3447:
3425:
3347:
2491:
1244:
The complex relationship between HaRNAV and other viruses capable of infecting
915:
324:, which was the first report of a single-stranded RNA virus capable of causing
245:
127:
3165:
3083:
2467:
1997:
1832:
1105:
species identified thus far that has demonstrated this broad of a host range.
960:
Various marine phytoplankton species serve as hosts for members of the family
809:. However, these sequences are not assigned under any of the existing genera.
209:
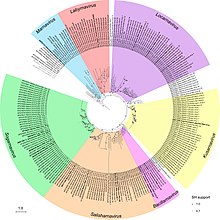
Maximum-likelihood phylogeny of the
Picornavirales RdRp amino acid sequences.
7183:
6984:
6879:
6865:
6693:
6686:
6665:
6506:
6455:
6448:
6312:
6208:
6186:
6159:
6135:
6095:
6052:
6017:
6010:
5982:
5968:
5961:
5862:
5798:
5769:
5748:
5727:
5716:
5696:
5628:
5450:
5419:
5408:
5386:
5253:
5120:
5106:
5096:
5063:
4987:
4939:
4882:
4875:
4722:
4622:
4496:
4410:
4392:
4371:
4357:
4257:
4194:
4144:
4066:
4044:
4017:
3980:
3882:
3849:
3697:
3675:
3475:
3458:
3436:
3414:
3336:
3325:
2978:
2862:"Role of Phylogenetic Structure in the Dynamics of Coastal Viral Assemblages"
2548:
2499:
2452:
1711:
1407:
family, and was found to contain three ORFs within its 9.895 kb long genome.
1126:
91:
1289:
In terms of ecological impact and species diversity in the ocean, the genus
7006:
6951:
6886:
6855:
6822:
6672:
6644:
6462:
6434:
6413:
6399:
6392:
6382:
6218:
6152:
5929:
5899:
5888:
5835:
5791:
5781:
5762:
5738:
5689:
5601:
5514:
5472:
5311:
5243:
5221:
5201:
5191:
5001:
4917:
4828:
4799:
4787:
4777:
4765:
4755:
4733:
4691:
4681:
4654:
4633:
4611:
4587:
4577:
4555:
4452:
4403:
4364:
4350:
4329:
4098:
4076:
4054:
3921:
3823:
3811:
3768:
3757:
3664:
3653:
3618:
3524:
3376:
3183:
3091:
3056:
2997:
2903:
2846:
2789:
2730:
2689:
2654:
2597:
2411:
2354:
2313:
2251:
2187:
2066:
2015:
1914:
1850:
1786:
1730:
1625:
1565:
1368:
sp. thraustochytrids (which decompose organic matter during the decline of
834:
801:
Additionally, there are 653 virus sequences that have been identified (via
591:
438:
349:
261:
170:
103:
32:
3208:
2721:
2704:
2345:
2328:
7094:
6872:
6651:
6637:
6550:
6406:
6196:
6109:
6102:
6045:
5951:
5872:
5755:
5672:
5638:
5564:
5526:
5507:
5443:
5436:
5267:
5127:
4816:
4644:
4544:
4439:
4289:
4271:
4247:
4110:
4088:
3914:
3892:
3860:
3801:
3590:
3502:
3364:
3314:
3290:
2885:
2681:
2636:
2393:
2295:
2270:
Shirai Y, Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K (July 2008).
2168:
1607:
1291:
1154:
877:: The MCPs are coloured in green, magenta and cyan and the mCP in yellow.
802:
373:
2564:"Virus-induced spore formation as a defense mechanism in marine diatoms"
1896:
1586:
Lang AS, Vlok M, Culley AI, Suttle CA, Takao Y, Tomaru Y (August 2021).
1097:
Meunier strain 2–10, however it is also able to infect at least 4 other
220:
Phylogeny of environmental sequences of genera within the
Marnaviridae.
6744:
6700:
6594:
6355:
5164:
5154:
4906:
4506:
3833:
3583:
668:
452:
325:
256:
184:
3011:
Takao Y, Tomaru Y, Nagasaki K, Honda D (23 July 2015). Anil AC (ed.).
2770:
2579:
2232:
2047:
850:
6940:
6811:
6345:
6301:
5375:
5052:
5024:
4463:
3303:
3275:
2539:
2522:
2443:
1324:
372:
as the only genus and HaRNAV as the only species. After the usage of
240:
79:
7127:
7056:
2955:
2561:
1688:
1133:, with the highest number of the picornaviruses being placed in the
7079:
1749:
Xu A, Xu S, Tu Q, Qiao H, Lin W, Li J, et al. (January 2023).
1516:. International Committee on Taxonomy of Viruses (ICTV). March 2021
307:), together with the prefix "ma" being derived from the Latin word
1250:
is important in terms of understanding how viruses play a role in
1058:
1213:
strains originating from Japan, thus suggesting that members of
2370:"Virus-specific responses of Heterosigma akashiwo to infection"
1513:
1462:
862:
339:, Canada, from a concentrated virus assemblage using the host
1267:
population over its blooming period. Studies have shown that
66:
2703:
Short SM, Staniewski MA, Chaban YV, Long AM, Wang D (2020).
2465:
2327:
Takao Y, Mise K, Nagasaki K, Okuno T, Honda D (March 2006).
1236:
1144:
1085:
2667:
2214:
Wang H, Munke A, Li S, Tomaru Y, Okamoto K (October 2022).
1351:
are considered to be important decomposers in ecosystems.
986:
2802:
2702:
1007:
HaRNAV is known to be able to infect multiple strains of
3010:
2145:
2916:
2367:
2269:
2148:"Capsid Structure of a Marine Algal Virus of the Order
1038:
2860:
Gustavsen JA, Suttle CA (May 2021). Johnson KN (ed.).
2611:
Kimura K, Tomaru Y (February 2015). Wommack KE (ed.).
2368:
Lawrence JE, Brussaard CP, Suttle CA (December 2006).
2326:
1340:
222:
https://academic.oup.com/ve/article/5/2/vez056/5692928
211:
https://academic.oup.com/ve/article/5/2/vez056/5692928
2803:
Tarutani K, Nagasaki K, Yamaguchi M (November 2000).
2424:
2146:
Munke A, Kimura K, Tomaru Y, Okamoto K (April 2020).
2094:
International Committee on Taxonomy of Viruses (ICTV)
1493:
International Committee on Taxonomy of Viruses (ICTV)
1467:
International Committee on Taxonomy of Viruses (ICTV)
3261:
1585:
1093:
Like CtenRNAV01, CtenRNAVII was first isolated from
938:
One exception to the typical genome organization of
1649:
2425:Tomaru Y, Kimura K, Yamaguchi H (2 October 2014).
2028:
1927:
4564:
2705:"Diversity of Viruses Infecting Eukaryotic Algae"
2213:
1310:
7181:
2751:
1537:
331:HaRNAV was isolated from water collected in the
5230:
4926:
3968:
3069:
2859:
2031:"RNA Viruses in Aquatic Unicellular Eukaryotes"
1979:
6971:
6842:
6538:
6494:
6321:
5589:
4175:
3684:
2265:
2263:
2261:
2029:Sadeghi M, Tomaru Y, Ahola T (February 2021).
1874:
1322:species that infects the bloom-forming diatom
805:analysis) as possibly belonging to the family
6960:
6831:
6732:
5823:
4863:
4483:
4472:
4164:
4153:
3511:
3445:
3423:
3247:
2141:
2139:
1921:
1414:
407:Further metagenomic studies have yielded 653
6310:
5860:
5626:
5384:
5094:
5061:
4937:
4494:
3673:
3456:
3334:
3323:
3139:
2610:
2520:
2209:
2207:
2205:
2203:
2201:
2199:
2197:
2116:
2114:
2112:
2110:
1975:
1973:
1971:
1870:
1868:
1866:
1864:
1862:
1860:
1814:
1538:Lang AS, Culley AI, Suttle CA (March 2004).
1403:has been suggested to be a member under the
1381:
1376:
1046:CtenRNAV01 infects the bloom-forming diatom
994:HaRNAV is the first identified virus in the
854:Schematic drawing of a virion of the family
376:analysis on the amino acid sequences of the
6949:
6853:
6820:
6380:
5897:
5241:
5189:
4915:
4679:
4575:
4553:
3662:
3522:
2958:"First Viruses Infecting the Marine Diatom
2258:
1810:
1808:
1806:
1804:
1691:"First Viruses Infecting the Marine Diatom
1581:
1579:
1577:
1575:
1533:
1531:
1438:, and MRV5 and MRV6 belonging to the genus
303:" is based on its genome type (RNA virus -
5670:
4642:
3312:
3254:
3240:
2136:
1748:
1645:
1643:
1641:
1639:
1637:
1635:
1188:When looking at a structural phylogeny of
31:
6925:
5152:
3173:
3127:10.14184/j.cnki.issn1004-843x.2014.01.034
3046:
3036:
2987:
2977:
2893:
2836:
2779:
2769:
2720:
2644:
2587:
2538:
2442:
2401:
2344:
2303:
2241:
2231:
2194:
2177:
2167:
2107:
2056:
2046:
2005:
1968:
1904:
1857:
1840:
1776:
1766:
1744:
1742:
1740:
1720:
1710:
1615:
1555:
1145:Host-Specific Receptor-Binding Mechanisms
907:Chaetoceros tenuissimus RNA virus type II
6938:
6809:
6796:
6343:
6299:
5373:
5360:
5050:
2752:Yau S, Seth-Pasricha M (February 2019).
1875:Vlok M, Lang AS, Suttle CA (July 2019).
1801:
1572:
1528:
1506:
1053:Nautella, Polaribacter, and Sulitobacter
861:
849:
833:
821:
260:, which infects the toxic bloom-forming
215:
204:
6286:
2754:"Viruses of Polar Aquatic Environments"
2084:
2082:
2080:
2078:
2076:
1632:
7182:
5037:
2866:Applied and Environmental Microbiology
2809:Applied and Environmental Microbiology
2617:Applied and Environmental Microbiology
2374:Applied and Environmental Microbiology
2276:Applied and Environmental Microbiology
1737:
1429:family. These viruses, referred to as
7061:
7060:
6924:
6795:
6285:
5359:
5036:
4437:
4436:
3287:
3235:
3112:
2521:Kimura K, Tomaru Y (29 August 2014).
1108:
830:(HaRNAV) particles. Scale 50 nm.
419:The following genera are recognised:
400:were also classified as genera under
347:(HaV-1, isolate: HaV53) in the genus
3288:
3146:Associated with Sexual Precocity in
2073:
1954:10.1046/j.1529-8817.2001.037002216.x
1282:
1031:
950:
494:are designated based on analysis of
273:Interactions between members of the
2709:Current Issues in Molecular Biology
384:domains on viruses under the order
13:
812:
687:
501:
14:
7206:
3194:
3072:Journal of Invertebrate Pathology
1254:bloom dynamics. When compared to
1078:
473:
2935:10.1111/j.1440-1835.2012.00670.x
2829:10.1128/AEM.66.11.4916-4920.2000
1668:10.1046/j.1529-8817.2003.01162.x
1120:
368:existed for multiple years with
53:
3133:
3106:
3063:
3004:
2949:
2910:
2853:
2796:
2745:
2696:
2670:The Journal of General Virology
2661:
2604:
2555:
2514:
2459:
2418:
2361:
2333:The Journal of General Virology
2320:
2022:
1596:The Journal of General Virology
1463:"Virus Taxonomy: 2018b Release"
1204:
998:family. When the virus infects
979:
737:
610:
583:
559:
1768:10.1016/j.virusres.2022.199026
1682:
1588:"ICTV Virus Taxonomy Profile:
1514:"Virus Taxonomy: 2020 Release"
1481:
1455:
988:Heterosigma akashiwo RNA virus
858:, cross section and side view.
828:Heterosigma akashiwo RNA virus
252:Heterosigma akashiwo RNA virus
1:
1448:
955:
933:internal ribosome entry sites
660:
3038:10.1371/journal.pone.0133395
817:
496:RNA-dependent RNA polymerase
7:
1557:10.1016/j.virol.2003.10.015
414:
10:
7211:
2492:10.1038/s41561-021-00711-6
1415:Growth Retardation Disease
1399:sample infected with GPD,
1337:are also likely involved.
1299:have been shown to target
1199:
605:single-strand RNA virus 01
294:
44:under electron microscope
40:Low-resolution image of a
7069:
7004:
6982:
6934:
6920:
6805:
6791:
6782:
6721:
6625:
6575:
6564:
6527:
6483:
6369:
6332:
6295:
6281:
6236:
6206:
6184:
6173:
6133:
6062:
5949:
5938:
5927:
5886:
5849:
5812:
5779:
5736:
5725:
5714:
5659:
5615:
5578:
5524:
5460:
5417:
5406:
5395:
5369:
5355:
5299:
5219:
5178:
5141:
5083:
5072:
5046:
5032:
5023:
4975:
4904:
4852:
4826:
4797:
4775:
4753:
4731:
4720:
4709:
4668:
4631:
4620:
4609:
4542:
4461:
4450:
4446:
4432:
4303:
4281:
4245:
4234:
4227:
4142:
4108:
4086:
4064:
4042:
4031:
4005:
3994:
3957:
3946:
3902:
3880:
3858:
3847:
3821:
3799:
3788:
3777:
3766:
3755:
3717:
3695:
3651:
3500:
3434:
3412:
3374:
3345:
3301:
3297:
3283:
3274:
3166:10.1128/mSystems.00003-21
3148:Macrobrachium rosenbergii
3084:10.1016/j.jip.2022.107752
2966:Frontiers in Microbiology
2527:Aquatic Microbial Ecology
2431:Aquatic Microbial Ecology
1998:10.1038/s41564-020-0755-4
1833:10.1128/spectrum.01462-22
1699:Frontiers in Microbiology
1431:Macrobrachium rosenbergii
1422:Macrobrachium rosenbergii
1393:glass post-larvae disease
1382:Glass post-larvae disease
1377:Prevalence in aquaculture
922:
901:RNA virus 1 (CsfrRNAV01,
845:Modified after ViralZone.
152:
147:
48:
39:
30:
23:
2979:10.3389/fmicb.2018.03235
1712:10.3389/fmicb.2018.03235
1360:Sicyoidochytrium minutum
1101:species. It is the only
3115:Scientific Fish Farming
1261:nuclear inclusion virus
1087:Chaetoceros tenuissimus
1040:Chaetoceros tenuissimus
867:Chaetoceros tenuissimus
826:Electron micrograph of
781:Chaetenuissarnavirus II
540:Chaetoceros tenuissimus
360:nuclear inclusion virus
2090:"Family: Marnaviridae"
1048:Chaetoceros tenussimus
878:
859:
847:
831:
254:(HaRNAV) in the genus
224:
213:
2923:Phycological Research
2722:10.21775/cimb.039.029
2346:10.1099/vir.0.81204-0
1821:Microbiology Spectrum
1165:Unlike viruses under
865:
853:
837:
825:
550:Rhizosolenia setigera
219:
208:
3267:virus classification
2960:Guinardia delicatula
2886:10.1128/AEM.02704-20
2682:10.1099/jgv.0.001772
2637:10.1128/AEM.02380-14
2394:10.1128/AEM.01207-06
2296:10.1128/AEM.00509-08
2169:10.1128/JVI.01855-19
1934:Journal of Phycology
1930:Heterosigma akashiwo
1693:Guinardia delicatula
1656:Journal of Phycology
1652:Heterosigma akashiwo
1608:10.1099/jgv.0.001633
1312:Guinardia delicatula
1257:Heterosigma akashiwo
1238:Heterosigma akashiwo
1060:Chaetoceros socialis
895:Chaetoceros socialis
678:Heterosigma akashiwo
524:Chaetoceros socialis
356:Heterosigma akashiwo
345:H. akashiwo virus 01
320:off of the coast of
267:Heterosigma akashiwo
50:Virus classification
6249:Permutotetraviridae
4008:Orthopolintovirales
3483:Malacoherpesviridae
3224:Taxonomy History –
3029:2015PLoSO..1033395T
2878:2021ApEnM..87E2704G
2821:2000ApEnM..66.4916T
2629:2015ApEnM..81.1120K
2568:The New Phytologist
2484:2021NatGe..14..231K
2386:2006ApEnM..72.7829L
2288:2008ApEnM..74.4022S
2156:Journal of Virology
1986:Nature Microbiology
1946:2001JPcgy..37..216L
1932:(Raphidophyceae)".
1654:(Raphidophyceae)".
4567:Malgrandaviricetes
4202:Sphaerolipoviridae
4188:Matshushitaviridae
3780:Nucleocytoviricota
3142:"A Novel Virus of
1344:RNA virus (AuRNAV)
1240:RNA virus (HaRNAV)
1109:Unassigned viruses
893:The structures of
879:
860:
848:
832:
486:order, members of
328:in phytoplankton.
225:
214:
7177:
7176:
7063:Taxon identifiers
7054:
7053:
7050:
7049:
7046:
7045:
7042:
7041:
7038:
7037:
7034:
7033:
7030:
7029:
7026:
7025:
6916:
6915:
6912:
6911:
6908:
6907:
6904:
6903:
6900:
6899:
6896:
6895:
6778:
6777:
6774:
6773:
6770:
6769:
6766:
6765:
6762:
6761:
6758:
6757:
6754:
6753:
6724:Yunchangviricetes
6717:
6716:
6560:
6559:
6523:
6522:
6479:
6478:
6365:
6364:
6277:
6276:
6273:
6272:
6269:
6268:
6265:
6264:
6232:
6231:
6228:
6227:
6169:
6168:
5923:
5922:
5919:
5918:
5882:
5881:
5845:
5844:
5808:
5807:
5710:
5709:
5706:
5705:
5683:Carmotetraviridae
5655:
5654:
5611:
5610:
5574:
5573:
5558:Gammaflexiviridae
5551:Deltaflexiviridae
5537:Alphaflexiviridae
5430:Alphatetraviridae
5351:
5350:
5347:
5346:
5343:
5342:
5339:
5338:
5295:
5294:
5291:
5290:
5215:
5214:
5211:
5210:
5174:
5173:
5137:
5136:
5086:Chrymotiviricetes
5019:
5018:
5015:
5014:
5011:
5010:
4971:
4970:
4967:
4966:
4963:
4962:
4959:
4958:
4900:
4899:
4896:
4895:
4892:
4891:
4848:
4847:
4744:Bacilladnaviridae
4705:
4704:
4701:
4700:
4664:
4663:
4605:
4604:
4601:
4600:
4597:
4596:
4538:
4537:
4534:
4533:
4530:
4529:
4428:
4427:
4424:
4423:
4420:
4419:
4386:Portogloboviridae
4299:
4298:
4223:
4222:
4219:
4218:
4215:
4214:
4211:
4210:
4138:
4137:
4134:
4133:
4130:
4129:
4027:
4026:
3990:
3989:
3949:Preplasmiviricota
3942:
3941:
3938:
3937:
3843:
3842:
3751:
3750:
3747:
3746:
3743:
3742:
3739:
3738:
3647:
3646:
3643:
3642:
3639:
3638:
3635:
3634:
3542:Autographiviridae
3496:
3495:
3492:
3491:
3469:Alloherpesviridae
3408:
3407:
3404:
3403:
3400:
3399:
3396:
3395:
2872:(11): e02704–20.
2815:(11): 4916–4920.
2771:10.3390/v11020189
2580:10.1111/nph.16951
2472:Nature Geoscience
2380:(12): 7829–7834.
2339:(Pt 3): 723–733.
2282:(13): 4022–4027.
2233:10.3390/v14112369
2048:10.3390/v13030362
1992:(10): 1262–1270.
1897:10.1093/ve/vez056
1180:
1176:
1089:RNA virus type II
1075:viral particles.
951:Host interactions
846:
795:Palmarnavirus 156
731:Palmarnavirus 473
724:Palmarnavirus 128
333:Strait of Georgia
238:positive-stranded
229:
228:
16:Family of viruses
7202:
7170:
7169:
7157:
7156:
7144:
7143:
7131:
7130:
7118:
7117:
7105:
7104:
7103:
7090:
7089:
7088:
7058:
7057:
6980:
6979:
6969:
6968:
6958:
6957:
6947:
6946:
6936:
6935:
6928:dsDNA-RT viruses
6922:
6921:
6851:
6850:
6840:
6839:
6829:
6828:
6818:
6817:
6807:
6806:
6799:ssRNA-RT viruses
6793:
6792:
6789:
6788:
6730:
6729:
6573:
6572:
6541:Serpentovirales
6536:
6535:
6514:Orthomyxoviridae
6492:
6491:
6442:Peribunyaviridae
6378:
6377:
6341:
6340:
6335:Chunqiuviricetes
6330:
6329:
6319:
6318:
6308:
6307:
6297:
6296:
6289:(–)ssRNA viruses
6283:
6282:
6182:
6181:
6025:Nanghoshaviridae
5947:
5946:
5936:
5935:
5910:Botourmiaviridae
5895:
5894:
5858:
5857:
5852:Howeltoviricetes
5821:
5820:
5815:Amabiliviricetes
5734:
5733:
5723:
5722:
5668:
5667:
5624:
5623:
5587:
5586:
5544:Betaflexiviridae
5415:
5414:
5404:
5403:
5393:
5392:
5382:
5381:
5371:
5370:
5363:(+)ssRNA viruses
5357:
5356:
5275:Picobirnaviridae
5239:
5238:
5233:Duplopiviricetes
5228:
5227:
5187:
5186:
5181:Vidaverviricetes
5150:
5149:
5144:Resentoviricetes
5114:Megabirnaviridae
5092:
5091:
5081:
5080:
5075:Duplornaviricota
5070:
5069:
5059:
5058:
5048:
5047:
5034:
5033:
5030:
5029:
4935:
4934:
4929:Huolimaviricetes
4924:
4923:
4913:
4912:
4861:
4860:
4855:Repensiviricetes
4729:
4728:
4718:
4717:
4712:Cressdnaviricota
4677:
4676:
4640:
4639:
4629:
4628:
4618:
4617:
4573:
4572:
4562:
4561:
4551:
4550:
4492:
4491:
4481:
4480:
4470:
4469:
4459:
4458:
4448:
4447:
4434:
4433:
4243:
4242:
4232:
4231:
4173:
4172:
4162:
4161:
4151:
4150:
4040:
4039:
4034:Tectiliviricetes
4003:
4002:
3997:Polintoviricetes
3971:Priklausovirales
3966:
3965:
3955:
3954:
3929:Marseilleviridae
3856:
3855:
3797:
3796:
3786:
3785:
3775:
3774:
3764:
3763:
3730:Papillomaviridae
3720:Zurhausenvirales
3693:
3692:
3682:
3681:
3671:
3670:
3660:
3659:
3535:Ackermannviridae
3520:
3519:
3509:
3508:
3454:
3453:
3443:
3442:
3432:
3431:
3421:
3420:
3387:Tristromaviridae
3358:Lipothrixviridae
3343:
3342:
3332:
3331:
3321:
3320:
3310:
3309:
3299:
3298:
3285:
3284:
3281:
3280:
3256:
3249:
3242:
3233:
3232:
3188:
3187:
3177:
3137:
3131:
3130:
3110:
3104:
3103:
3067:
3061:
3060:
3050:
3040:
3008:
3002:
3001:
2991:
2981:
2953:
2947:
2946:
2914:
2908:
2907:
2897:
2857:
2851:
2850:
2840:
2800:
2794:
2793:
2783:
2773:
2749:
2743:
2742:
2724:
2700:
2694:
2693:
2665:
2659:
2658:
2648:
2623:(3): 1120–1131.
2608:
2602:
2601:
2591:
2574:(4): 2251–2259.
2559:
2553:
2552:
2542:
2540:10.3354/ame01705
2518:
2512:
2511:
2463:
2457:
2456:
2446:
2444:10.3354/ame01713
2422:
2416:
2415:
2405:
2365:
2359:
2358:
2348:
2324:
2318:
2317:
2307:
2267:
2256:
2255:
2245:
2235:
2211:
2192:
2191:
2181:
2171:
2143:
2134:
2133:
2131:
2129:
2118:
2105:
2104:
2102:
2100:
2086:
2071:
2070:
2060:
2050:
2026:
2020:
2019:
2009:
1977:
1966:
1965:
1925:
1919:
1918:
1908:
1872:
1855:
1854:
1844:
1812:
1799:
1798:
1780:
1770:
1746:
1735:
1734:
1724:
1714:
1686:
1680:
1679:
1647:
1630:
1629:
1619:
1583:
1570:
1569:
1559:
1535:
1526:
1525:
1523:
1521:
1510:
1504:
1503:
1501:
1499:
1485:
1479:
1478:
1476:
1474:
1459:
1388:Penaeus vannamei
1366:Aurantiochytrium
1354:Aurantiochytrium
1349:Thraustochytrids
1342:Aurantiochytrium
1178:
1174:
944:Aurantiochytrium
844:
788:Jericarnavirus A
774:Chaetarnavirus 2
633:Jericarnavirus B
601:Aurantiochytrium
516:Bacillarnavirus.
337:British Columbia
322:British Columbia
58:
57:
35:
21:
20:
7210:
7209:
7205:
7204:
7203:
7201:
7200:
7199:
7180:
7179:
7178:
7173:
7165:
7160:
7152:
7147:
7139:
7134:
7126:
7121:
7113:
7108:
7099:
7098:
7093:
7084:
7083:
7078:
7065:
7055:
7022:
7000:
6974:Revtraviricetes
6930:
6892:
6845:Revtraviricetes
6801:
6750:
6713:
6680:Paramyxoviridae
6628:Mononegavirales
6621:
6602:Crepuscuviridae
6556:
6519:
6497:Articulavirales
6486:Insthoviricetes
6475:
6361:
6324:Negarnaviricota
6291:
6261:
6224:
6202:
6176:Stelpaviricetes
6165:
6129:
6124:Polycipiviridae
6117:Dicistroviridae
6058:
5941:Pisoniviricetes
5915:
5878:
5841:
5804:
5775:
5702:
5662:Tolucaviricetes
5651:
5607:
5592:Amarillovirales
5570:
5520:
5480:Closteroviridae
5463:Martellivirales
5456:
5398:Kitrinoviricota
5365:
5335:
5319:Polymycoviridae
5287:
5207:
5170:
5133:
5042:
5007:
4995:Finnlakeviridae
4955:
4950:Pleolipoviridae
4888:
4844:
4822:
4793:
4771:
4749:
4697:
4671:Quintoviricetes
4660:
4593:
4526:
4442:
4416:
4295:
4277:
4207:
4178:Halopanivirales
4126:
4104:
4082:
4060:
4023:
3986:
3960:Maveriviricetes
3934:
3898:
3876:
3871:Phycodnaviridae
3839:
3817:
3791:Pokkesviricetes
3735:
3713:
3687:Papovaviricetes
3631:
3598:Rountreeviridae
3488:
3392:
3370:
3293:
3270:
3260:
3197:
3192:
3191:
3160:(3): e0000321.
3138:
3134:
3111:
3107:
3068:
3064:
3023:(7): e0133395.
3009:
3005:
2954:
2950:
2915:
2911:
2858:
2854:
2801:
2797:
2750:
2746:
2701:
2697:
2666:
2662:
2609:
2605:
2560:
2556:
2519:
2515:
2464:
2460:
2423:
2419:
2366:
2362:
2325:
2321:
2268:
2259:
2212:
2195:
2144:
2137:
2127:
2125:
2120:
2119:
2108:
2098:
2096:
2088:
2087:
2074:
2027:
2023:
1978:
1969:
1926:
1922:
1885:Virus Evolution
1873:
1858:
1827:(6): e0146222.
1813:
1802:
1747:
1738:
1687:
1683:
1648:
1633:
1584:
1573:
1536:
1529:
1519:
1517:
1512:
1511:
1507:
1497:
1495:
1487:
1486:
1482:
1472:
1470:
1461:
1460:
1456:
1451:
1417:
1384:
1379:
1346:
1320:Bacillarnavirus
1316:
1297:Bacillarnavirus
1287:
1284:Bacillarnavirus
1242:
1227:Limnopolar Lake
1207:
1202:
1147:
1123:
1111:
1091:
1083:
1068:
1044:
1036:
1033:Bacillarnavirus
992:
984:
971:
967:
958:
953:
925:
903:Bacillarnavirus
888:Jelly roll fold
875:Bacillarnavirus
843:
820:
815:
813:Characteristics
767:Britarnavirus 3
760:Britarnavirus 2
742:
717:Britarnavirus 4
710:Britarnavirus 1
702:Salisharnavirus
697:Salisharnavirus
692:
689:Salisharnavirus
665:
654:Sanfarnavirus 3
647:Sanfarnavirus 2
640:Sanfarnavirus 1
615:
588:
564:
511:Bacillarnavirus
506:
503:Bacillarnavirus
476:
460:Salisharnavirus
425:Bacillarnavirus
417:
398:Bacillarnavirus
378:capsid proteins
297:
280:Bacillarnavirus
236:is a family of
192:Salisharnavirus
157:Bacillarnavirus
117:Pisoniviricetes
52:
17:
12:
11:
5:
7208:
7198:
7197:
7195:Virus families
7192:
7190:Picornavirales
7175:
7174:
7172:
7171:
7158:
7145:
7132:
7119:
7106:
7091:
7075:
7073:
7067:
7066:
7052:
7051:
7048:
7047:
7044:
7043:
7040:
7039:
7036:
7035:
7032:
7031:
7028:
7027:
7024:
7023:
7021:
7020:
7017:Caulimoviridae
7012:
7010:
7002:
7001:
6999:
6998:
6995:Hepadnaviridae
6990:
6988:
6977:
6966:
6963:Artverviricota
6955:
6944:
6932:
6931:
6918:
6917:
6914:
6913:
6910:
6909:
6906:
6905:
6902:
6901:
6898:
6897:
6894:
6893:
6891:
6890:
6883:
6876:
6869:
6861:
6859:
6848:
6837:
6834:Artverviricota
6826:
6815:
6803:
6802:
6786:
6780:
6779:
6776:
6775:
6772:
6771:
6768:
6767:
6764:
6763:
6760:
6759:
6756:
6755:
6752:
6751:
6749:
6748:
6740:
6738:
6735:Goujianvirales
6727:
6719:
6718:
6715:
6714:
6712:
6711:
6704:
6697:
6690:
6683:
6676:
6669:
6662:
6655:
6648:
6641:
6633:
6631:
6623:
6622:
6620:
6619:
6612:
6605:
6598:
6591:
6583:
6581:
6578:Jingchuvirales
6570:
6567:Monjiviricetes
6562:
6561:
6558:
6557:
6555:
6554:
6546:
6544:
6533:
6530:Milneviricetes
6525:
6524:
6521:
6520:
6518:
6517:
6510:
6502:
6500:
6489:
6481:
6480:
6477:
6476:
6474:
6473:
6466:
6459:
6452:
6445:
6438:
6431:
6424:
6421:Leishbuviridae
6417:
6410:
6403:
6396:
6388:
6386:
6375:
6372:Ellioviricetes
6367:
6366:
6363:
6362:
6360:
6359:
6351:
6349:
6338:
6327:
6316:
6305:
6293:
6292:
6279:
6278:
6275:
6274:
6271:
6270:
6267:
6266:
6263:
6262:
6260:
6259:
6256:Sarthroviridae
6252:
6240:
6238:
6234:
6233:
6230:
6229:
6226:
6225:
6223:
6222:
6214:
6212:
6204:
6203:
6201:
6200:
6192:
6190:
6179:
6171:
6170:
6167:
6166:
6164:
6163:
6156:
6149:
6146:Alvernaviridae
6141:
6139:
6131:
6130:
6128:
6127:
6120:
6113:
6106:
6099:
6092:
6089:Solinviviridae
6085:
6078:
6075:Picornaviridae
6070:
6068:
6065:Picornavirales
6060:
6059:
6057:
6056:
6049:
6042:
6035:
6032:Nanhypoviridae
6028:
6021:
6014:
6007:
6004:Medioniviridae
6000:
5993:
5986:
5979:
5976:Cremegaviridae
5972:
5965:
5957:
5955:
5944:
5933:
5925:
5924:
5921:
5920:
5917:
5916:
5914:
5913:
5905:
5903:
5892:
5884:
5883:
5880:
5879:
5877:
5876:
5868:
5866:
5855:
5847:
5846:
5843:
5842:
5840:
5839:
5831:
5829:
5826:Wolframvirales
5818:
5810:
5809:
5806:
5805:
5803:
5802:
5795:
5787:
5785:
5777:
5776:
5774:
5773:
5766:
5759:
5752:
5744:
5742:
5731:
5720:
5712:
5711:
5708:
5707:
5704:
5703:
5701:
5700:
5693:
5686:
5678:
5676:
5665:
5657:
5656:
5653:
5652:
5650:
5649:
5646:Sinhaliviridae
5642:
5634:
5632:
5621:
5618:Magsaviricetes
5613:
5612:
5609:
5608:
5606:
5605:
5597:
5595:
5584:
5581:Flasuviricetes
5576:
5575:
5572:
5571:
5569:
5568:
5561:
5554:
5547:
5540:
5532:
5530:
5522:
5521:
5519:
5518:
5511:
5504:
5497:
5490:
5487:Endornaviridae
5483:
5476:
5468:
5466:
5458:
5457:
5455:
5454:
5447:
5440:
5433:
5425:
5423:
5412:
5401:
5390:
5379:
5367:
5366:
5353:
5352:
5349:
5348:
5345:
5344:
5341:
5340:
5337:
5336:
5334:
5333:
5330:Botybirnavirus
5322:
5315:
5303:
5301:
5297:
5296:
5293:
5292:
5289:
5288:
5286:
5285:
5282:Partitiviridae
5278:
5271:
5264:
5261:Curvulaviridae
5257:
5249:
5247:
5236:
5225:
5217:
5216:
5213:
5212:
5209:
5208:
5206:
5205:
5197:
5195:
5184:
5176:
5175:
5172:
5171:
5169:
5168:
5160:
5158:
5147:
5139:
5138:
5135:
5134:
5132:
5131:
5124:
5117:
5110:
5102:
5100:
5089:
5078:
5067:
5056:
5044:
5043:
5027:
5021:
5020:
5017:
5016:
5013:
5012:
5009:
5008:
5006:
5005:
4998:
4991:
4979:
4977:
4973:
4972:
4969:
4968:
4965:
4964:
4961:
4960:
4957:
4956:
4954:
4953:
4945:
4943:
4932:
4921:
4910:
4902:
4901:
4898:
4897:
4894:
4893:
4890:
4889:
4887:
4886:
4879:
4871:
4869:
4866:Geplafuvirales
4858:
4850:
4849:
4846:
4845:
4843:
4842:
4839:Redondoviridae
4834:
4832:
4824:
4823:
4821:
4820:
4813:
4805:
4803:
4795:
4794:
4792:
4791:
4783:
4781:
4773:
4772:
4770:
4769:
4761:
4759:
4751:
4750:
4748:
4747:
4739:
4737:
4726:
4715:
4707:
4706:
4703:
4702:
4699:
4698:
4696:
4695:
4687:
4685:
4674:
4666:
4665:
4662:
4661:
4659:
4658:
4650:
4648:
4637:
4626:
4615:
4607:
4606:
4603:
4602:
4599:
4598:
4595:
4594:
4592:
4591:
4583:
4581:
4570:
4559:
4548:
4540:
4539:
4536:
4535:
4532:
4531:
4528:
4527:
4525:
4524:
4521:Plectroviridae
4517:
4514:Paulinoviridae
4510:
4502:
4500:
4489:
4486:Faserviricetes
4478:
4475:Hofneiviricota
4467:
4456:
4444:
4443:
4430:
4429:
4426:
4425:
4422:
4421:
4418:
4417:
4415:
4414:
4407:
4396:
4389:
4382:
4379:Polydnaviridae
4375:
4368:
4361:
4354:
4347:
4344:Globuloviridae
4340:
4337:Fuselloviridae
4333:
4326:
4323:Bicaudaviridae
4319:
4316:Ampullaviridae
4307:
4305:
4301:
4300:
4297:
4296:
4294:
4293:
4285:
4283:
4279:
4278:
4276:
4275:
4268:
4265:Hytrosaviridae
4261:
4253:
4251:
4240:
4237:Naldaviricetes
4229:
4225:
4224:
4221:
4220:
4217:
4216:
4213:
4212:
4209:
4208:
4206:
4205:
4198:
4191:
4183:
4181:
4170:
4167:Laserviricetes
4159:
4156:Dividoviricota
4148:
4140:
4139:
4136:
4135:
4132:
4131:
4128:
4127:
4125:
4124:
4121:Corticoviridae
4116:
4114:
4106:
4105:
4103:
4102:
4094:
4092:
4084:
4083:
4081:
4080:
4072:
4070:
4062:
4061:
4059:
4058:
4050:
4048:
4037:
4029:
4028:
4025:
4024:
4022:
4021:
4013:
4011:
4000:
3992:
3991:
3988:
3987:
3985:
3984:
3976:
3974:
3963:
3952:
3944:
3943:
3940:
3939:
3936:
3935:
3933:
3932:
3925:
3918:
3910:
3908:
3905:Pimascovirales
3900:
3899:
3897:
3896:
3888:
3886:
3878:
3877:
3875:
3874:
3866:
3864:
3853:
3845:
3844:
3841:
3840:
3838:
3837:
3829:
3827:
3819:
3818:
3816:
3815:
3807:
3805:
3794:
3783:
3772:
3761:
3753:
3752:
3749:
3748:
3745:
3744:
3741:
3740:
3737:
3736:
3734:
3733:
3725:
3723:
3715:
3714:
3712:
3711:
3708:Polyomaviridae
3703:
3701:
3690:
3679:
3668:
3657:
3649:
3648:
3645:
3644:
3641:
3640:
3637:
3636:
3633:
3632:
3630:
3629:
3622:
3615:
3608:
3605:Salasmaviridae
3601:
3594:
3587:
3580:
3577:Herelleviridae
3573:
3566:
3563:Drexlerviridae
3559:
3556:Demerecviridae
3552:
3545:
3538:
3530:
3528:
3517:
3514:Caudoviricetes
3506:
3498:
3497:
3494:
3493:
3490:
3489:
3487:
3486:
3479:
3472:
3464:
3462:
3451:
3448:Herviviricetes
3440:
3429:
3426:Heunggongvirae
3418:
3410:
3409:
3406:
3405:
3402:
3401:
3398:
3397:
3394:
3393:
3391:
3390:
3382:
3380:
3372:
3371:
3369:
3368:
3361:
3353:
3351:
3348:Ligamenvirales
3340:
3329:
3318:
3307:
3295:
3294:
3278:
3272:
3271:
3259:
3258:
3251:
3244:
3236:
3230:
3229:
3220:
3213:
3206:
3204:: Marnaviridae
3196:
3195:External links
3193:
3190:
3189:
3132:
3105:
3062:
3003:
2948:
2909:
2852:
2795:
2744:
2695:
2660:
2603:
2554:
2513:
2478:(4): 231–237.
2458:
2437:(2): 171–183.
2417:
2360:
2319:
2257:
2193:
2150:Picornavirales
2135:
2106:
2072:
2021:
1967:
1940:(2): 216–222.
1920:
1879:Picornavirales
1856:
1800:
1755:Virus Research
1736:
1681:
1662:(2): 206–217.
1631:
1571:
1550:(2): 206–217.
1527:
1505:
1480:
1453:
1452:
1450:
1447:
1416:
1413:
1383:
1380:
1378:
1375:
1345:
1339:
1331:G. delicatula.
1318:GdelRNAV is a
1315:
1309:
1286:
1281:
1241:
1235:
1206:
1203:
1201:
1198:
1167:Picornaviridae
1146:
1143:
1131:Picornavirales
1122:
1119:
1110:
1107:
1090:
1084:
1082:
1077:
1067:
1057:
1043:
1037:
1035:
1030:
991:
985:
983:
978:
969:
965:
957:
954:
952:
949:
924:
921:
916:Picornaviridae
838:Genome map of
819:
816:
814:
811:
799:
798:
791:
784:
777:
770:
763:
741:
736:
735:
734:
727:
720:
713:
691:
686:
685:
684:
664:
659:
658:
657:
650:
643:
636:
614:
609:
608:
607:
587:
582:
581:
580:
563:
558:
557:
556:
546:
536:
505:
500:
484:Picornavirales
475:
474:Classification
472:
471:
470:
463:
456:
449:
442:
435:
428:
416:
413:
386:Picornavirales
296:
293:
246:Picornavirales
227:
226:
203:
202:
195:
188:
181:
174:
167:
160:
150:
149:
145:
144:
137:
133:
132:
129:Picornavirales
125:
121:
120:
113:
109:
108:
101:
97:
96:
89:
85:
84:
77:
70:
69:
64:
60:
59:
46:
45:
37:
36:
28:
27:
15:
9:
6:
4:
3:
2:
7207:
7196:
7193:
7191:
7188:
7187:
7185:
7168:
7163:
7159:
7155:
7150:
7146:
7142:
7137:
7133:
7129:
7124:
7120:
7116:
7111:
7107:
7102:
7096:
7092:
7087:
7081:
7077:
7076:
7074:
7072:
7068:
7064:
7059:
7019:
7018:
7014:
7013:
7011:
7009:
7008:
7003:
6997:
6996:
6992:
6991:
6989:
6987:
6986:
6985:Blubervirales
6981:
6978:
6976:
6975:
6970:
6967:
6965:
6964:
6959:
6956:
6954:
6953:
6948:
6945:
6943:
6942:
6937:
6933:
6929:
6923:
6919:
6889:
6888:
6884:
6882:
6881:
6880:Pseudoviridae
6877:
6875:
6874:
6870:
6868:
6867:
6866:Belpaoviridae
6863:
6862:
6860:
6858:
6857:
6852:
6849:
6847:
6846:
6841:
6838:
6836:
6835:
6830:
6827:
6825:
6824:
6819:
6816:
6814:
6813:
6808:
6804:
6800:
6794:
6790:
6787:
6785:
6781:
6747:
6746:
6742:
6741:
6739:
6737:
6736:
6731:
6728:
6726:
6725:
6720:
6710:
6709:
6705:
6703:
6702:
6698:
6696:
6695:
6694:Rhabdoviridae
6691:
6689:
6688:
6687:Pneumoviridae
6684:
6682:
6681:
6677:
6675:
6674:
6670:
6668:
6667:
6666:Mymonaviridae
6663:
6661:
6660:
6656:
6654:
6653:
6649:
6647:
6646:
6642:
6640:
6639:
6635:
6634:
6632:
6630:
6629:
6624:
6618:
6617:
6616:Natareviridae
6613:
6611:
6610:
6606:
6604:
6603:
6599:
6597:
6596:
6592:
6590:
6589:
6585:
6584:
6582:
6580:
6579:
6574:
6571:
6569:
6568:
6563:
6553:
6552:
6548:
6547:
6545:
6543:
6542:
6537:
6534:
6532:
6531:
6526:
6516:
6515:
6511:
6509:
6508:
6507:Amnoonviridae
6504:
6503:
6501:
6499:
6498:
6493:
6490:
6488:
6487:
6482:
6472:
6471:
6470:Wupedeviridae
6467:
6465:
6464:
6460:
6458:
6457:
6456:Phenuiviridae
6453:
6451:
6450:
6449:Phasmaviridae
6446:
6444:
6443:
6439:
6437:
6436:
6432:
6430:
6429:
6425:
6423:
6422:
6418:
6416:
6415:
6411:
6409:
6408:
6404:
6402:
6401:
6397:
6395:
6394:
6390:
6389:
6387:
6385:
6384:
6379:
6376:
6374:
6373:
6368:
6358:
6357:
6353:
6352:
6350:
6348:
6347:
6342:
6339:
6337:
6336:
6331:
6328:
6326:
6325:
6320:
6317:
6315:
6314:
6313:Orthornavirae
6309:
6306:
6304:
6303:
6298:
6294:
6290:
6284:
6280:
6258:
6257:
6253:
6251:
6250:
6245:
6242:
6241:
6239:
6235:
6221:
6220:
6216:
6215:
6213:
6211:
6210:
6209:Stellavirales
6205:
6199:
6198:
6194:
6193:
6191:
6189:
6188:
6187:Patatavirales
6183:
6180:
6178:
6177:
6172:
6162:
6161:
6160:Solemoviridae
6157:
6155:
6154:
6150:
6148:
6147:
6143:
6142:
6140:
6138:
6137:
6136:Sobelivirales
6132:
6126:
6125:
6121:
6119:
6118:
6114:
6112:
6111:
6107:
6105:
6104:
6100:
6098:
6097:
6096:Caliciviridae
6093:
6091:
6090:
6086:
6084:
6083:
6079:
6077:
6076:
6072:
6071:
6069:
6067:
6066:
6061:
6055:
6054:
6053:Tobaniviridae
6050:
6048:
6047:
6043:
6041:
6040:
6036:
6034:
6033:
6029:
6027:
6026:
6022:
6020:
6019:
6018:Mononiviridae
6015:
6013:
6012:
6011:Mesoniviridae
6008:
6006:
6005:
6001:
5999:
5998:
5997:Gresnaviridae
5994:
5992:
5991:
5990:Euroniviridae
5987:
5985:
5984:
5983:Coronaviridae
5980:
5978:
5977:
5973:
5971:
5970:
5969:Arteriviridae
5966:
5964:
5963:
5962:Abyssoviridae
5959:
5958:
5956:
5954:
5953:
5948:
5945:
5943:
5942:
5937:
5934:
5932:
5931:
5926:
5912:
5911:
5907:
5906:
5904:
5902:
5901:
5896:
5893:
5891:
5890:
5885:
5875:
5874:
5870:
5869:
5867:
5865:
5864:
5863:Cryppavirales
5859:
5856:
5854:
5853:
5848:
5838:
5837:
5833:
5832:
5830:
5828:
5827:
5822:
5819:
5817:
5816:
5811:
5801:
5800:
5799:Steitzviridae
5796:
5794:
5793:
5789:
5788:
5786:
5784:
5783:
5778:
5772:
5771:
5770:Solspiviridae
5767:
5765:
5764:
5760:
5758:
5757:
5753:
5751:
5750:
5749:Atkinsviridae
5746:
5745:
5743:
5741:
5740:
5735:
5732:
5730:
5729:
5728:Leviviricetes
5724:
5721:
5719:
5718:
5717:Lenarviricota
5713:
5699:
5698:
5697:Tombusviridae
5694:
5692:
5691:
5687:
5685:
5684:
5680:
5679:
5677:
5675:
5674:
5669:
5666:
5664:
5663:
5658:
5648:
5647:
5643:
5641:
5640:
5636:
5635:
5633:
5631:
5630:
5629:Nodamuvirales
5625:
5622:
5620:
5619:
5614:
5604:
5603:
5599:
5598:
5596:
5594:
5593:
5588:
5585:
5583:
5582:
5577:
5567:
5566:
5562:
5560:
5559:
5555:
5553:
5552:
5548:
5546:
5545:
5541:
5539:
5538:
5534:
5533:
5531:
5529:
5528:
5523:
5517:
5516:
5512:
5510:
5509:
5505:
5503:
5502:
5498:
5496:
5495:
5491:
5489:
5488:
5484:
5482:
5481:
5477:
5475:
5474:
5470:
5469:
5467:
5465:
5464:
5459:
5453:
5452:
5451:Matonaviridae
5448:
5446:
5445:
5441:
5439:
5438:
5434:
5432:
5431:
5427:
5426:
5424:
5422:
5421:
5420:Hepelivirales
5416:
5413:
5411:
5410:
5409:Alsuviricetes
5405:
5402:
5400:
5399:
5394:
5391:
5389:
5388:
5387:Orthornavirae
5383:
5380:
5378:
5377:
5372:
5368:
5364:
5358:
5354:
5332:
5331:
5326:
5323:
5321:
5320:
5316:
5314:
5313:
5308:
5305:
5304:
5302:
5298:
5284:
5283:
5279:
5277:
5276:
5272:
5270:
5269:
5265:
5263:
5262:
5258:
5256:
5255:
5254:Amalgaviridae
5251:
5250:
5248:
5246:
5245:
5240:
5237:
5235:
5234:
5229:
5226:
5224:
5223:
5218:
5204:
5203:
5199:
5198:
5196:
5194:
5193:
5188:
5185:
5183:
5182:
5177:
5167:
5166:
5162:
5161:
5159:
5157:
5156:
5151:
5148:
5146:
5145:
5140:
5130:
5129:
5125:
5123:
5122:
5121:Quadriviridae
5118:
5116:
5115:
5111:
5109:
5108:
5107:Chrysoviridae
5104:
5103:
5101:
5099:
5098:
5097:Ghabrivirales
5093:
5090:
5088:
5087:
5082:
5079:
5077:
5076:
5071:
5068:
5066:
5065:
5064:Orthornavirae
5060:
5057:
5055:
5054:
5049:
5045:
5041:
5040:dsRNA viruses
5035:
5031:
5028:
5026:
5022:
5004:
5003:
4999:
4997:
4996:
4992:
4990:
4989:
4988:Anelloviridae
4984:
4981:
4980:
4978:
4974:
4952:
4951:
4947:
4946:
4944:
4942:
4941:
4940:Haloruvirales
4936:
4933:
4931:
4930:
4925:
4922:
4920:
4919:
4914:
4911:
4909:
4908:
4903:
4885:
4884:
4883:Genomoviridae
4880:
4878:
4877:
4876:Geminiviridae
4873:
4872:
4870:
4868:
4867:
4862:
4859:
4857:
4856:
4851:
4841:
4840:
4836:
4835:
4833:
4831:
4830:
4825:
4819:
4818:
4814:
4812:
4811:
4810:Metaxyviridae
4807:
4806:
4804:
4802:
4801:
4796:
4790:
4789:
4785:
4784:
4782:
4780:
4779:
4774:
4768:
4767:
4763:
4762:
4760:
4758:
4757:
4752:
4746:
4745:
4741:
4740:
4738:
4736:
4735:
4730:
4727:
4725:
4724:
4723:Arfiviricetes
4719:
4716:
4714:
4713:
4708:
4694:
4693:
4689:
4688:
4686:
4684:
4683:
4678:
4675:
4673:
4672:
4667:
4657:
4656:
4652:
4651:
4649:
4647:
4646:
4641:
4638:
4636:
4635:
4630:
4627:
4625:
4624:
4623:Cossaviricota
4619:
4616:
4614:
4613:
4608:
4590:
4589:
4585:
4584:
4582:
4580:
4579:
4574:
4571:
4569:
4568:
4563:
4560:
4558:
4557:
4552:
4549:
4547:
4546:
4541:
4523:
4522:
4518:
4516:
4515:
4511:
4509:
4508:
4504:
4503:
4501:
4499:
4498:
4497:Tubulavirales
4493:
4490:
4488:
4487:
4482:
4479:
4477:
4476:
4471:
4468:
4466:
4465:
4460:
4457:
4455:
4454:
4449:
4445:
4441:
4440:ssDNA viruses
4435:
4431:
4413:
4412:
4411:Rhizidiovirus
4408:
4406:
4405:
4400:
4397:
4395:
4394:
4393:Thaspiviridae
4390:
4388:
4387:
4383:
4381:
4380:
4376:
4374:
4373:
4372:Plasmaviridae
4369:
4367:
4366:
4362:
4360:
4359:
4358:Halspiviridae
4355:
4353:
4352:
4348:
4346:
4345:
4341:
4339:
4338:
4334:
4332:
4331:
4327:
4325:
4324:
4320:
4318:
4317:
4312:
4309:
4308:
4306:
4302:
4292:
4291:
4287:
4286:
4284:
4280:
4274:
4273:
4269:
4267:
4266:
4262:
4260:
4259:
4258:Baculoviridae
4255:
4254:
4252:
4250:
4249:
4244:
4241:
4239:
4238:
4233:
4230:
4226:
4204:
4203:
4199:
4197:
4196:
4195:Simuloviridae
4192:
4190:
4189:
4185:
4184:
4182:
4180:
4179:
4174:
4171:
4169:
4168:
4163:
4160:
4158:
4157:
4152:
4149:
4147:
4146:
4145:Helvetiavirae
4141:
4123:
4122:
4118:
4117:
4115:
4113:
4112:
4107:
4101:
4100:
4096:
4095:
4093:
4091:
4090:
4085:
4079:
4078:
4074:
4073:
4071:
4069:
4068:
4067:Kalamavirales
4063:
4057:
4056:
4052:
4051:
4049:
4047:
4046:
4045:Belfryvirales
4041:
4038:
4036:
4035:
4030:
4020:
4019:
4018:Adintoviridae
4015:
4014:
4012:
4010:
4009:
4004:
4001:
3999:
3998:
3993:
3983:
3982:
3981:Lavidaviridae
3978:
3977:
3975:
3973:
3972:
3967:
3964:
3962:
3961:
3956:
3953:
3951:
3950:
3945:
3931:
3930:
3926:
3924:
3923:
3919:
3917:
3916:
3912:
3911:
3909:
3907:
3906:
3901:
3895:
3894:
3890:
3889:
3887:
3885:
3884:
3883:Imitervirales
3879:
3873:
3872:
3868:
3867:
3865:
3863:
3862:
3857:
3854:
3852:
3851:
3850:Megaviricetes
3846:
3836:
3835:
3831:
3830:
3828:
3826:
3825:
3820:
3814:
3813:
3809:
3808:
3806:
3804:
3803:
3798:
3795:
3793:
3792:
3787:
3784:
3782:
3781:
3776:
3773:
3771:
3770:
3765:
3762:
3760:
3759:
3754:
3732:
3731:
3727:
3726:
3724:
3722:
3721:
3716:
3710:
3709:
3705:
3704:
3702:
3700:
3699:
3698:Sepolyvirales
3694:
3691:
3689:
3688:
3683:
3680:
3678:
3677:
3676:Cossaviricota
3672:
3669:
3667:
3666:
3661:
3658:
3656:
3655:
3650:
3628:
3627:
3626:Zobellviridae
3623:
3621:
3620:
3616:
3614:
3613:
3612:Schitoviridae
3609:
3607:
3606:
3602:
3600:
3599:
3595:
3593:
3592:
3588:
3586:
3585:
3581:
3579:
3578:
3574:
3572:
3571:
3570:Guelinviridae
3567:
3565:
3564:
3560:
3558:
3557:
3553:
3551:
3550:
3546:
3544:
3543:
3539:
3537:
3536:
3532:
3531:
3529:
3527:
3526:
3521:
3518:
3516:
3515:
3510:
3507:
3505:
3504:
3499:
3485:
3484:
3480:
3478:
3477:
3476:Herpesviridae
3473:
3471:
3470:
3466:
3465:
3463:
3461:
3460:
3459:Herpesvirales
3455:
3452:
3450:
3449:
3444:
3441:
3439:
3438:
3437:Peploviricota
3433:
3430:
3428:
3427:
3422:
3419:
3417:
3416:
3415:Duplodnaviria
3411:
3389:
3388:
3384:
3383:
3381:
3379:
3378:
3373:
3367:
3366:
3362:
3360:
3359:
3355:
3354:
3352:
3350:
3349:
3344:
3341:
3339:
3338:
3337:Tokiviricetes
3333:
3330:
3328:
3327:
3326:Taleaviricota
3322:
3319:
3317:
3316:
3311:
3308:
3306:
3305:
3300:
3296:
3292:
3291:dsDNA viruses
3286:
3282:
3279:
3277:
3273:
3268:
3264:
3257:
3252:
3250:
3245:
3243:
3238:
3237:
3234:
3228:
3227:
3221:
3218:
3214:
3212:
3211:
3207:
3205:
3203:
3199:
3198:
3185:
3181:
3176:
3171:
3167:
3163:
3159:
3155:
3151:
3149:
3145:
3136:
3128:
3124:
3120:
3116:
3109:
3101:
3097:
3093:
3089:
3085:
3081:
3077:
3073:
3066:
3058:
3054:
3049:
3044:
3039:
3034:
3030:
3026:
3022:
3018:
3014:
3007:
2999:
2995:
2990:
2985:
2980:
2975:
2971:
2967:
2963:
2961:
2952:
2944:
2940:
2936:
2932:
2928:
2924:
2920:
2913:
2905:
2901:
2896:
2891:
2887:
2883:
2879:
2875:
2871:
2867:
2863:
2856:
2848:
2844:
2839:
2834:
2830:
2826:
2822:
2818:
2814:
2810:
2806:
2799:
2791:
2787:
2782:
2777:
2772:
2767:
2763:
2759:
2755:
2748:
2740:
2736:
2732:
2728:
2723:
2718:
2714:
2710:
2706:
2699:
2691:
2687:
2683:
2679:
2676:(6): 001772.
2675:
2671:
2664:
2656:
2652:
2647:
2642:
2638:
2634:
2630:
2626:
2622:
2618:
2614:
2607:
2599:
2595:
2590:
2585:
2581:
2577:
2573:
2569:
2565:
2558:
2550:
2546:
2541:
2536:
2532:
2528:
2524:
2517:
2509:
2505:
2501:
2497:
2493:
2489:
2485:
2481:
2477:
2473:
2469:
2462:
2454:
2450:
2445:
2440:
2436:
2432:
2428:
2421:
2413:
2409:
2404:
2399:
2395:
2391:
2387:
2383:
2379:
2375:
2371:
2364:
2356:
2352:
2347:
2342:
2338:
2334:
2330:
2323:
2315:
2311:
2306:
2301:
2297:
2293:
2289:
2285:
2281:
2277:
2273:
2266:
2264:
2262:
2253:
2249:
2244:
2239:
2234:
2229:
2225:
2221:
2217:
2210:
2208:
2206:
2204:
2202:
2200:
2198:
2189:
2185:
2180:
2175:
2170:
2165:
2161:
2157:
2153:
2151:
2142:
2140:
2123:
2117:
2115:
2113:
2111:
2095:
2091:
2085:
2083:
2081:
2079:
2077:
2068:
2064:
2059:
2054:
2049:
2044:
2040:
2036:
2032:
2025:
2017:
2013:
2008:
2003:
1999:
1995:
1991:
1987:
1983:
1976:
1974:
1972:
1963:
1959:
1955:
1951:
1947:
1943:
1939:
1935:
1931:
1924:
1916:
1912:
1907:
1902:
1898:
1894:
1891:(2): vez056.
1890:
1886:
1882:
1880:
1871:
1869:
1867:
1865:
1863:
1861:
1852:
1848:
1843:
1838:
1834:
1830:
1826:
1822:
1818:
1811:
1809:
1807:
1805:
1796:
1792:
1788:
1784:
1779:
1774:
1769:
1764:
1760:
1756:
1752:
1745:
1743:
1741:
1732:
1728:
1723:
1718:
1713:
1708:
1704:
1700:
1696:
1694:
1685:
1677:
1673:
1669:
1665:
1661:
1657:
1653:
1646:
1644:
1642:
1640:
1638:
1636:
1627:
1623:
1618:
1613:
1609:
1605:
1602:(8): 001633.
1601:
1597:
1593:
1591:
1582:
1580:
1578:
1576:
1567:
1563:
1558:
1553:
1549:
1545:
1541:
1534:
1532:
1515:
1509:
1494:
1490:
1484:
1468:
1464:
1458:
1454:
1446:
1445:
1444:Kusarnavirus.
1441:
1440:Sogarnavirus.
1437:
1432:
1428:
1424:
1423:
1412:
1410:
1406:
1402:
1398:
1394:
1390:
1389:
1374:
1371:
1367:
1363:
1361:
1356:
1355:
1350:
1343:
1338:
1336:
1335:G. delicatula
1332:
1328:
1326:
1321:
1313:
1308:
1306:
1302:
1298:
1294:
1293:
1285:
1280:
1279:
1274:
1270:
1266:
1262:
1259:
1258:
1253:
1249:
1248:
1239:
1234:
1232:
1228:
1224:
1220:
1216:
1212:
1197:
1195:
1191:
1186:
1182:
1172:
1168:
1163:
1162:
1161:Marnaviridae.
1157:
1156:
1151:
1142:
1140:
1139:Marnaviridae,
1136:
1132:
1128:
1127:Echinodermata
1121:Echinodermata
1118:
1116:
1106:
1104:
1100:
1096:
1095:C. tenussimus
1088:
1081:
1076:
1073:
1065:
1061:
1056:
1054:
1049:
1041:
1034:
1029:
1027:
1023:
1019:
1015:
1010:
1005:
1003:
1002:
997:
989:
982:
977:
973:
963:
962:Marnaviridae.
948:
945:
941:
936:
934:
929:
920:
918:
917:
912:
909:(CtenRNAVII,
908:
904:
900:
896:
891:
889:
884:
876:
872:
868:
864:
857:
852:
841:
836:
829:
824:
810:
808:
804:
797:
796:
792:
790:
789:
785:
783:
782:
778:
776:
775:
771:
769:
768:
764:
762:
761:
757:
756:
755:
753:
749:
748:
740:
733:
732:
728:
726:
725:
721:
719:
718:
714:
712:
711:
707:
706:
705:
703:
699:
698:
690:
683:
680:
679:
675:
674:
673:
671:
670:
663:
656:
655:
651:
649:
648:
644:
642:
641:
637:
635:
634:
630:
629:
628:
626:
622:
621:
613:
606:
603:
602:
598:
597:
596:
594:
593:
586:
579:
578:
574:
573:
572:
570:
569:
562:
555:
552:
551:
547:
545:
542:
541:
537:
535:
533:
529:
526:
525:
521:
520:
519:
517:
513:
512:
504:
499:
497:
493:
489:
485:
481:
469:
468:
464:
462:
461:
457:
455:
454:
450:
448:
447:
443:
441:
440:
436:
434:
433:
429:
427:
426:
422:
421:
420:
412:
410:
405:
403:
399:
395:
391:
387:
383:
379:
375:
371:
367:
363:
361:
358:
357:
352:
351:
346:
342:
338:
334:
329:
327:
323:
319:
318:
312:
310:
306:
302:
292:
291:
290:Marnaviridae.
286:
282:
281:
276:
271:
269:
268:
263:
259:
258:
253:
248:
247:
243:in the order
242:
239:
235:
234:
223:
218:
212:
207:
201:
200:
196:
194:
193:
189:
187:
186:
182:
180:
179:
175:
173:
172:
168:
166:
165:
161:
159:
158:
154:
153:
151:
146:
143:
142:
138:
135:
134:
131:
130:
126:
123:
122:
119:
118:
114:
111:
110:
107:
106:
102:
99:
98:
95:
94:
93:Orthornavirae
90:
87:
86:
83:
82:
78:
75:
72:
71:
68:
65:
62:
61:
56:
51:
47:
43:
38:
34:
29:
26:
22:
19:
7101:Marnaviridae
7071:Marnaviridae
7070:
7015:
7007:Ortervirales
7005:
6993:
6983:
6972:
6961:
6952:Pararnavirae
6950:
6939:
6887:Retroviridae
6885:
6878:
6871:
6864:
6856:Ortervirales
6854:
6843:
6832:
6823:Pararnavirae
6821:
6810:
6743:
6733:
6722:
6708:Xinmoviridae
6706:
6699:
6692:
6685:
6678:
6673:Nyamiviridae
6671:
6664:
6659:Lispiviridae
6657:
6650:
6645:Bornaviridae
6643:
6636:
6626:
6614:
6609:Myriaviridae
6607:
6600:
6593:
6588:Aliusviridae
6586:
6576:
6565:
6549:
6539:
6528:
6512:
6505:
6495:
6484:
6468:
6463:Tospoviridae
6461:
6454:
6447:
6440:
6435:Nairoviridae
6433:
6426:
6419:
6414:Hantaviridae
6412:
6405:
6400:Arenaviridae
6398:
6393:Cruliviridae
6391:
6383:Bunyavirales
6381:
6370:
6354:
6344:
6333:
6322:
6311:
6300:
6254:
6247:
6243:
6219:Astroviridae
6217:
6207:
6195:
6185:
6174:
6158:
6153:Barnaviridae
6151:
6144:
6134:
6122:
6115:
6108:
6101:
6094:
6087:
6082:Marnaviridae
6081:
6080:
6073:
6063:
6051:
6044:
6039:Olifoviridae
6037:
6030:
6023:
6016:
6009:
6002:
5995:
5988:
5981:
5974:
5967:
5960:
5950:
5939:
5930:Pisuviricota
5928:
5908:
5900:Ourlivirales
5898:
5889:Miaviricetes
5887:
5871:
5861:
5850:
5836:Narnaviridae
5834:
5824:
5813:
5797:
5792:Blumeviridae
5790:
5782:Timlovirales
5780:
5768:
5763:Fiersviridae
5761:
5754:
5747:
5739:Norzivirales
5737:
5726:
5715:
5695:
5690:Luteoviridae
5688:
5681:
5671:
5660:
5644:
5637:
5627:
5616:
5602:Flaviviridae
5600:
5590:
5579:
5563:
5556:
5549:
5542:
5535:
5525:
5515:Virgaviridae
5513:
5506:
5499:
5492:
5485:
5478:
5473:Bromoviridae
5471:
5461:
5449:
5442:
5435:
5428:
5418:
5407:
5396:
5385:
5374:
5328:
5324:
5317:
5312:Birnaviridae
5310:
5306:
5280:
5273:
5266:
5259:
5252:
5244:Durnavirales
5242:
5231:
5222:Pisuviricota
5220:
5202:Cystoviridae
5200:
5192:Mindivirales
5190:
5179:
5163:
5153:
5142:
5126:
5119:
5112:
5105:
5095:
5084:
5073:
5062:
5051:
5002:Spiraviridae
5000:
4993:
4986:
4982:
4948:
4938:
4927:
4918:Saleviricota
4916:
4905:
4881:
4874:
4864:
4853:
4837:
4829:Recrevirales
4827:
4815:
4808:
4800:Mulpavirales
4798:
4788:Smacoviridae
4786:
4778:Cremevirales
4776:
4766:Circoviridae
4764:
4756:Cirlivirales
4754:
4742:
4734:Baphyvirales
4732:
4721:
4710:
4692:Parvoviridae
4690:
4682:Piccovirales
4680:
4669:
4655:Bidnaviridae
4653:
4643:
4634:Mouviricetes
4632:
4621:
4612:Shotokuvirae
4610:
4588:Microviridae
4586:
4578:Petitvirales
4576:
4565:
4556:Phixviricota
4554:
4543:
4519:
4512:
4505:
4495:
4484:
4473:
4462:
4453:Monodnaviria
4451:
4409:
4404:Dinodnavirus
4402:
4398:
4391:
4384:
4377:
4370:
4365:Ovaliviridae
4363:
4356:
4351:Guttaviridae
4349:
4342:
4335:
4330:Clavaviridae
4328:
4321:
4314:
4310:
4288:
4270:
4263:
4256:
4246:
4235:
4200:
4193:
4186:
4176:
4165:
4154:
4143:
4119:
4109:
4099:Adenoviridae
4097:
4087:
4077:Tectiviridae
4075:
4065:
4055:Turriviridae
4053:
4043:
4032:
4016:
4006:
3995:
3979:
3969:
3958:
3947:
3927:
3922:Iridoviridae
3920:
3913:
3903:
3891:
3881:
3869:
3859:
3848:
3832:
3824:Chitovirales
3822:
3812:Asfarviridae
3810:
3800:
3789:
3778:
3769:Bamfordvirae
3767:
3758:Varidnaviria
3756:
3728:
3718:
3706:
3696:
3685:
3674:
3665:Shotokuvirae
3663:
3654:Monodnaviria
3652:
3624:
3619:Siphoviridae
3617:
3610:
3603:
3596:
3589:
3582:
3575:
3568:
3561:
3554:
3549:Chaseviridae
3547:
3540:
3533:
3525:Caudovirales
3523:
3512:
3501:
3481:
3474:
3467:
3457:
3446:
3435:
3424:
3413:
3385:
3377:Primavirales
3375:
3363:
3356:
3346:
3335:
3324:
3313:
3302:
3226:Marnaviridae
3225:
3217:Marnaviridae
3209:
3201:
3157:
3153:
3147:
3144:Flaviviridae
3143:
3135:
3118:
3114:
3108:
3075:
3071:
3065:
3020:
3016:
3006:
2969:
2965:
2959:
2951:
2929:(1): 27–36.
2926:
2922:
2912:
2869:
2865:
2855:
2812:
2808:
2798:
2761:
2757:
2747:
2712:
2708:
2698:
2673:
2669:
2663:
2620:
2616:
2606:
2571:
2567:
2557:
2533:(1): 69–80.
2530:
2526:
2516:
2475:
2471:
2461:
2434:
2430:
2420:
2377:
2373:
2363:
2336:
2332:
2322:
2279:
2275:
2226:(11): 2369.
2223:
2219:
2159:
2155:
2149:
2126:. Retrieved
2122:"Viral Zone"
2097:. Retrieved
2093:
2038:
2034:
2024:
1989:
1985:
1937:
1933:
1929:
1923:
1888:
1884:
1878:
1824:
1820:
1758:
1754:
1702:
1698:
1692:
1684:
1659:
1655:
1651:
1599:
1595:
1590:Marnaviridae
1589:
1547:
1543:
1518:. Retrieved
1508:
1496:. Retrieved
1492:
1483:
1471:. Retrieved
1469:. March 2019
1466:
1457:
1443:
1439:
1436:Labyrnavirus
1435:
1430:
1427:Marnaviridae
1426:
1420:
1418:
1408:
1405:Marnaviridae
1404:
1400:
1396:
1386:
1385:
1369:
1365:
1359:
1352:
1347:
1341:
1334:
1330:
1323:
1319:
1317:
1311:
1304:
1300:
1296:
1290:
1288:
1283:
1278:H. akashiwo.
1277:
1272:
1268:
1264:
1255:
1251:
1245:
1243:
1237:
1231:Marnaviridae
1230:
1223:Marnaviridae
1222:
1219:Marnaviridae
1218:
1215:Marnaviridae
1214:
1210:
1208:
1205:Distribution
1194:Marnaviridae
1193:
1190:Marnaviridae
1189:
1187:
1183:
1171:Marnaviridae
1170:
1166:
1164:
1160:
1153:
1150:Marnaviridae
1149:
1148:
1138:
1135:Marnaviridae
1134:
1130:
1124:
1115:Marnaviridae
1114:
1112:
1103:Marnaviridae
1102:
1098:
1094:
1092:
1086:
1080:Sogarnavirus
1079:
1071:
1069:
1063:
1059:
1052:
1047:
1045:
1042:RNA virus 01
1039:
1032:
1025:
1021:
1017:
1013:
1008:
1006:
999:
996:Marnaviridae
995:
993:
987:
980:
974:
961:
959:
943:
940:Marnaviridae
939:
937:
928:Marnaviridae
927:
926:
914:
911:Sogarnavirus
910:
906:
902:
898:
894:
892:
883:Marnaviridae
882:
880:
874:
870:
866:
856:Marnaviridae
855:
840:Marnaviridae
839:
827:
807:Marnaviridae
806:
800:
793:
786:
779:
772:
765:
758:
752:Sogarnavirus
751:
747:Sogarnavirus
745:
743:
739:Sogarnavirus
738:
729:
722:
715:
708:
701:
695:
693:
688:
676:
667:
666:
661:
652:
645:
638:
631:
625:Locarnavirus
624:
620:Locarnavirus
618:
616:
612:Locarnavirus
611:
599:
592:Labyrnavirus
590:
589:
585:Labyrnavirus
584:
577:Astarnavirus
575:
568:Kusarnavirus
566:
565:
561:Kusarnavirus
560:
554:RNA virus 01
548:
544:RNA virus 01
538:
531:
522:
515:
509:
507:
502:
492:Marnaviridae
491:
488:Marnaviridae
487:
483:
480:Marnaviridae
479:
477:
467:Sogarnavirus
465:
458:
451:
446:Locarnavirus
444:
439:Labyrnavirus
437:
432:Kusarnavirus
430:
423:
418:
408:
406:
402:Marnaviridae
401:
397:
394:Labyrnavirus
393:
390:Marnaviridae
389:
385:
369:
366:Marnaviridae
365:
364:
354:
350:Raphidovirus
348:
344:
340:
330:
315:
313:
308:
304:
301:marnaviridae
300:
298:
289:
285:Baishivirus,
284:
278:
275:Marnaviridae
274:
272:
265:
262:Raphidophyte
255:
251:
244:
233:Marnaviridae
232:
231:
230:
199:Sogarnavirus
197:
190:
183:
178:Locarnavirus
176:
171:Labyrnavirus
169:
164:Kusarnavirus
162:
155:
141:Marnaviridae
140:
139:
128:
116:
105:Pisuviricota
104:
92:
80:
73:
63:(unranked):
41:
25:Marnaviridae
24:
18:
7095:Wikispecies
6873:Metaviridae
6652:Filoviridae
6638:Artoviridae
6551:Aspiviridae
6428:Mypoviridae
6407:Fimoviridae
6197:Potyviridae
6110:Secoviridae
6103:Iflaviridae
6046:Roniviridae
5952:Nidovirales
5873:Mitoviridae
5756:Duinviridae
5673:Tolivirales
5639:Nodaviridae
5565:Tymoviridae
5527:Tymovirales
5508:Togaviridae
5501:Mayoviridae
5494:Kitaviridae
5444:Hepeviridae
5437:Benyviridae
5268:Hypoviridae
5128:Totiviridae
4817:Nanoviridae
4645:Polivirales
4545:Sangervirae
4290:Nimaviridae
4272:Nudiviridae
4248:Lefavirales
4111:Vinavirales
4089:Rowavirales
3915:Ascoviridae
3893:Mimiviridae
3861:Algavirales
3802:Asfuvirales
3591:Podoviridae
3503:Uroviricota
3365:Rudiviridae
3315:Zilligvirae
1409:Baishivirus
1401:Baishivirus
1397:P. vannamei
1370:H. akashiwo
1305:Chaetoceros
1301:Chaetoceros
1292:Chaetoceros
1273:H. akashiwo
1269:H. akashiwo
1265:H. akashiwo
1252:H. akashiwo
1247:H. akashiwo
1211:H. akashiwo
1155:Chaetoceros
1099:Chaetoceros
1072:C. socialis
1070:The diatom
1066:RNA virus 1
1026:H. akashiwo
1022:H. akashiwo
1018:H. akashiwo
1016:than other
1014:H. akashiwo
1009:H. akashiwo
1001:H. akashiwo
881:Virions in
869:RNA virus 0
803:metagenomic
534:RNA virus 1
374:metagenomic
341:H. akashiwo
317:H. akashiwo
241:RNA viruses
7184:Categories
6745:Yueviridae
6701:Sunviridae
6595:Chuviridae
6356:Qinviridae
6237:Unassigned
5300:Unassigned
5165:Reoviridae
5155:Reovirales
4976:Unassigned
4907:Trapavirae
4507:Inoviridae
4304:Unassigned
4282:Unassigned
4228:Unassigned
3834:Poxviridae
3584:Myoviridae
3078:: 107752.
2764:(2): 189.
2041:(3): 362.
1761:: 199026.
1449:References
1327:delicatula
981:Marnavirus
956:Life cycle
669:Marnavirus
662:Marnavirus
453:Marnavirus
409:Marnavirus
370:Marnavirus
326:cell lysis
305:rnaviridae
299:The name "
257:Marnavirus
185:Marnavirus
42:Marnavirus
7086:Q15642831
6941:Riboviria
6812:Riboviria
6346:Muvirales
6302:Riboviria
5376:Riboviria
5053:Riboviria
4464:Loebvirae
3304:Adnaviria
3263:Baltimore
3202:Viralzone
3121:: 56–58.
3100:247896446
2739:211193770
2715:: 29–62.
2549:0948-3055
2508:232766599
2500:1752-0908
2453:0948-3055
1795:254772235
1362:DNA virus
1325:Guinardia
1314:RNA virus
1196:viruses.
818:Structure
682:RNA virus
362:(HaNIV).
88:Kingdom:
81:Riboviria
7080:Wikidata
6244:Families
5307:Families
4983:Families
4311:Families
3219:(family)
3184:34100644
3154:mSystems
3092:35367462
3057:26203654
3017:PLOS ONE
2998:30687251
2972:: 3235.
2943:83839588
2904:33741635
2847:11055943
2790:30813316
2731:32073403
2690:35766975
2655:25452289
2598:32978816
2412:17041155
2355:16476996
2314:18469125
2252:36366467
2188:32024776
2124:. ExPASy
2099:20 March
2067:33668994
2016:32690954
1962:86563435
1915:31908848
1851:36445118
1787:36529302
1778:10194266
1731:30687251
1705:: 3235.
1676:86464306
1626:34356002
1566:15016544
1544:Virology
1498:27 April
1473:16 March
990:(HaRNAV)
873:, genus
415:Taxonomy
136:Family:
100:Phylum:
3175:8269200
3048:4512727
3025:Bibcode
2989:6334475
2895:8208158
2874:Bibcode
2817:Bibcode
2781:6410135
2758:Viruses
2646:4292505
2625:Bibcode
2589:7894508
2480:Bibcode
2403:1694243
2382:Bibcode
2305:2446501
2284:Bibcode
2243:9697754
2220:Viruses
2179:7163153
2128:15 June
2058:7996518
2035:Viruses
2007:7508674
1942:Bibcode
1906:6938265
1842:9769563
1722:6334475
1617:8513639
1200:Ecology
1064:radians
899:radians
532:radians
311:(sea).
295:History
148:Genera
124:Order:
112:Class:
7167:600083
7154:324901
7141:106874
5325:Genera
4399:Genera
3222:ICTV:
3215:NCBI:
3182:
3172:
3098:
3090:
3055:
3045:
2996:
2986:
2941:
2902:
2892:
2845:
2835:
2788:
2778:
2737:
2729:
2688:
2653:
2643:
2596:
2586:
2547:
2506:
2498:
2451:
2410:
2400:
2353:
2312:
2302:
2250:
2240:
2186:
2176:
2065:
2055:
2014:
2004:
1960:
1913:
1903:
1849:
1839:
1793:
1785:
1775:
1729:
1719:
1674:
1624:
1614:
1564:
1520:20 May
1062:forma
923:Genome
905:) and
897:forma
478:While
353:, and
264:alga,
7162:WoRMS
7136:IRMNG
7115:624LM
6926:VII:
5038:III:
3096:S2CID
2939:S2CID
2838:92399
2735:S2CID
2504:S2CID
2162:(9).
1958:S2CID
1791:S2CID
1672:S2CID
1592:2021"
528:forma
74:Realm
67:Virus
7149:NCBI
7128:4516
7123:GBIF
6797:VI:
5361:IV:
4438:II:
3210:ICTV
3180:PMID
3088:PMID
3053:PMID
2994:PMID
2900:PMID
2843:PMID
2786:PMID
2727:PMID
2686:PMID
2651:PMID
2594:PMID
2545:ISSN
2496:ISSN
2449:ISSN
2408:PMID
2351:PMID
2310:PMID
2248:PMID
2184:PMID
2130:2015
2101:2023
2063:PMID
2012:PMID
1911:PMID
1847:PMID
1783:PMID
1727:PMID
1622:PMID
1562:PMID
1522:2021
1500:2017
1475:2019
744:The
694:The
617:The
508:The
396:and
382:RdRp
380:and
309:mare
7110:CoL
6287:V:
5025:RNA
3289:I:
3276:DNA
3170:PMC
3162:doi
3123:doi
3080:doi
3076:190
3043:PMC
3033:doi
2984:PMC
2974:doi
2931:doi
2890:PMC
2882:doi
2833:PMC
2825:doi
2776:PMC
2766:doi
2717:doi
2678:doi
2674:103
2641:PMC
2633:doi
2584:PMC
2576:doi
2572:229
2535:doi
2488:doi
2439:doi
2398:PMC
2390:doi
2341:doi
2300:PMC
2292:doi
2238:PMC
2228:doi
2174:PMC
2164:doi
2053:PMC
2043:doi
2002:PMC
1994:doi
1950:doi
1901:PMC
1893:doi
1837:PMC
1829:doi
1773:PMC
1763:doi
1759:324
1717:PMC
1707:doi
1664:doi
1612:PMC
1604:doi
1600:102
1552:doi
1548:320
942:is
335:in
7186::
7164::
7151::
7138::
7125::
7112::
7097::
7082::
6784:RT
6246::
5327::
5309::
4985::
4401::
4313::
3178:.
3168:.
3156:.
3152:.
3117:.
3094:.
3086:.
3074:.
3051:.
3041:.
3031:.
3021:10
3019:.
3015:.
2992:.
2982:.
2968:.
2964:.
2937:.
2927:61
2925:.
2921:.
2898:.
2888:.
2880:.
2870:87
2868:.
2864:.
2841:.
2831:.
2823:.
2813:66
2811:.
2807:.
2784:.
2774:.
2762:11
2760:.
2756:.
2733:.
2725:.
2713:39
2711:.
2707:.
2684:.
2672:.
2649:.
2639:.
2631:.
2621:81
2619:.
2615:.
2592:.
2582:.
2570:.
2566:.
2543:.
2531:73
2529:.
2525:.
2502:.
2494:.
2486:.
2476:14
2474:.
2470:.
2447:.
2435:73
2433:.
2429:.
2406:.
2396:.
2388:.
2378:72
2376:.
2372:.
2349:.
2337:87
2335:.
2331:.
2308:.
2298:.
2290:.
2280:74
2278:.
2274:.
2260:^
2246:.
2236:.
2224:14
2222:.
2218:.
2196:^
2182:.
2172:.
2160:94
2158:.
2154:.
2138:^
2109:^
2092:.
2075:^
2061:.
2051:.
2039:13
2037:.
2033:.
2010:.
2000:.
1988:.
1984:.
1970:^
1956:.
1948:.
1938:37
1936:.
1909:.
1899:.
1887:.
1883:.
1859:^
1845:.
1835:.
1825:10
1823:.
1819:.
1803:^
1789:.
1781:.
1771:.
1757:.
1753:.
1739:^
1725:.
1715:.
1701:.
1697:.
1670:.
1660:39
1658:.
1634:^
1620:.
1610:.
1598:.
1594:.
1574:^
1560:.
1546:.
1542:.
1530:^
1491:.
1465:.
1169:,
404:.
388:,
76::
3269:)
3265:(
3255:e
3248:t
3241:v
3186:.
3164::
3158:6
3150:"
3129:.
3125::
3119:1
3102:.
3082::
3059:.
3035::
3027::
3000:.
2976::
2970:9
2962:"
2945:.
2933::
2906:.
2884::
2876::
2849:.
2827::
2819::
2792:.
2768::
2741:.
2719::
2692:.
2680::
2657:.
2635::
2627::
2600:.
2578::
2551:.
2537::
2510:.
2490::
2482::
2455:.
2441::
2414:.
2392::
2384::
2357:.
2343::
2316:.
2294::
2286::
2254:.
2230::
2190:.
2166::
2152:"
2132:.
2103:.
2069:.
2045::
2018:.
1996::
1990:5
1964:.
1952::
1944::
1917:.
1895::
1889:5
1881:"
1853:.
1831::
1797:.
1765::
1733:.
1709::
1703:9
1695:"
1678:.
1666::
1628:.
1606::
1568:.
1554::
1524:.
1502:.
1477:.
1179:2
1177:E
1175:1
970:2
968:E
966:1
871:1
842:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.