123:
recombination is thought to occur by the Double
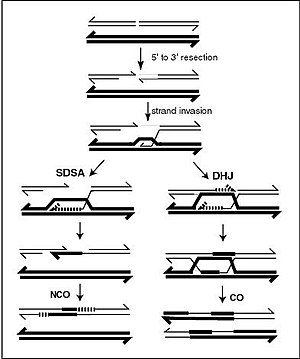
Holliday Junction (DHJ) model, illustrated on the right, above. NCO recombinants are thought to occur primarily by the Synthesis Dependent Strand Annealing (SDSA) model, illustrated on the left, above. Most recombination events appear to be the SDSA type. At various steps of the recombination process, heteroduplex DNA (double-stranded DNA consisting of single strands from each of the two homologous chromosomes) is formed.
22:
203:(see Figure). One of these 3’ single stranded DNA segments then invades a homologous sequence on the homologous chromosome, forming an intermediate which can be repaired through different pathways resulting either in crossovers (CO) or noncrossovers (NCO) as illustrated in the Figure. By one pathway, a structure called a double
119:
215:
which may or may not be perfectly complementary) is formed. During meiosis non-crossover recombinants occur frequently and these appear to arise mainly by the SDSA pathway. Non-crossover recombination events occurring during meiosis likely reflect instances of repair of DNA double-strand damages or
238:
enzyme makes staggered nicks in a pair of sister chromatid strands (in a tetrad organization of prophase). Subsequent enzymes trim back the 5' ends of the strand and a protein complex binds to the 3' single-stranded ends. Rad51 protein is recruited and binds in a protein complex to search for a
122:
A current model of meiotic recombination, initiated by a double-strand break or gap, followed by pairing with a homologous chromosome and strand invasion to initiate the recombinational repair process. Repair of the gap can lead to crossover (CO) or non-crossover (NCO) of the flanking regions. CO
276:"Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes"
211:, there is information exchange but not physical exchange. At various steps of these recombination processes, heteroduplex DNA (double-stranded DNA consisting of single strands from each of the two
179:
227:
pathway for repairing DNA double-strand breaks appears to be the SDSA pathway (rather than the DSBR pathway). The SDSA pathway produces non-crossover recombinants (see Figure).
239:
complementary sequence analogous to double-strand-break repair. The filament searches for the homologous chromosome, strand invasion occurs where the new chromosome forms a
216:
other types of DNA damages. When mismatches occur in heteroduplex DNA, the sequence of one strand can be repaired to bind the other strand with perfect complementarity.
170:
analyses. Another is the heteroduplexes formed when non-natural analogs of nucleic acids are used to bind with nucleic acids; these heteroduplexes result from performing
234:, the process of crossing-over occurs between non-sister chromatids, which results in new allelic combinations in the gametes. In crossing-over, a
208:
86:
58:
105:
65:
43:
72:
39:
54:
132:
224:
192:
32:
207:(DHJ) is formed, leading to the exchange of DNA strands. By the other pathway, referred to as
247:
that when cut in a transversal pattern by endonucleases form 2 heteroduplex strand products.
243:
over the bottom sister chromatid, then the ends are annealed. This process can yield double
212:
140:
325:"Differential timing and control of noncrossover and crossover recombination during meiosis"
175:
79:
8:
354:
300:
275:
244:
341:
324:
199:. The 5’ ends of the break are degraded, leaving long 3’ overhangs of several hundred
346:
305:
204:
159:
358:
336:
295:
287:
372:
378:
350:
309:
291:
167:
163:
136:
118:
148:
200:
183:
251:
171:
152:
21:
231:
220:
240:
186:
235:
158:
One such example is the heteroduplex DNA strand formed in
196:
254:(smRNAs), causing post-transcriptional gene silencing.
195:can be initiated by a double-strand break (DSB) in
46:. Unsourced material may be challenged and removed.
370:
273:
269:
267:
322:
143:of single complementary strands derived from
264:
316:
209:Synthesis-dependent strand annealing (SDSA)
147:sources, such as from different homologous
340:
299:
274:Andersen SL, Sekelsky J (December 2010).
106:Learn how and when to remove this message
117:
371:
250:Heteroduplex DNA is also a source of
44:adding citations to reliable sources
15:
13:
14:
390:
323:Allers T, Lichten M (July 2001).
174:techniques using single-stranded
20:
31:needs additional citations for
1:
342:10.1016/s0092-8674(01)00416-0
257:
180:2'-O-methyl phosphorothioate
7:
10:
395:
225:homologous recombination
162:processes, usually for
151:or even from different
139:originated through the
292:10.1002/bies.201000087
213:homologous chromosomes
131:is a double-stranded (
124:
193:Meiotic recombination
141:genetic recombination
121:
176:peptide nucleic acid
40:improve this article
245:Holliday junctions
189:to bind with RNA.
125:
286:(12): 1058–1066.
205:Holliday junction
116:
115:
108:
90:
386:
363:
362:
344:
320:
314:
313:
303:
271:
111:
104:
100:
97:
91:
89:
48:
24:
16:
394:
393:
389:
388:
387:
385:
384:
383:
369:
368:
367:
366:
321:
317:
272:
265:
260:
112:
101:
95:
92:
49:
47:
37:
25:
12:
11:
5:
392:
382:
381:
365:
364:
315:
262:
261:
259:
256:
135:) molecule of
114:
113:
55:"Heteroduplex"
28:
26:
19:
9:
6:
4:
3:
2:
391:
380:
377:
376:
374:
360:
356:
352:
348:
343:
338:
334:
330:
326:
319:
311:
307:
302:
297:
293:
289:
285:
281:
277:
270:
268:
263:
255:
253:
248:
246:
242:
237:
233:
228:
226:
222:
217:
214:
210:
206:
202:
198:
194:
190:
188:
185:
181:
177:
173:
169:
165:
161:
160:hybridization
156:
154:
150:
146:
142:
138:
134:
130:
120:
110:
107:
99:
88:
85:
81:
78:
74:
71:
67:
64:
60:
57: –
56:
52:
51:Find sources:
45:
41:
35:
34:
29:This article
27:
23:
18:
17:
335:(1): 47–57.
332:
328:
318:
283:
279:
249:
229:
223:, the major
218:
191:
168:phylogenetic
164:biochemistry
157:
144:
137:nucleic acid
129:heteroduplex
128:
126:
102:
93:
83:
76:
69:
62:
50:
38:Please help
33:verification
30:
201:nucleotides
149:chromosomes
258:References
252:small RNAs
184:Morpholino
96:April 2023
66:newspapers
280:BioEssays
172:antisense
153:organisms
145:different
373:Category
351:11461701
310:20967781
359:1878863
301:3090628
232:meiosis
221:mitosis
219:During
166:-based
80:scholar
357:
349:
308:
298:
241:D-loop
187:oligos
133:duplex
82:
75:
68:
61:
53:
355:S2CID
236:Spo11
87:JSTOR
73:books
347:PMID
329:Cell
306:PMID
59:news
379:DNA
337:doi
333:106
296:PMC
288:doi
230:In
197:DNA
182:or
42:by
375::
353:.
345:.
331:.
327:.
304:.
294:.
284:32
282:.
278:.
266:^
178:,
155:.
127:A
361:.
339::
312:.
290::
109:)
103:(
98:)
94:(
84:·
77:·
70:·
63:·
36:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.