991:; this allows the small-molecule ligands to stack on the planar terminal tetrads within the G-quadruplex regions. A disadvantage of using small-molecule ligands as a therapeutic technique is that specificity is difficult to manage due to the variability of G-quadruplexes in their primary sequences, orientation, thermodynamic stability, and nucleic acid strand stoichiometry. As of now, no single small-molecule ligand has been able to be perfectly specific for a single G-quadruplex sequence. However, a cationic porphyrin known as TMPyP4 is able to bind to the C9orf72 GGGGCC repeat region, which causes the G-quadruplex repeat region to unfold and lose its interactions with proteins causing it to lose its functionality. Small-molecule ligands, composed primarily of lead, can target GGGGCC repeat regions as well and ultimately decreased both repeat-associated non-ATG translation and RNA foci in neuron cells derived from patients with
413:, has also been identified as a key G-quadruplex resolvase. In 2009, a metastasis suppressor protein NM23H2 (also known as NME2) was found to directly interact with G-quadruplex in the promoter of the c-myc gene, and transcriptionally regulate c-myc. More recently, NM23H2 was reported to interact with G-quadruplex in the promoter of the human telomerase (hTERT) gene and regulate hTERT expression In 2019, the telomere-binding-factor-2 (TRF2 or TERF2) was shown to bind to thousands of non-telomeric G-quadruplexes in the human genome by TRF2 ChIP-seq. There are many studies that implicate quadruplexes in both positive and negative transcriptional regulation, including epigenetic regulation of genes like hTERT. Function of G-quadruplexes have also been reported in allowing programmed recombination of immunologlobin heavy genes and the pilin
292:. Analysis of human, chimpanzee, mouse and rat genomes showed enormous number of potential G-quadruplex (pG4)-forming sequences in non-telomeric regions. A large number of the non-telomeric G-quadruplexes were found within gene promoters, and were conserved across the species. Similarly, large number of G-quadruplexes were found in the E. coli and hundreds of other microbial genomes. Here also, like vertebrates G-quadruplexes were enriched within gene promoters. In addition, there was found more than one-billion-year conserved G-quadruplex locus in plants and algae, in gene encoding large subunit of RNA polymerase II. Although these studies predicted G-quadruplex-mediated gene regulation, it is unlikely that all pG4s would form in vivo. The
162:. However, as currently used in molecular biology, the term G4 can mean G-quadruplexes of any molecularity. Longer sequences, which contain two contiguous runs of three or more guanine bases, where the guanine regions are separated by one or more bases, only require two such sequences to provide enough guanine bases to form a quadruplex. These structures, formed from two separate G-rich strands, are termed bimolecular quadruplexes. Finally, sequences which contain four distinct runs of guanine bases can form stable quadruplex structures by themselves, and a quadruplex formed entirely from a single strand is called an intramolecular quadruplex.
130:, these structures hold a biologically relevant role through interactions with the promoter regions of oncogenes and the telomeric regions of DNA strands. Current research consists of identifying the biological function of these G-Quadruplex structures for specific oncogenes and discovering effective therapeutic treatments for cancer based on interactions with G-quadruplexes. Early evidence for the formation of G-quadruplexes in vivo in cells was established by isolating them from cells, and later by the observation that specific DNA helicases could be identified where small molecules specific for these DNA structures accumulated in cells.
798:
these signals at 262 and 295 nm, respectively. To verify G-quadruplex formation, one should also perform the CD experiments under non-G-quadruplex stabilizing (Li+) and G-quadruplex stabilizing conditions (such as K+ or with G-quadruplex ligands), and scan toward the far-UV region (180–230 nm). Likewise, the thermostability of the G-quadruplex structure can be identified by observing the UV signal at 295 nm. Upon G-quadruplex melting, the UV absorbance at 295 nm decreases, leading to a hypochromic shift that is a distinctive feature of G-quadruplex structure. Another approach for detection of G-quadruplexes includes
547:
behind the loading of these TFs, where APE1 dissociates from the G-quadruplex structures. When a study downregulated the presence of APE1 and AcAPE1 in the cell, the formation of G-quadruplex structures was inhibited, which proves the importance of APE1 for the formation of these structures. However, not all G-quadruplex structures require APE1 for formation, in fact some of them formed greater G-quadruplex structures in its absence. Therefore, we can conclude that APE1 has two important roles in genome regulation- Stabilizing the formation of g-quadruplex structures and loading the transcriptional factors onto the AP site
453:(BER). Base excision repair processes in cells have been proved to be reduced with aging as its components in the mitochondria begin to decline, which can lead to the formation of many diseases such as Alzheimer's disease (AD). These G-quadruplex structures are said to be formed in the promoter regions of DNA through superhelicity, which favors the unwinding of the double helical structure of DNA and in turn loops the strands to form G-quadruplex structures in guanine rich regions. The BER pathway is signalled when it indicates an oxidative DNA base damage, where structures like,
166:
intramolecular quadruplexes, this means that any loop regions present must be of the propeller type, positioned to the sides of the quadruplex. If one or more of the runs of guanine bases has a 5’-3’ direction opposite to the other runs of guanine bases, the quadruplex is said to have adopted an antiparallel topology. The loops joining runs of guanine bases in intramolecular antiparallel quadruplexes are either diagonal, joining two diagonally opposite runs of guanine bases, or lateral (edgewise) type loops, joining two adjacent runs of guanine base pairs.
423:. The roles of quadruplex structure in translation control are not as well explored. The direct visualization of G-quadruplex structures in human cells as well as the co-crystal structure of an RNA helicase bound to a G-quadruplex have provided important confirmations of their relevance to cell biology. The potential positive and negative roles of quadruplexes in telomere replication and function remains controversial. T-loops and G-quadruplexes are described as the two tertiary DNA structures that protect telomere ends and regulate telomere length.
763:. Although the rule effectively identifies sites of G-quadruplex formation, it also identifies a subset of the imperfect homopurine mirror repeats capable of triplex formation and C-strand i-motif formation. Moreover, these sequences also have the capacity to form slipped and foldback structures that are implicit intermediates in the formation of both quadruplex and triplex DNA structures. In one study, it was found that the observed number per base pair (i.e. the frequency) of these motifs has increased rapidly in the
542:
G-quadruplex structures in that region. This promotes formation of G-quadruplex structures by the folding of the stand. This looping process brings four bases in close proximity that will be held together by
Hoogsteen base pairing. After this stage the APE1 gets acetylated by multiple lysine residues on the chromatin, forming acetylated APE1 (AcAPE1). AcAPE1 is very crucial to the BER pathway as it acts as a transcriptional coactivator or corepressor, functioning to load
20:
654:
regions to inhibit the production of both the c-Myc protein product and the human telomerase reverse transcriptase (hTERT). This main pathway of targeting this region results in the lack of telomerase elongation, leading to arrested cell development. Further research remains necessary for the discovery of a single gene target to minimize unwanted reactivity with more efficient antitumor activity.
38:. They are helical in shape and contain guanine tetrads that can form from one, two or four strands. The unimolecular forms often occur naturally near the ends of the chromosomes, better known as the telomeric regions, and in transcriptional regulatory regions of multiple genes, both in microbes and across vertebrates including oncogenes in humans. Four guanine bases can associate through
614:
through the abundant guanine sequence in the promoter region of this specific pathway. The cyclin-dependent cell cycle checkpoint kinase inhibitor-1 CDKN1A (also known as p21) gene harbours promoter G-quadruplex. Interaction of this G-quadruplex with TRF2 (also known as TERF2) resulted in epigenetic regulation of p21, which was tested using the G-quadruplex-binding ligand 360A.
621:. Through recent research into this specific gene pathway, the polypurine and polypyrimidine region allows for the transcription of this specific gene and the formation of an intramolecular G-quadruplex structure. However, more research is necessary to determine whether the formation of G-quadruplex regulates the expression of this gene in a positive or negative manner.
89:. G-quadruplex structures can be computationally predicted from DNA or RNA sequence motifs, but their actual structures can be quite varied within and between the motifs, which can number over 100,000 per genome. Their activities in basic genetic processes are an active area of research in telomere, gene regulation, and functional genomics research.
637:
the promoter region of PDGF-A has exhibited the ability to form intramolecular parallel G-quadruplex structures and remains suggested to play a role in transcriptional regulation of PDGF-A. However, research has also identified the presence of G-quadruplex structures within this region due to the interaction of TMPyP4 with this promoter sequence.
568:. However, in cancer cells that have mutated helicase these complexes cannot be unwound and leads to potential damage of the cell. This causes replication of damaged and cancerous cells. For therapeutic advances, stabilizing the G-quadruplexes of cancerous cells can inhibit cell growth and replication leading to the
714:. Furthermore, the hydrogen bonds of ligands with smaller side chains are shorter and therefore stronger. Ligands with mobile side chains, ones that are able to rotate around its center chromophore, associate more strongly to G-quadruplexes because conformation of the G4 loops and the ligand side chains can align.
653:
Ligand design and development remains an important field of research into therapeutic reagents due to the abundance of G-quadruplexes and their multiple conformational differences. One type of ligand involving a
Quindoline derivative, SYUIQ-05, utilizes the stabilization of G-quadruplexes in promoter
636:
Another oncogene pathway involving PDGF-A, platelet-derived growth factor, involves the process of wound healing and function as mitogenic growth factors for cells. High levels of expression of PDGF have been associated with increased cell growth and cancer. The presence of a guanine-rich sequence in
528:
AP endonuclease 1 (APE1) is an enzyme responsible for the promotion and the formation of G-quadruplex structures. APE1 is mainly in charge of repairing damage caused to AP sites through the BER pathway. APE1 is considered to be very crucial as AP site damage is known to be the most recurring type of
541:
does so by cleaving the oxidized base and thus creating an AP site, primarily through the process of negative superhelicity. This AP site then signals cells to engage APE1 binding, which binds to the open duplex region. The binding of APE1 then plays an important role by stabilizing the formation of
998:
Metal complexes have a number of features that make them particularly suitable as G4 DNA binders and therefore as potential drugs. While the metal plays largely a structural role in most G4 binders, there are also examples where it interacts directly with G4s by electrostatic interactions or direct
546:
into the site of damage allowing it to regulate the gene expression. AcAPE1 is also very important since it allows APE1 to bind for longer periods of time by delay of its dissociation from the sequence, allowing the repair process to be more efficient. Deacetylation of AcAPE1 is the driving force
448:
and ARP, have indicated that AP site damage occurrence is nonrandom. AP site damage was also more prevalent in certain regions of the genome that contain specific active promoter and enhancer markers, some of which were linked to regions responsible for lung adenocarcinoma and colon cancer. AP site
5572:
Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C, Knopman DS, Kretzschmar HA, Neumann M, Caselli RJ, White CL, Mackenzie IR, Petersen RC, Strong MJ, Miller
797:
The topology of the G-quadruplex structure can be determined by monitoring the positive or negative circular dichroism (CD) signals at specific wavelengths. Parallel G-quadruplexes have negative and positive CD signals at 240 and 262 nm, respectively, whereas antiparallel G-quadruplexes place
613:
of the VEGF gene. Through recent research on the role of G-quadruplex function in vivo, the stabilization of G-quadruplex structures was shown to regulate VEGF gene transcription, with inhibition of transcription factors in this pathway. The intramolecular G-quadruplex structures are formed mostly
952:
are common strategies used to target neurological diseases linked to G-quadruplex expansion repeats. Therefore, these techniques are especially advantageous for targeting neurological diseases that have a gain-of-function mechanism, which is when the altered gene product has a new function or new
788:
Biochemical techniques were employed to interrogate G-quadruplex formation in a longer sequence context. In the DNA polymerase stop assay, the formation of a G-quadruplex in a DNA template can act as a roadblock and cause polymerase stalling, which halts the primer extension. The dimethyl sulfate
649:
which makes them effective anticancer agents. However, TMPyP4 has been limited for used due to its non-selectivity toward cancer cell telomeres and normal double stranded DNA (dsDNA). To address this issue analog of TMPyP4, it was synthesized known as 5Me which targets only G quadruplex DNA which
632:
which has been abundant in certain types of cancer. The guanine rich sequence in the promoter region for this pathway exudes a necessity for baseline transcription of this receptor tyrosine kinase. In certain types of cancers, the RET protein has shown increased expression levels. The research on
592:
and VEGF, showing the importance of this secondary structure in cancer growth and development. While the formation of G-quadruplex structure vary to some extent for the different promoter regions of oncogenes, the consistent stabilization of these structures have been found in cancer development.
125:
function of G-quadruplexes surged after large scale genome-wide analysis showed the prevalence of potential G-quadruplex (pG4)-forming sequences within gene promoters of human, chimpanzee, mouse, and rat - presented in the First
International G-quadruplex Meeting held in April 2007 in Louisville,
963:
is the process by which synthesized strands of nucleic acids are used to bind directly and specifically to the mRNA produced by a certain gene, which will inactivate it. Antisense oligonucleotides (ASOs) are commonly used to target C9orf72 RNA of the G-quadruplex GGGGCC expansion repeat region,
507:
Furthermore, the concentration of 8-oxo-dG is a known biomarker of oxidative stress within a cell, and excessive amount of oxidative stress has been linked to carcinogenesis and other diseases. When produced, 8-oxo-dG, has the ability to inactivate OGG1, thus preventing the repair of DNA damage
6167:
Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, Qiu J, Sun Y, Ling SC, Zhu Q, Polymenidou M, Drenner K, Artates JW, McAlonis-Downes M, Markmiller S, Hutt KR, Pizzo DP, Cady J, Harms MB, Baloh RH, Vandenberg SR, Yeo GW, Fu XD, Bennett CF, Cleveland DW,
608:
Another gene pathway deals with the VEGF gene, Vascular
Endothelial Growth Factor, which remains involved in the process of angiogenesis or the formation of new blood vessels. The formation of an intramolecular G-quadruplex structure has been shown through studies on the polypurine tract of the
624:
The c-kit oncogene deals with a pathway that encodes an RTK, which was shown to have elevated expression levels in certain types of cancer. The rich guanine sequence of this promoter region has shown the ability to form a variety of quadruplexes. Current research on this pathway is focusing on
604:
Regulation of c-myc through Human telomerase reverse transcriptase (hTERT) is also directly regulated through promoter G-quadruplex by interaction with the transcription factor NM23H2 where epigenetic modifications were dependent on NM23H2-G-quadruplex association. Recently, hTERT epigenetic
165:
Depending on how the individual runs of guanine bases are arranged in a bimolecular or intramolecular quadruplex, a quadruplex can adopt one of a number of topologies with varying loop configurations. If all strands of DNA proceed in the same direction, the quadruplex is termed parallel. For
226:
adds TTAGGG repeats at the 3’ end of DNA strands. At these 3’ end protrusions, the G-rich overhang can form secondary structures such as G-quadruplexes if the overhang is longer than four TTAGGG repeats. The presence of these structures prevent telomere elongation by the telomerase complex.
971:
that mimic the G-quadruplex structure and compete with the endogenous G-quadruplexes for binding to transcription factors. These decoys are typically composed of a G-rich sequence that can form a stable G-quadruplex structure and a short linker region that can be modified to optimize their
964:
which has lowered the toxicity in cellular models of C9orf72. ASOs have previously been used to restore normal phenotypes in other neurological diseases that have gain-of-function mechanisms, the only difference is that it was used in the absence of G-quadruplex expansion repeat regions.
494:
to bind and remove the oxidative damage with the help of APE1, resulting in an AP site. Moreover, an AP site is a location in DNA that has neither a purine or a pyrimidine base due to DNA damage, they are the most prevalent type of endogenous DNA damage in cells. AP sites can be generated
157:
The length of the nucleic acid sequences involved in tetrad formation determines how the quadruplex folds. Short sequences, consisting of only a single contiguous run of three or more guanine bases, require four individual strands to form a quadruplex. Such a quadruplex is described as
435:
A new technique to map AP sites has been developed known as AP-seq which utilizes a biotin-labeled aldehyde-reactive probe (ARP) to tag certain regions of the genome where AP site damage occurrence has been significant. Another genome-wide mapping sequencing method known as
822:
G-quadruplexes have been implicated in neurological disorders through two main mechanisms. The first is through expansions of G-repeats within genes that lead to the formation of G-quadruplex structures that directly cause disease, as is the case with the C9orf72 gene and
596:
One particular gene region, the c-myc pathway, plays an integral role in the regulation of a protein product, c-Myc. With this product, the c-Myc protein functions in the processes of apoptosis and cell growth or development and as a transcriptional control on human
709:
When designing ligands to be bound to G-quadruplexes, the ligands have a higher affinity for parallel folded G-quadruplexes. It has been found that ligands with smaller side chains bind better to the quadruplex because smaller ligands have more concentrated
137:
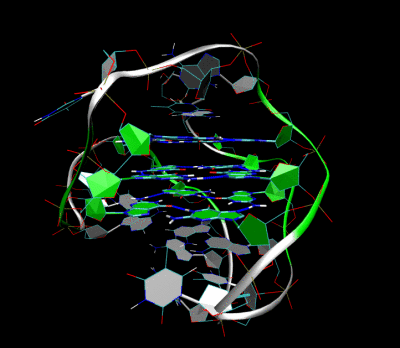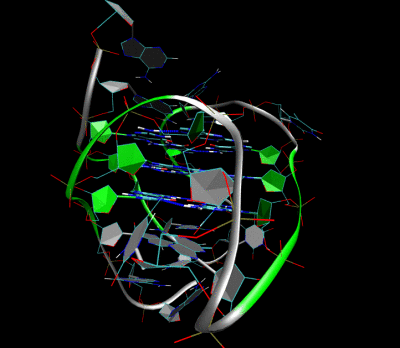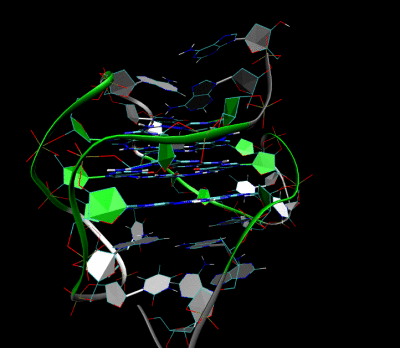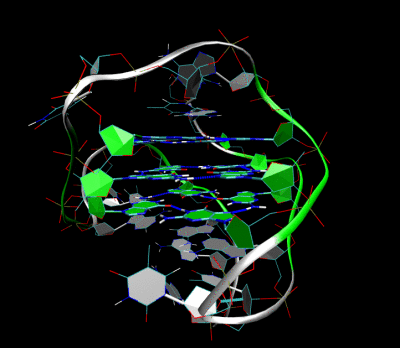
3D Structure of the intramolecular human telomeric G-quadruplex in potassium solution. The backbone is represented by a tube. The center of this structure contains three layers of G-tetrads. The hydrogen bonds in these layers are represented by blue dashed lines.
789:(DMS) followed by the piperidine cleavage assay is based on the fact that the formation of a G-quadruplex will prohibit the N7 guanine methylation caused by DMS, leading to a protection pattern observed at the DNA G-quadruplex region after piperidine cleavage.
771:
More recently, advanced web-based toolboxes for identifying G-quadruplex forming sequences were developed, including user-friendly and open access version of G4Hunter based on sliding window approach or G4RNA Screener based on machine learning algorithm.
217:
function to provide this signaling. Telomeres, rich in guanine and with a propensity to form g-quadruplexes, are located at the terminal ends of chromosomes and help maintain genome integrity by protecting these vulnerable terminal ends from instability.
529:
endogenous damage to DNA. The oxidation of certain purine bases, like guanine, forms oxidized nucleotides that impairs DNA function by mismatching nucleotides in the sequences. This is more common in PQS sequences which form oxidized structures, such as
6117:
Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD (October 2013).
722:
Identifying and predicting sequences which have the capacity to form quadruplexes is an important tool in further understanding their role. Generally, a simple pattern match is used for searching for possible intrastrand quadruplex forming sequences:
4620:
Chilakamarthi U, Koteshwar D, Jinka S, Vamsi
Krishna N, Sridharan K, Nagesh N, Giribabu L (November 2018). "Novel Amphiphilic G-Quadruplex Binding Synthetic Derivative of TMPyP4 and Its Effect on Cancer Cell Proliferation and Apoptosis Induction".
6491:
Cogoi S, Paramasivam M, Filichev V, Géci I, Pedersen EB, Xodo LE (January 2009). "Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells".
1372:
Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S (2008). "Genome-wide
Computational and Expression Analyses Reveal G-quadruplex DNA Motifs as Conserved Cis-Regulatory Elements in Human and Related Species".
7067:
Hou X, Guo W, Xia F, Nie FQ, Dong H, Tian Y, Wen L, Wang L, Cao L, Yang Y, Xue J, Song Y, Wang Y, Liu D, Jiang L (June 2009). "A biomimetic potassium responsive nanochannel: G-quadruplex DNA conformational switching in a synthetic nanopore".
221:
These telomeric regions are characterized by long regions of double-stranded CCCTAA:TTAGGG repeats. The repeats end with a 3’ protrusion of between 10 and 50 single-stranded TTAGGG repeats. The heterodimeric complex ribonucleoprotein enzyme
694:
holding it together. When bound, MM41's central chromophore is situated on top of the 3’ terminal G-quartet and the side chains of the ligand associate to the loops of the quadruplex. The quartet and the chromophore are bound with a
4305:
Ou TM, Lin J, Lu YJ, Hou JQ, Tan JH, Chen SH, Li Z, Li YP, Li D, Gu LQ, Huang ZS (August 2011). "Inhibition of cell proliferation by quindoline derivative (SYUIQ-05) through its preferential interaction with c-myc promoter G-quadruplex".
508:
caused by the oxidation of guanine. The possible inactivation allows for un-repaired DNA damages to gather in non-replicating cells, like muscle, and can cause aging as well. Moreover, oxidative DNA damage like 8-oxo-dG contributes to
212:
Because repair enzymes would naturally recognize ends of linear chromosomes as damaged DNA and would process them as such to harmful effect for the cell, clear signaling and tight regulation is needed at the ends of linear chromosomes.
6227:
Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, Gendron T, Petrucelli L, Baughn M, Ravits J, Harms MB, Rigo F, Bennett CF, Otis TS, Svendsen CN, Baloh RH (October 2013).
995:(ALS). This provides evidence that small-molecule ligands are an effective and efficient process to target GGGGCC regions, and that specificity for small-molecule ligand binding is a feasible goal for the scientific community.
4428:
Hussain T, Saha D, Purohit G, Mukherjee AK, Sharma S, Sengupta S, Dhapola P, Maji B, Vedagopuram S, Horikoshi NT, Horikoshi N, Pandita RK, Bhattacharya S, Bajaj A, Riou JF, Pandita TK, Chowdhury S (September 2017).
495:
spontaneously or after the cleavage of modified bases, like 8-OH-Gua. The generation of an AP site enables the melting of the duplex DNA to unmask the PQS, adopting a G-quadruplex fold. With the use of genome-wide
1825:
Rodriguez, Raphaël; Miller, Kyle M; Forment, Josep V; Bradshaw, Charles R; Nikan, Mehran; Britton, Sébastien; Oelschlaegel, Tobias; Xhemalce, Blerta; Balasubramanian, Shankar; Jackson, Stephen P (5 February 2012).
684:
The binding of ligands to G-quadruplexes is vital for anti-cancer pursuits because G-quadruplexes are found typically at translocation hot spots. MM41, a ligand that binds selectively for a quadruplex on the
4486:
De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW (December 2005). "Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter".
689:
promoter, is shaped with a central core and 4 side chains branching sterically out. The shape of the ligand is vital because it closely matches the quadruplex which has stacked quartets and the loops of
5622:
Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, Campbell T, Uphill J, Borg A, Fratta P, Orrell RW, Malaspina A, Rowe J, Brown J, Hodges J, Sidle K, Polke JM, Houlden H, Schott JM, Fox NC,
4381:"Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents"
469:
Guanine (G) bases in G-quadruplex have the lowest redox potential causing it to be more susceptible to the formation of 8-oxoguanine (8-oxoG), an endogenous oxidized DNA base damage in the genome. Due to
126:
Kentucky. In 2006, the prevalence of G-quadruplexes within gene promoters of several bacterial genomes was reported predicting G-quadruplex-mediated gene regulation. With the abundance of G-quadruplexes
767:
for which complete genomic sequences are available. This suggests that the sequences may be under positive selection enabled by the evolution of systems capable of suppressing non-B structure formation.
482:
within a cell. When DNA undergoes oxidative damage, a possible structural change in guanine, after ionizing radiation, gives rise to an enol form, 8-OH-Gua. This oxidative product is formed through a
431:
Many of the genome regulatory processes have been linked to the formation of G-quadruplex structures, attributable to the huge role it plays in DNA repair of apurinic/apyrimidinic sites also known as
699:
while the side chains and loops are not bound but are in close proximity. What makes this binding strong is the fluidity in the position of the loops to better associate with the ligand side chains.
967:
The G-quadruplex decoy strategy is another promising approach for targeting cancer cells by exploiting the unique structural features of the G-quadruplex. The strategy involves designing synthetic
254:) consists of many repeats of the sequenced (TTAGGG), and the quadruplexes formed by this structure can be in bead-like structures of 5 nm to 8 nm in size and have been well studied by
588:
promoter regions. The structures most present in the promoter regions of these oncogenes tend to be parallel-stranded G-quadruplex DNA structures. Some of these oncogenes include c-KIT, PDGF-A,
3647:"The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression"
169:
In quadruplexes formed from double-stranded DNA, possible interstrand topologies have also been discussed . Interstrand quadruplexes contain guanines that originate from both strands of dsDNA.
564:
cells, especially in telomeres, 5` untranslated strands, and translocation hot spots. G-quadruplexes can inhibit normal cell function, and in healthy cells, are easily and readily unwound by
670:, can bind to the G-quadruplex. These ligands can be naturally occurring or synthetic. This has become an increasingly large field of research in genetics, biochemistry, and pharmacology.
512:
through the modulation of gene expression, or the induction of mutations. On the condition that 8-oxo-dG is repaired by BER, parts of the repair protein is left behind which can lead to
987:. These can be used to target G-quadruplex regions that cause neurological disorders. Approximately 1,000 various G-quadruplex ligands exist in which they are able to interact via their
4664:
Ohnmacht SA, Marchetti C, Gunaratnam M, Besser RJ, Haider SM, Di Vita G, Lowe HL, Mellinas-Gomez M, Diocou S, Robson M, Šponer J, Islam B, Pedley RB, Hartley JA, Neidle S (June 2015).
101:
association became apparent in the early 1960s, through the identification of gel-like substances associated with guanines. More specifically, this research detailed the four-stranded
7239:
134:
1725:
Henderson E, Hardin CC, Walk SK, Tinoco I, Blackburn EH (December 1987). "Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs".
7122:
Rowland GB, Barnett K, Dupont JI, Akurathi G, Le VH, Lewis EA (December 2013). "The effect of pyridyl substituents on the thermodynamics of porphyrin binding to G-quadruplex DNA".
181:, many guanine-rich sequences that had the potential to form quadruplexes were discovered. Depending on cell type and cell cycle, mediating factors such as DNA-binding proteins on
617:
Hypoxia inducible factor 1ɑ, HIF-1ɑ, remains involved in cancer signaling through its binding to
Hypoxia Response Element, HRE, in the presence of hypoxia to begin the process of
6442:
Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, Montaillier C, Lemaitre M, Corey DR (November 2010).
6529:"MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice"
912:
Fragile X mental retardation protein (FMRP) is a widely expressed protein coded by the FMR1 gene that binds to G-quadruplex secondary structures in neurons and is involved in
401:
switching. As cells have evolved mechanisms for resolving (i.e., unwinding) quadruplexes that form. Quadruplex formation may be potentially damaging for a cell; the helicases
49:
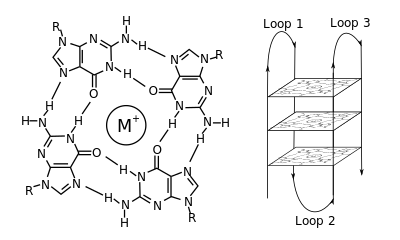
The placement and bonding to form G-quadruplexes is not random and serve very unusual functional purposes. The quadruplex structure is further stabilized by the presence of a
499:
analyses, cell-based assays, and in vitro biochemical analyses, a connection has been made between oxidized DNA base-derived AP sites, and the formation of the G-quadruplex.
197:
indicate that the antiparallel g-quadruplex is stabilized by molecular crowding. This effect seems to be mediated by alteration of the hydration of the DNA and its effect on
780:
A number of experimental methods have been developed to identify G-quadruplexes. These methods can be broadly defined into two classes: biophysical and biochemical methods.
1126:
Largy E, Mergny J, Gabelica V (2016). "Chapter 7. Role of Alkali Metal Ions in G-Quadruplex
Nucleic Acid Structure and Stability". In Astrid S, Helmut S, Roland KO (eds.).
6395:"Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin"
158:
tetramolecular, reflecting the requirement of four separate strands. The term G4 DNA was originally reserved for these tetramolecular structures that might play a role in
6770:
Ren J, Wang J, Han L, Wang E, Wang J (October 2011). "Kinetically grafting G-quadruplexes onto DNA nanostructures for structure and function encoding via a DNA machine".
6648:"TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins"
2349:
Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ (October 2005). "Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo".
706: The way in which TMPyP4 binds to G4's is similar to MM41, with the ring stacking onto the external G-quartet and side chains associating to the loops of G4's.
6033:
Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST (September 1992). "DNA methylation represses FMR-1 transcription in fragile X syndrome".
5225:
Sun D, Hurley LH (2009-10-23). "Biochemical
Techniques for the Characterization of G-Quadruplex Structures: EMSA, DMS Footprinting, and DNA Polymerase Stop Assay".
625:
discovering the biological function of this specific quadruplex formation on the c-kit pathway, while this quadruplex sequence has been noticed in various species.
4573:"Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4"
645:
Telomeres are generally made up of G-quadruplexes and remain important targets for therapeutic research and discoveries. These complexes have a high affinity for
807:
449:
damage was found to be predominant in PQS regions of the genome, where formation of G-quadruplex structures is regulated and promoted by the DNA repair process,
4952:
Mirkin SM, Lyamichev VI, Drushlyak KN, Dobrynin VN, Filippov SA, Frank-Kamenetskii MD (1987). "DNA H form requires a homopurine-homopyrimidine mirror repeat".
803:
520:
gene of humans, it was determine that if 8-oxo-dG was left unchecked and not repaired by BER, it can lead to frequent mutations and eventually carcinogenesis.
46:(G-tetrad or G-quartet), and two or more guanine tetrads (from G-tracts, continuous runs of guanine) can stack on top of each other to form a G-quadruplex.
5629:"Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population"
5276:
Paramasivan S, Rujan I, Bolton PH (December 2007). "Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding".
109:
regions of DNA in the 1980s. The importance of discovering G-quadruplex structure was described through the statement, “If G-quadruplexes form so readily
3842:"8-Oxo-7,8-dihydro-2'-deoxyguanosine and abasic site tandem lesions are oxidation prone yielding hydantoin products that strongly destabilize duplex DNA"
593:
Current therapeutic research actively focuses on targeting this stabilization of G-quadruplex structures to arrest unregulated cell growth and division.
5990:
Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL (August 1991). "Absence of expression of the FMR-1 gene in fragile X syndrome".
857:
terminals. Mutations of the C9orf72 gene have been linked to the development of FTD and ALS. These two diseases have a causal relationship to GGGGCC (G
189:
proteins, and other environmental conditions and stresses affect the dynamic formation of quadruplexes. For instance, quantitative assessments of the
5030:
Brázda, Václav; Kolomazník, Jan; Lýsek, Jiří; Bartas, Martin; Fojta, Miroslav; Šťastný, Jiří; Mergny, Jean-Louis (2019-09-15). Hancock, John (ed.).
3126:"Telomere Repeat-Binding Factor 2 Binds Extensively to Extra-Telomeric G-quadruplexes and Regulates the Epigenetic Status of Several Gene Promoters"
3946:"Human Apurinic/Apyrimidinic Endonuclease (APE1) Is Acetylated at DNA Damage Sites in Chromatin, and Acetylation Modulates Its DNA Repair Activity"
2116:
Miyoshi D, Karimata H, Sugimoto N (June 2006). "Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions".
5784:
Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J (March 2014).
5735:"The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures"
2994:"Metastases Suppressor NM23-H2 Interaction With G-quadruplex DNA Within c-MYC Promoter Nuclease Hypersensitive Element Induces c-MYC Expression"
916:. FMRP acts as a negative regulator of translation, and its binding stabilizes G-quadruplex structures in mRNA transcripts, inhibiting ribosome
3500:
806:
can detect G-quadruplexes based on size exclusion and specific interaction of G-quadruplex and protein nanocavity. The novel approach combines
662:
One way of inducing or stabilizing G-quadruplex formation is to introduce a molecule which can bind to the G-quadruplex structure. A number of
350:-wide surveys based on a quadruplex folding rule have been performed, which have identified 376,000 Putative Quadruplex Sequences (PQS) in the
1231:
Sen D, Gilbert W (July 1988). "Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis".
486:
shift from the original damage guanine, 8-oxo-Gua, and represents DNA damage that causes changes in the structure. This form allows for the
2917:"The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates"
205:. In addition, the propensity of g-quadruplex formation during transcription in RNA sequences with the potential to form mutually exclusive
3699:"Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair"
2659:"An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution"
977:
81:, or tetramolecular. Depending on the direction of the strands or parts of a strand that form the tetrads, structures may be described as
6576:
Campbell NH, Patel M, Tofa AB, Ghosh R, Parkinson GN, Neidle S (March 2009). "Selectivity in ligand recognition of G-quadruplex loops".
5419:
Bošković F, Zhu J, Chen K, Keyser UF (November 2019). "Monitoring G-Quadruplex
Formation with DNA Carriers and Solid-State Nanopores".
3075:"Epigenetic Suppression of Human Telomerase ( hTERT) Is Mediated by the Metastasis Suppressor NME2 in a G-quadruplex-dependent Fashion"
909:
within the nucleus, and separation of nucleolin by the mutated RNA transcripts impairs nucleolar function and ribosomal RNA synthesis.
893:
repeats in DNA have the ability to form mixed parallel-antiparallel G-quadruplex structures as well. These RNA transcripts containing G
7098:
1494:"QuadBase: Genome-Wide Database of G4 DNA--occurrence and Conservation in Human, Chimpanzee, Mouse and Rat Promoters and 146 Microbes"
4905:"Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene"
2858:"The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures"
928:, and other neurological disorders. Specifically, Fragile X Syndrome is caused by an increase from 50 to over 200 CGG repeats within
457:(OGG1), APE1 and G-quadruplex play a huge role in its repair. These enzymes participate in BER to repair certain DNA lesions such as
3041:
Borman S (November 2009). "Promoter Quadruplexes Folded DNA structures in gene-activation sites may be useful cancer drug targets".
1639:
Borman S (November 2009). "Promoter quadruplexes folded DNA structures in gene-activation sites may be useful cancer drug targets".
976:
and bind them leading to the regulation of gene expression. Decoys have been successfully demonstrated to inhibit oncogenic KRAS in
2018:"G-Quadruplexes Involving Both Strands of Genomic DNA Are Highly Abundant and Colocalize with Functional Sites in the Human Genome"
1017:"G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae"
924:
and controlling the timing of the transcript's expression. Mutations of this gene can cause the development of Fragile X Syndrome,
633:
this pathway suggested the formation of a G-quadruplex in the promoter region and an applicable target for therapeutic treatments.
5678:"C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes"
2243:"G-quadruplex formation at the 3' end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase"
6973:"Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription"
4723:"Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription"
2551:"Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription"
7230:
Quadfinder: Tool for Prediction and Analysis of G Quadruplex Motifs in DNA/RNA Sequences from Maiti's group, IGIB, Delhi, India
6697:
Vilar R (2018). "Chapter 12. Nucleic Acid Quadruplexes and Metallo-Drugs". In Sigel A, Sigel H, Freisinger E, Sigel RK (eds.).
6444:"Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat"
4341:
Sharma S, Mukherjee AK, Roy SS, Bagri S, Lier S, Verma M, Sengupta A, Kumar M, Nesse G, Pandey DP, Chowdhury S (January 2020).
7210:
7108:
6393:
Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR (December 2011).
3460:
1148:
885:
repeats. The transcribed RNA of these repeats have been shown to form stable G-quadruplexes, with evidence showing that the G
831:(FTD). The second mechanism is through mutations that affect the expression of G-quadruplex binding proteins, as seen in the
4431:"Transcription Regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 Is Epigenetically Controlled Through the REST Repressor Complex"
2153:"Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA"
3124:
Mukherjee AK, Sharma S, Bagri S, Kutum R, Kumar P, Hussain A, Singh P, Saha D, Kar A, Dash D, Chowdhury S (November 2019).
7195:
3995:"Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1"
389:
strand helps to maintain an open conformation of the coding DNA strand and enhance an expression of the respective gene.
6277:
Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA (August 2012).
4524:"Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene"
4199:
2292:"In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei"
1175:
Sundquist WI, Klug A (December 1989). "Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops".
333:
4342:
605:
regulation reported to be mediated through interaction of hTERT promoter G-quadruplex with the telomeric factor TRF2.
7172:– a website to discuss publications and other information of interest to those working in the field of G-quadruplexes
6714:
5242:
5573:
BL, Boeve BF, Uitti RJ, Boylan KB, Wszolek ZK, Graff-Radford NR, Dickson DW, Ross OA, Rademakers R (December 2012).
7213:- a web server to predict G-quadruplex forming motifs and other non-B DNA forming motifs from users' DNA sequences.
814:
for label-free detection of G-quadruplexes, for their mapping on dsDNA, and for monitoring G-quadruplex formation.
598:
31:
5135:
1969:"Computational detection and analysis of sequences with duplex-derived interstrand G-quadruplex forming potential"
6170:"Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration"
5511:
2709:
Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S (June 2006).
7241:
pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R.
7175:
7024:
Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S (May 2006).
4141:
2807:
Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S (May 2006).
398:
265:
determination. The formation of these quadruplexes in telomeres has been shown to decrease the activity of the
940:
modifications of FMR1 that prevent the transcription of the gene, leading to pathological low levels of FMRP.
601:. Interaction of c-Myc promoter G-quadruplex with NM23H2 was shown to regulate c-Myc in cancer cells in 2009
992:
824:
5362:"Single-molecule investigation of G-quadruplex folds of the human telomere sequence in a protein nanocavity"
3073:
Saha D, Singh A, Hussain T, Srivastava V, Sengupta S, Kar A, Dhapola P, Ummanni R, Chowdhury S (July 2017).
1289:
Rawal P, Kummarasetti VB, Ravindran R, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S (2006).
7181:
2992:
Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S (January 2009).
2915:
Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA (November 2005).
917:
7224:
QGRS Mapper: a web-based application for predicting G-quadruplexes in nucleotide sequences and NCBI genes
5933:"Translational regulation of the human achaete-scute homologue-1 by fragile X mental retardation protein"
4343:"Human Telomerase Expression is under Direct Transcriptional Control of the Telomere-binding-factor TRF2"
1345:
Borman S (May 28, 2007). "Ascent of quadruplexes nucleic acid structures become promising drug targets".
86:
4666:"A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer"
3529:"Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome"
475:
5529:
Ratnavalli E, Brayne C, Dawson K, Hodges JR (June 2002). "The prevalence of frontotemporal dementia".
3283:
Chen MC, Tippana R, Demeshkina NA, Murat P, Balasubramanian S, Myong S, Ferré-D'Amaré AR (June 2018).
2400:
single-stranded DNA self-condenses into compact beaded filaments stabilized by G-quadruplex formation"
5031:
4044:"Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene"
2711:"A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene"
478:(8-oxo-dG), is a known major product of DNA oxidation. Its concentration is used as a measurement of
6752:
6230:"Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion"
4239:
4091:
Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, et al. (January 2010).
303:
hypersensitive region critical for gene activity. Other genes shown to form G-quadruplexes in their
7026:"Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation"
3746:
Burrows CJ, Muller JG (May 1998). "Oxidative Nucleobase Modifications Leading to Strand Scission".
2809:"Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation"
1768:
Müller, Sebastian; Kumari, Sunita; Rodriguez, Raphaël; Balasubramanian, Shankar (10 October 2010).
1408:
Han H, Hurley LH (April 2000). "G-quadruplex DNA: a potential target for anti-cancer drug design".
1291:"Genome-wide Prediction of G4 DNA as Regulatory Motifs: Role in Escherichia Coli Global Regulation"
382:
3389:
Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. (October 2016).
2451:"G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story"
369:. There are several possible models for how quadruplexes could influence gene activity, either by
5088:
1127:
828:
272:, which is responsible for maintaining length of telomeres and is involved in around 85% of all
5931:
Fähling M, Mrowka R, Steege A, Kirschner KM, Benko E, Förstera B, et al. (February 2009).
5676:
Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM (December 2012).
5472:"G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome"
3527:
Roychoudhury S, Pramanik S, Harris HL, Tarpley M, Sarkar A, Spagnol G, et al. (May 2020).
538:
534:
491:
454:
406:
6070:"C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function?"
5843:"Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function"
1076:"A G-quadruplex DNA-affinity Approach for Purification of Enzymaticacvly Active G4 Resolvase1"
7259:
4781:
440:, was utilized to map both; damage in AP sites, and the enzyme responsible for its repair,
209:
or G-quadruplex structures depends heavily on the position of the hairpin-forming sequence.
7114:
6984:
6347:
6290:
6181:
5797:
5689:
5428:
5373:
4961:
4734:
4677:
4442:
3993:
Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, et al. (December 2008).
3540:
3296:
3239:
2869:
2562:
2449:
Volná A, Bartas M, Karlický V, Nezval J, Kundrátová K, Pečinka P, et al. (July 2021).
2303:
2029:
1781:
1679:
1240:
1184:
1028:
973:
702:
TMPyP4, a cationic porphyrin, is a more well known G4 binding ligand that helps to repress
543:
487:
450:
74:
70:
5972:
3944:
Roychoudhury S, Nath S, Song H, Hegde ML, Bellot LJ, Mantha AK, et al. (March 2017).
8:
5590:
3610:
913:
610:
414:
198:
105:
structures with a high association of guanines, which was later identified in eukaryotic
82:
66:
39:
6988:
6611:
Ohnmacht SA, Neidle S (June 2014). "Small-molecule quadruplex-targeted drug discovery".
6351:
6294:
6185:
6120:"RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention"
5801:
5693:
5432:
5377:
4965:
4881:
4738:
4681:
4446:
3544:
3300:
3243:
2873:
2566:
2307:
2033:
1828:"Small-molecule–induced DNA damage identifies alternative DNA structures in human genes"
1785:
1683:
1244:
1188:
1032:
7050:
7025:
6954:
6929:
6911:
6886:
6868:
6843:
6825:
6800:
6740:
6728:
6674:
6647:
6553:
6528:
6468:
6443:
6419:
6394:
6370:
6335:
6311:
6278:
6254:
6229:
6204:
6169:
6144:
6119:
6094:
6069:
6015:
5872:
5818:
5785:
5761:
5734:
5710:
5677:
5653:
5628:
5599:
5574:
5554:
5452:
5396:
5361:
5342:
5253:
5116:
5064:
4985:
4929:
4904:
4845:
4820:
4698:
4665:
4646:
4597:
4572:
4548:
4523:
4463:
4430:
4405:
4380:
4361:
4279:
4254:
4227:
4210:
4117:
4092:
4019:
3994:
3970:
3945:
3918:
3893:
3866:
3841:
3814:
3789:
3671:
3646:
3619:
3594:
3563:
3528:
3494:
3482:
3418:
3366:
3341:
3317:
3284:
3260:
3227:
3203:
3176:
3152:
3125:
3101:
3074:
3018:
2993:
2892:
2857:
2833:
2808:
2784:
2759:
2735:
2710:
2683:
2658:
2634:
2609:
2477:
2450:
2426:
2393:
2374:
2267:
2242:
2177:
2152:
2052:
2017:
1993:
1968:
1944:
1919:
1900:
1852:
1827:
1802:
1769:
1750:
1616:
1591:
1567:
1542:
1518:
1493:
1469:
1444:
1315:
1290:
1264:
1208:
1100:
1075:
1051:
1016:
836:
811:
378:
304:
194:
7007:
6972:
5859:
5842:
5575:"Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype"
5329:
5312:
5202:
5177:
4782:"Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands"
4780:
Collie GW, Promontorio R, Hampel SM, Micco M, Neidle S, Parkinson GN (February 2012).
4757:
4722:
4184:
Sequence, stability, and structure of G-quadruplexes and their interactions with drugs
4068:
4043:
3390:
2585:
2550:
2526:
2501:
1702:
1667:
1421:
238:
repeats in a variety of organisms have been shown to form these quadruplex structures
57:, which sits in a central channel between each pair of tetrads. They can be formed of
7139:
7104:
7085:
7055:
7012:
6959:
6916:
6873:
6830:
6787:
6720:
6710:
6679:
6628:
6593:
6558:
6509:
6473:
6424:
6375:
6316:
6259:
6209:
6149:
6099:
6050:
6007:
6003:
5954:
5913:
5864:
5823:
5766:
5715:
5658:
5604:
5546:
5493:
5456:
5444:
5401:
5334:
5293:
5258:
5238:
5207:
5158:
5108:
5069:
5051:
5012:
4977:
4934:
4885:
4850:
4801:
4762:
4703:
4638:
4602:
4553:
4504:
4468:
4410:
4365:
4323:
4284:
4270:
4215:
4195:
4164:
4122:
4073:
4024:
3975:
3923:
3871:
3819:
3763:
3728:
3723:
3698:
3676:
3624:
3568:
3486:
3474:
3466:
3456:
3410:
3371:
3322:
3265:
3208:
3157:
3106:
3023:
2938:
2897:
2838:
2789:
2740:
2688:
2639:
2590:
2531:
2482:
2431:
2366:
2331:
2326:
2291:
2272:
2223:
2182:
2133:
2098:
2057:
1998:
1949:
1892:
1857:
1807:
1742:
1738:
1707:
1621:
1572:
1523:
1474:
1425:
1390:
1320:
1256:
1200:
1154:
1144:
1105:
1056:
960:
677:
have been shown to bind intercalatively with G-quadruplexes, as well as the molecule
262:
140:
6732:
6019:
5558:
5120:
5047:
4650:
3422:
1904:
1754:
584:
regions of DNA, G-quadruplex structures have been identified in various human proto-
385:
of the gene, and hence de-activating it. In another model, quadruplex formed at the
7131:
7077:
7045:
7037:
7002:
6992:
6949:
6941:
6906:
6898:
6863:
6855:
6820:
6812:
6779:
6702:
6669:
6659:
6620:
6585:
6548:
6540:
6501:
6463:
6455:
6414:
6406:
6365:
6355:
6306:
6298:
6249:
6241:
6199:
6189:
6139:
6131:
6089:
6081:
6042:
5999:
5944:
5903:
5892:"Phosphorylation influences the translation state of FMRP-associated polyribosomes"
5876:
5854:
5813:
5805:
5756:
5746:
5705:
5697:
5648:
5640:
5594:
5586:
5538:
5483:
5436:
5391:
5381:
5346:
5324:
5285:
5248:
5230:
5197:
5189:
5150:
5100:
5089:"G4RNA screener web server: User focused interface for RNA G-quadruplex prediction"
5059:
5043:
4989:
4969:
4924:
4916:
4877:
4840:
4832:
4793:
4752:
4742:
4693:
4685:
4630:
4592:
4584:
4543:
4535:
4496:
4458:
4450:
4400:
4392:
4353:
4315:
4274:
4266:
4205:
4187:
4156:
4112:
4104:
4063:
4055:
4014:
4006:
3965:
3957:
3913:
3905:
3861:
3853:
3809:
3801:
3755:
3718:
3710:
3666:
3658:
3614:
3606:
3558:
3548:
3448:
3402:
3361:
3353:
3312:
3304:
3255:
3247:
3198:
3188:
3147:
3137:
3096:
3086:
3050:
3013:
3005:
2969:
2928:
2887:
2877:
2828:
2820:
2779:
2771:
2730:
2722:
2678:
2670:
2657:
Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D (February 2006).
2629:
2621:
2580:
2570:
2521:
2513:
2472:
2462:
2421:
2411:
2378:
2358:
2321:
2311:
2262:
2254:
2213:
2172:
2164:
2125:
2088:
2047:
2037:
1988:
1980:
1939:
1931:
1884:
1847:
1839:
1797:
1789:
1734:
1697:
1687:
1648:
1611:
1603:
1562:
1554:
1543:"QuadBase2: Web Server for Multiplexed Guanine Quadruplex Mining and Visualization"
1513:
1505:
1464:
1456:
1417:
1382:
1354:
1310:
1302:
1268:
1248:
1212:
1192:
1136:
1095:
1087:
1046:
1036:
854:
711:
517:
516:
alterations, or the modulation of gene expression. Upon insertion of 8-oxo-dG into
479:
23:
Structure of a G-quadruplex. Left: a G-tetrad. Right: an intramolecular G4 complex.
6801:"In vivo veritas: using yeast to probe the biological functions of G-quadruplexes"
5488:
5471:
5154:
2200:
Endoh T, Rode AB, Takahashi S, Kataoka Y, Kuwahara M, Sugimoto N (February 2016).
1074:
Routh ED, Creacy SD, Beerbower PE, Akman SA, Vaughn JP, Smaldino PJ (March 2017).
7199:
6816:
6245:
6135:
6085:
5440:
5104:
5003:
Smith SS (2010). "Evolutionary expansion of structurally complex DNA sequences".
4191:
4160:
3595:"Base excision DNA repair levels in mitochondrial lysates of Alzheimer's disease"
3193:
2882:
2290:
Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Plückthun A (July 2001).
2218:
2201:
2042:
1041:
968:
937:
933:
496:
445:
441:
437:
426:
5289:
5234:
4634:
3452:
1984:
1875:
Simonsson T (April 2001). "G-quadruplex DNA structures--variations on a theme".
1140:
7234:
6977:
Proceedings of the National Academy of Sciences of the United States of America
6624:
6340:
Proceedings of the National Academy of Sciences of the United States of America
6174:
Proceedings of the National Academy of Sciences of the United States of America
5644:
5512:"C9orf72 chromosome 9 open reading frame 72 [Homo sapiens] - Gene]"
5366:
Proceedings of the National Academy of Sciences of the United States of America
4727:
Proceedings of the National Academy of Sciences of the United States of America
4454:
3892:
Kitsera N, Rodriguez-Alvarez M, Emmert S, Carell T, Khobta A (September 2019).
3805:
3533:
Proceedings of the National Academy of Sciences of the United States of America
3447:. Methods in Molecular Biology. Vol. 2175. Clifton, N.J. pp. 95–108.
3441:"AP-Seq: A Method to Measure Apurinic Sites and Small Base Adducts Genome-Wide"
3357:
2555:
Proceedings of the National Academy of Sciences of the United States of America
2296:
Proceedings of the National Academy of Sciences of the United States of America
1672:
Proceedings of the National Academy of Sciences of the United States of America
1358:
972:
properties. When introduced to cancer cells the decoy can intercept associated
509:
409:
have a high affinity for resolving DNA G-quadruplexes. The DEAH/RHA helicase,
386:
374:
311:
190:
43:
7192:
7135:
6706:
5841:
Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB (November 2001).
5627:, Tabrizi SJ, Isaacs AM, Hardy J, Warren JD, Collinge J, Mead S (March 2013).
4357:
3308:
3054:
2093:
2076:
1652:
901:
repeats were shown to bind and separate a wide variety of proteins, including
464:
7253:
5786:"C9orf72 nucleotide repeat structures initiate molecular cascades of disease"
5624:
5229:. Methods in Molecular Biology. Vol. 608. Humana Press. pp. 65–79.
5055:
4522:
Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH (August 2007).
3470:
3142:
2416:
1445:"DNA secondary structures: stability and function of G-quadruplex structures"
474:
having a lower electron reduction potential than the other nucleotides bases,
341:
337:
322:
293:
6664:
6360:
6194:
5890:
Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST (December 2003).
5751:
5386:
3553:
3440:
3091:
2517:
1692:
877:
repeats, but individuals with FTD or ALS have from 500 to several thousand G
7269:
7264:
7186:
7143:
7089:
7059:
7016:
6997:
6963:
6920:
6877:
6834:
6791:
6724:
6683:
6632:
6597:
6562:
6513:
6477:
6428:
6379:
6320:
6263:
6213:
6153:
6103:
5958:
5949:
5932:
5917:
5868:
5827:
5770:
5719:
5662:
5608:
5550:
5542:
5497:
5448:
5405:
5297:
5262:
5193:
5162:
5112:
5073:
5016:
4938:
4854:
4805:
4766:
4747:
4707:
4642:
4606:
4557:
4508:
4472:
4414:
4327:
4288:
4219:
4168:
4126:
4077:
4059:
4028:
3979:
3927:
3875:
3823:
3767:
3732:
3714:
3680:
3628:
3572:
3478:
3414:
3375:
3326:
3285:"Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36"
3269:
3212:
3161:
3110:
3027:
2942:
2933:
2916:
2901:
2842:
2793:
2744:
2692:
2643:
2594:
2575:
2486:
2435:
2370:
2335:
2316:
2276:
2227:
2186:
2137:
2102:
2061:
2002:
1953:
1896:
1861:
1811:
1711:
1625:
1576:
1527:
1478:
1429:
1394:
1324:
1158:
1109:
1060:
691:
678:
618:
530:
458:
402:
370:
277:
6054:
6046:
6011:
5338:
5211:
5136:"G-Quadruplexes: Prediction, Characterization, and Biological Application"
4981:
4889:
4255:"Making sense of G-quadruplex and i-motif functions in oncogene promoters"
4108:
3228:"Quantitative visualization of DNA G-quadruplex structures in human cells"
2625:
2535:
1746:
1260:
1204:
7223:
7205:
6945:
6902:
6859:
6544:
6527:
Cogoi S, Zorzet S, Rapozzi V, Géci I, Pedersen EB, Xodo LE (April 2013).
5908:
5891:
4920:
4836:
4588:
4396:
4010:
3961:
3909:
3790:"The genomics of oxidative DNA damage, repair, and resulting mutagenesis"
3009:
2974:
2957:
2775:
2467:
2258:
2168:
1935:
1843:
1607:
1558:
1509:
1135:. Metal Ions in Life Sciences. Vol. 16. Springer. pp. 203–258.
988:
362:
78:
6410:
6336:"RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1"
6302:
5809:
5087:
Garant, Jean-Michel; Perreault, Jean-Pierre; Scott, Michelle S. (2018).
4619:
2241:
Wang Q, Liu JQ, Chen Z, Zheng KW, Chen CY, Hao YH, Tan Z (August 2011).
1888:
7041:
6783:
5571:
3857:
3251:
3225:
2824:
1306:
752:
569:
561:
513:
269:
251:
223:
202:
118:
7081:
6589:
6505:
6459:
5733:
Reddy K, Zamiri B, Stanley SY, Macgregor RB, Pearson CE (April 2013).
5701:
4797:
4689:
4539:
4500:
4319:
3759:
3662:
3068:
3066:
3064:
2987:
2985:
2726:
2674:
2129:
1793:
1386:
144:
7246:
pqsfinder: online search tool using the latest R/Bioconductor package
4973:
4093:"SIRT1 deacetylates APE1 and regulates cellular base excision repair"
2202:"Real-Time Monitoring of G-Quadruplex Formation during Transcription"
1340:
1338:
1336:
1334:
1284:
1282:
1280:
1278:
1252:
1196:
983:
Another commonly used technique is the utilization of small-molecule
980:
leading to reduced tumour growth and increased median survival time.
902:
850:
764:
760:
674:
646:
533:. Once the cell is aware of oxidative stress and damage, it recruits
419:
377:. One model is shown below, with G-quadruplex formation in or near a
318:
214:
182:
54:
7169:
6279:"Targeting nuclear RNA for in vivo correction of myotonic dystrophy"
5178:"A DNA polymerase stop assay for G-quadruplex-interactive compounds"
5032:"G4Hunter web application: a web server for G-quadruplex prediction"
3891:
3406:
3226:
Biffi G, Tannahill D, McCafferty J, Balasubramanian S (March 2013).
2362:
1767:
1460:
133:
7229:
6887:"Highly prevalent putative quadruplex sequence motifs in human DNA"
5470:
Simone R, Fratta P, Neidle S, Parkinson GN, Isaacs AM (June 2015).
4821:"Highly prevalent putative quadruplex sequence motifs in human DNA"
3388:
3061:
2982:
921:
906:
799:
585:
581:
565:
483:
300:
289:
240:
235:
201:
bonding. These quadruplexes seemed to readily occur at the ends of
106:
6971:
Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH (September 2002).
4868:
Frank-Kamenetskii MD, Mirkin SM (1995). "Triplex DNA structures".
4721:
Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH (September 2002).
2549:
Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH (September 2002).
1331:
1275:
1091:
869:
of C9orf72 gene. Normal individuals typically have around 2 to 8 G
4663:
3526:
1770:"Small-molecule-mediated G-quadruplex isolation from human cells"
1492:
Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S (2008).
984:
954:
949:
843:
756:
696:
667:
537:
to the site, whose main function is to initiate the BER pathway.
471:
432:
308:
246:
206:
186:
159:
98:
35:
7236:
G4Hunter from Mergny's group but user need to run the code in R.
4951:
4042:
Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S (December 2003).
397:
It has been suggested that quadruplex formation plays a role in
7244:
7023:
6441:
6226:
2806:
2348:
1288:
925:
866:
847:
663:
629:
461:(8-oxoG), which forms under oxidative stress to guanine bases.
361:. A similar studies have identified putative G-quadruplexes in
354:
347:
273:
266:
178:
50:
34:(G4) are formed in nucleic acids by sequences that are rich in
19:
7163:
6970:
4720:
2548:
2289:
2077:"Existence and consequences of G-quadruplex structures in DNA"
1824:
444:(APE1). Both of these genome-wide mapping sequencing methods,
427:
Genome Regulation through formation of G-quadruplex structures
6646:
Zamiri B, Reddy K, Macgregor RB, Pearson CE (February 2014).
4521:
686:
410:
351:
329:
325:
296:
259:
7121:
7096:
6490:
6166:
6032:
5930:
5889:
5528:
4779:
4571:
Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH (2007-11-26).
3992:
2914:
2392:
Kar A, Jones N, Arat NÖ, Fishel R, Griffith JD (June 2018).
42:
hydrogen bonding to form a square planar structure called a
7178:– a database listing potential G-quadruplex regulated genes
6645:
5732:
5675:
5469:
5360:
An N, Fleming AM, Middleton EG, Burrows CJ (October 2014).
5029:
4427:
4090:
3592:
2708:
1491:
929:
905:. Nucleolin is involved in the synthesis and maturation of
832:
465:
Role of Endogenous Oxidized DNA Base Damage on G4 formation
314:
6928:
Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006).
6799:
Johnson JE, Smith JS, Kozak ML, Johnson FB (August 2008).
6699:
Metallo-Drugs: Development and Action of Anticancer Agents
5783:
3593:
Canugovi C, Shamanna RA, Croteau DL, Bohr VA (June 2014).
3282:
3072:
1918:
Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006).
172:
7189:- a database of G-quadruplexes near RNA processing sites.
6392:
6276:
6116:
5359:
3943:
3391:"G-quadruplex structures mark human regulatory chromatin"
3123:
2856:
Kamath-Loeb A, Loeb LA, Fry M (2012). Cotterill S (ed.).
2610:"G-quadruplexes in promoters throughout the human genome"
2448:
2199:
1724:
1073:
255:
102:
62:
58:
6798:
5621:
2502:"DNA tetraplex formation in the control region of c-myc"
1014:
288:
Quadruplexes are present in locations other than at the
276:. This is an active target of drug discovery, including
250:. The human telomeric repeat (which is the same for all
7217:
6927:
4867:
4340:
4142:"Quadruplex Nucleic Acids as Novel Therapeutic Targets"
2499:
2115:
1917:
7182:
Database on Quadruplex information: QuadBase from IGIB
6841:
6575:
6526:
5275:
4485:
3434:
3432:
2991:
2757:
2656:
2607:
1592:"G-quadruplexes and their regulatory roles in biology"
1371:
1015:
Capra JA, Paeschke K, Singh M, Zakian VA (July 2010).
775:
121:, Nobel Prize Winner in Chemistry (1982). Interest in
5989:
5840:
5418:
3342:"Structure and function of the telomeric CST complex"
3175:
Maizels N, Gray LT (April 2013). Rosenberg SM (ed.).
2955:
1442:
650:
inhibits cancer growth more effectively than TMPyP4.
244:, and subsequently they have also been shown to form
5086:
4252:
4041:
3697:
Hill JW, Hazra TK, Izumi T, Mitra S (January 2001).
2074:
1665:
953:
expression of a gene; this has been detected in the
948:
Antisense-mediated interventions and small-molecule
932:
13 of the FMR1 gene. This repeat expansion promotes
4902:
4570:
3696:
3429:
1443:Bochman ML, Paeschke K, Zakian VA (November 2012).
1125:
717:
628:The RET oncogene functions in the transcription of
6930:"Quadruplex DNA: sequence, topology and structure"
6884:
5313:"Following G-quartet formation by UV-spectroscopy"
4903:Guo K, Gokhale V, Hurley LH, Sun D (August 2008).
4818:
4378:
4253:Brooks TA, Kendrick S, Hurley L (September 2010).
3894:"Nucleotide excision repair of abasic DNA lesions"
3794:Computational and Structural Biotechnology Journal
3346:Computational and Structural Biotechnology Journal
2855:
2608:Huppert JL, Balasubramanian S (14 December 2006).
2391:
1920:"Quadruplex DNA: sequence, topology and structure"
1666:Gellert M, Lipsett MN, Davies DR (December 1962).
657:
6067:
5310:
4379:Sun D, Guo K, Rusche JJ, Hurley LH (2005-10-12).
4186:. Vol. Chapter 17. pp. 17.5.1–17.5.17.
817:
7251:
6844:"Prevalence of quadruplexes in the human genome"
6333:
5311:Mergny JL, Phan AT, Lacroix L (September 1998).
5175:
2958:"Structural Basis of DEAH/RHA Helicase Activity"
2760:"Prevalence of quadruplexes in the human genome"
2500:Simonsson T, Pecinka P, Kubista M (March 1998).
2240:
2151:Zheng KW, Chen Z, Hao YH, Tan Z (January 2010).
1966:
1540:
580:Along with the association of G-quadruplexes in
560:G-quadruplex forming sequences are prevalent in
3835:
3833:
3339:
2150:
523:
6769:
6610:
3839:
6639:
6604:
6569:
6435:
6386:
6327:
6270:
6220:
6160:
6110:
6061:
5516:National Center for Biotechnology Information
4300:
4298:
3745:
2704:
2702:
2081:Current Opinion in Genetics & Development
1174:
759:). This rule has been widely used in on-line
283:
113:, Nature will have found a way of using them
97:The identification of structures with a high
7066:
6613:Bioorganic & Medicinal Chemistry Letters
6334:Lee JE, Bennett CF, Cooper TA (March 2012).
5176:Han H, Hurley LH, Salazar M (January 1999).
3830:
3333:
2956:Chen MC, Ferré-D'Amaré AR (15 August 2017).
1226:
1224:
1222:
1170:
1168:
7202:- a database of G-quadruplexes in the UTRs.
5133:
3174:
2455:International Journal of Molecular Sciences
1589:
802:-based methods. Firstly, it was shown that
7097:Neidle & Balasubramanian, ed. (2006).
4304:
4295:
3499:: CS1 maint: location missing publisher (
2699:
1868:
1230:
1121:
1119:
357:, although not all of these probably form
7049:
7006:
6996:
6953:
6910:
6867:
6824:
6673:
6663:
6552:
6467:
6418:
6369:
6359:
6310:
6253:
6203:
6193:
6143:
6093:
6068:Mizielinska S, Isaacs AM (October 2014).
5948:
5907:
5858:
5817:
5760:
5750:
5709:
5652:
5598:
5487:
5395:
5385:
5328:
5252:
5224:
5201:
5063:
4928:
4844:
4756:
4746:
4697:
4596:
4547:
4462:
4404:
4278:
4209:
4116:
4067:
4018:
3969:
3917:
3865:
3813:
3722:
3670:
3644:
3618:
3562:
3552:
3365:
3316:
3259:
3202:
3192:
3151:
3141:
3100:
3090:
3017:
2973:
2932:
2891:
2881:
2832:
2783:
2734:
2682:
2633:
2584:
2574:
2525:
2476:
2466:
2425:
2415:
2351:Nature Structural & Molecular Biology
2325:
2315:
2266:
2217:
2176:
2092:
2075:Murat P, Balasubramanian S (April 2014).
2051:
2041:
1992:
1943:
1874:
1851:
1801:
1701:
1691:
1615:
1566:
1517:
1468:
1407:
1314:
1219:
1165:
1129:The Alkali Metal Ions: Their Role in Life
1099:
1050:
1040:
943:
230:
7070:Journal of the American Chemical Society
4786:Journal of the American Chemical Society
4528:Journal of the American Chemical Society
4181:
2663:Journal of the American Chemical Society
2118:Journal of the American Chemical Society
2015:
1967:Cao K, Ryvkin P, Johnson FB (May 2012).
666:, which can be both small molecules and
132:
18:
3840:Fleming AM, Burrows CJ (October 2017).
3787:
3438:
1116:
846:which is found throughout the brain in
842:The C9orf72 gene codes for the protein
185:, composed of DNA tightly wound around
173:Structure and functional role in genome
7252:
7157:
6885:Todd AK, Johnston M, Neidle S (2005).
6842:Huppert JL, Balasubramanian S (2005).
4819:Todd AK, Johnston M, Neidle S (2005).
4139:
3887:
3885:
3783:
3781:
3779:
3777:
3692:
3690:
3640:
3638:
3588:
3586:
3584:
3582:
3522:
3520:
3518:
3516:
3514:
3512:
3510:
3040:
2758:Huppert JL, Balasubramanian S (2005).
1638:
1344:
957:(chromosome 9 open reading frame 72).
792:
783:
503:DNA Oxidation Contribution to Diseases
6696:
5002:
3939:
3937:
7218:Tools to predict G-quadruplex motifs
7164:Nanopore and Aptamer Biosensor group
7124:Bioorganic & Medicinal Chemistry
5591:10.1016/j.neurobiolaging.2012.07.005
5518:. U.S. National Library of Medicine.
5134:Kwok CK, Merrick CJ (October 2017).
3846:Organic & Biomolecular Chemistry
3611:10.1016/j.neurobiolaging.2014.01.004
1541:Dhapola P, Chowdhury S (July 2016).
7211:non-B Motif Search Tool at non-B DB
6652:The Journal of Biological Chemistry
5937:The Journal of Biological Chemistry
5739:The Journal of Biological Chemistry
4882:10.1146/annurev.bi.64.070195.000433
3882:
3774:
3687:
3635:
3579:
3507:
3130:The Journal of Biological Chemistry
3079:The Journal of Biological Chemistry
2921:The Journal of Biological Chemistry
2404:The Journal of Biological Chemistry
1590:Rhodes D, Lipps HJ (October 2015).
833:fragile X mental retardation gene 1
776:Methods for studying G-quadruplexes
575:
13:
6762:
6701:. Vol. 18. pp. 325–349.
5633:American Journal of Human Genetics
3934:
1668:"Helix formation by guanylic acid"
1410:Trends in Pharmacological Sciences
177:Following sequencing of the human
14:
7281:
7152:
4182:Chen Y, Yang D (September 2012).
1080:Journal of Visualized Experiments
6690:
6520:
6484:
6026:
5983:
5965:
5924:
5883:
5834:
5005:Cancer Genomics & Proteomics
4271:10.1111/j.1742-4658.2010.07759.x
718:Quadruplex prediction techniques
599:telomerase reverse transcriptase
5777:
5726:
5669:
5615:
5565:
5522:
5504:
5463:
5412:
5353:
5304:
5269:
5218:
5169:
5127:
5080:
5023:
4996:
4945:
4896:
4861:
4812:
4773:
4714:
4657:
4613:
4564:
4515:
4479:
4421:
4372:
4334:
4246:
4175:
4133:
4084:
4035:
3986:
3739:
3382:
3276:
3219:
3168:
3117:
3034:
2949:
2908:
2849:
2800:
2751:
2650:
2601:
2542:
2493:
2442:
2385:
2342:
2283:
2234:
2193:
2144:
2109:
2068:
2009:
1960:
1911:
1818:
1761:
1718:
1659:
1632:
1583:
1534:
1485:
1436:
999:coordination with nucleobases.
658:Ligands which bind quadruplexes
640:
7103:. Royal Society of Chemistry.
6494:Journal of Medicinal Chemistry
6234:Science Translational Medicine
5973:"Fragile X Mental Retardation"
4308:Journal of Medicinal Chemistry
4149:Journal of Medicinal Chemistry
3999:Molecular and Cellular Biology
3950:Molecular and Cellular Biology
3651:Journal of Medicinal Chemistry
3340:Rice C, Skordalakes E (2016).
1401:
1375:Journal of Medicinal Chemistry
1365:
1067:
1008:
818:Role in neurological disorders
455:8-Oxoguanine-DNA glycosylase 1
16:Structure in molecular biology
1:
5860:10.1016/S0092-8674(01)00566-9
5489:10.1016/j.febslet.2015.05.003
5330:10.1016/s0014-5793(98)01043-6
5155:10.1016/j.tibtech.2017.06.012
5048:10.1093/bioinformatics/btz087
4870:Annual Review of Biochemistry
3645:Sun D, Hurley LH (May 2009).
3043:Chemical and Engineering News
1641:Chemical and Engineering News
1422:10.1016/s0165-6147(00)01457-7
1347:Chemical and Engineering News
1002:
993:Amyotrophic lateral sclerosis
825:amyotrophic lateral sclerosis
6817:10.1016/j.biochi.2008.02.013
6246:10.1126/scitranslmed.3007529
6136:10.1016/j.neuron.2013.10.015
6086:10.1097/WCO.0000000000000130
6074:Current Opinion in Neurology
6004:10.1016/0092-8674(91)90125-I
5441:10.1021/acs.nanolett.9b03184
5105:10.1016/j.biochi.2018.06.002
4192:10.1002/0471142700.nc1705s50
4161:10.1021/acs.jmedchem.5b01835
3194:10.1371/journal.pgen.1003468
2883:10.1371/journal.pone.0030189
2219:10.1021/acs.analchem.5b04396
2043:10.1371/journal.pone.0146174
1739:10.1016/0092-8674(87)90577-0
1042:10.1371/journal.pcbi.1000861
555:
524:APE1 role in Gene Regulation
7:
5290:10.1016/j.ymeth.2007.02.009
5235:10.1007/978-1-59745-363-9_5
4635:10.1021/acs.biochem.8b00843
3453:10.1007/978-1-0716-0763-3_8
1985:10.1016/j.ymeth.2012.05.002
1141:10.1007/978-3-319-21756-7_7
392:
152:
10:
7286:
7206:G-quadruplex Resource Site
6625:10.1016/j.bmcl.2014.04.029
6168:Ravits J (November 2013).
5645:10.1016/j.ajhg.2013.01.011
4455:10.1038/s41598-017-11177-1
3806:10.1016/j.csbj.2019.12.013
3358:10.1016/j.csbj.2016.04.002
1359:10.1021/cen-v085n009.p012a
1021:PLOS Computational Biology
544:transcription factors (TF)
488:base excision repair (BER)
399:immunoglobulin heavy chain
284:Non-telomeric quadruplexes
92:
7136:10.1016/j.bmc.2013.09.036
6707:10.1515/9783110470734-018
4358:10.1101/2020.01.15.907626
3788:Poetsch AR (2020-01-07).
3309:10.1038/s41586-018-0209-9
3055:10.1021/cen-v087n044.p028
2394:"Long repeating (TTAGGG)
2094:10.1016/j.gde.2013.10.012
1653:10.1021/cen-v087n044.p028
865:) repeats within the 1st
550:
417:system of the pathogenic
299:forms a quadruplex in a
7100:Quadruplex Nucleic Acids
6035:Human Molecular Genetics
5896:Human Molecular Genetics
3143:10.1074/jbc.RA119.008687
2417:10.1074/jbc.RA118.002158
1449:Nature Reviews. Genetics
920:of mRNA in the neuron's
459:7,8-dihydro-8-oxoguanine
6772:Chemical Communications
6665:10.1074/jbc.C113.502336
6361:10.1073/pnas.1117019109
6195:10.1073/pnas.1318835110
5752:10.1074/jbc.C113.452532
5387:10.1073/pnas.1415944111
5143:Trends in Biotechnology
3554:10.1073/pnas.1912355117
3092:10.1074/jbc.M117.792077
1832:Nature Chemical Biology
1693:10.1073/pnas.48.12.2013
1504:(Database): D381–D385.
829:frontotemporal dementia
476:8-oxo-2'-deoxyguanosine
365:, namely the bacterium
6998:10.1073/pnas.182256799
6934:Nucleic Acids Research
6891:Nucleic Acids Research
6848:Nucleic Acids Research
6533:Nucleic Acids Research
5950:10.1074/jbc.M807354200
5543:10.1212/WNL.58.11.1615
5182:Nucleic Acids Research
4909:Nucleic Acids Research
4825:Nucleic Acids Research
4748:10.1073/pnas.182256799
4577:Nucleic Acids Research
4385:Nucleic Acids Research
4140:Neidle S (July 2016).
4097:Nucleic Acids Research
3898:Nucleic Acids Research
3703:Nucleic Acids Research
2998:Nucleic Acids Research
2934:10.1074/jbc.C500348200
2764:Nucleic Acids Research
2614:Nucleic Acids Research
2576:10.1073/pnas.182256799
2506:Nucleic Acids Research
2317:10.1073/pnas.141229498
2247:Nucleic Acids Research
2157:Nucleic Acids Research
1924:Nucleic Acids Research
1596:Nucleic Acids Research
1547:Nucleic Acids Research
1498:Nucleic Acids Research
944:Therapeutic approaches
407:Bloom syndrome protein
321:-ligase RFP2, and the
231:Telomeric quadruplexes
149:
27:In molecular biology,
24:
5579:Neurobiology of Aging
3599:Neurobiology of Aging
2518:10.1093/nar/26.5.1167
974:transcription factors
936:and other epigenetic
808:solid-state nanopores
136:
22:
5194:10.1093/nar/27.2.537
4060:10.1093/emboj/cdg595
4011:10.1128/mcb.00244-08
3962:10.1128/mcb.00401-16
3715:10.1093/nar/29.2.430
2975:10.3390/cryst7080253
2468:10.3390/ijms22147381
2206:Analytical Chemistry
2016:Kudlicki AS (2016).
1877:Biological Chemistry
1844:10.1038/nchembio.780
804:biological nanopores
451:base excision repair
307:regions include the
32:secondary structures
7226:from Bagga's group.
7158:Quadruplex websites
6989:2002PNAS...9911593S
6411:10.1038/mt.2011.201
6352:2012PNAS..109.4221L
6303:10.1038/nature11362
6295:2012Natur.488..111W
6186:2013PNAS..110E4530L
6047:10.1093/hmg/1.6.397
5810:10.1038/nature13124
5802:2014Natur.507..195H
5694:2012NatSR...2E1016F
5433:2019NanoL..19.7996B
5378:2014PNAS..11114325A
5372:(40): 14325–14331.
4966:1987Natur.330..495M
4739:2002PNAS...9911593S
4682:2015NatSR...511385O
4447:2017NatSR...711541H
4109:10.1093/nar/gkp1039
3545:2020PNAS..11711409R
3539:(21): 11409–11420.
3439:Poetsch AR (2020).
3301:2018Natur.558..465C
3244:2013NatCh...5..182B
3136:(47): 17709–17722.
3085:(37): 15205–15215.
2874:2012PLoSO...730189K
2626:10.1093/nar/gkl1057
2567:2002PNAS...9911593S
2308:2001PNAS...98.8572S
2034:2016PLoSO..1146174K
1889:10.1515/BC.2001.073
1786:2010NatCh...2.1095M
1684:1962PNAS...48.2013G
1245:1988Natur.334..364S
1189:1989Natur.342..825S
1033:2010PLSCB...6E0861C
914:synaptic plasticity
793:Biophysical methods
784:Biochemical methods
415:antigenic variation
199:Hoogsteen base pair
7198:2011-07-20 at the
7170:G-Quadruplex World
7042:10.1101/gr.4508806
6946:10.1093/nar/gkl655
6903:10.1093/nar/gki553
6860:10.1093/nar/gki609
6784:10.1039/c1cc13973h
6545:10.1093/nar/gkt127
5909:10.1093/hmg/ddg350
5682:Scientific Reports
4921:10.1093/nar/gkn380
4837:10.1093/nar/gki553
4670:Scientific Reports
4589:10.1093/nar/gkm538
4435:Scientific Reports
4397:10.1093/nar/gki917
3910:10.1093/nar/gkz558
3858:10.1039/C7OB02096A
3252:10.1038/nchem.1548
3010:10.1093/nar/gkn919
2825:10.1101/gr.4508806
2776:10.1093/nar/gki609
2259:10.1093/nar/gkr164
2169:10.1093/nar/gkp898
1936:10.1093/nar/gkl655
1608:10.1093/nar/gkv862
1559:10.1093/nar/gkw425
1510:10.1093/nar/gkm781
1307:10.1101/gr.4508806
837:Fragile X Syndrome
812:DNA nanotechnology
751:), where N is any
195:molecular crowding
150:
25:
7110:978-0-85404-374-3
7082:10.1021/ja901574c
6590:10.1021/bi802233v
6506:10.1021/jm800874t
6460:10.1021/bi101208k
6399:Molecular Therapy
6240:(208): 208ra149.
5902:(24): 3295–3305.
5796:(7491): 195–200.
5702:10.1038/srep01016
5585:(12): 2950.e5–7.
5427:(11): 7996–8001.
5042:(18): 3493–3495.
4798:10.1021/ja2102423
4690:10.1038/srep11385
4629:(46): 6514–6527.
4540:10.1021/ja072185g
4501:10.1021/bi051618u
4320:10.1021/jm200062u
4155:(13): 5987–6011.
4054:(23): 6299–6309.
4005:(23): 7066–7080.
3904:(16): 8537–8547.
3852:(39): 8341–8353.
3760:10.1021/cr960421s
3663:10.1021/jm900055s
3462:978-1-0716-0762-6
3401:(10): 1267–1272.
3295:(7710): 465–469.
2727:10.1021/bi0601510
2675:10.1021/ja055636a
2410:(24): 9473–9485.
2130:10.1021/ja061267m
1794:10.1038/nchem.842
1780:(12): 1095–1098.
1553:(W1): W277–W283.
1387:10.1021/jm800448a
1381:(18): 5641–5649.
1150:978-3-319-21755-0
961:Antisense therapy
442:AP endonuclease 1
263:crystal structure
7277:
7147:
7118:
7113:. Archived from
7093:
7063:
7053:
7020:
7010:
7000:
6967:
6957:
6924:
6914:
6881:
6871:
6838:
6828:
6795:
6757:
6756:
6750:
6746:
6744:
6736:
6694:
6688:
6687:
6677:
6667:
6643:
6637:
6636:
6608:
6602:
6601:
6573:
6567:
6566:
6556:
6539:(7): 4049–4064.
6524:
6518:
6517:
6488:
6482:
6481:
6471:
6454:(47): 10166–78.
6439:
6433:
6432:
6422:
6390:
6384:
6383:
6373:
6363:
6331:
6325:
6324:
6314:
6274:
6268:
6267:
6257:
6224:
6218:
6217:
6207:
6197:
6164:
6158:
6157:
6147:
6114:
6108:
6107:
6097:
6065:
6059:
6058:
6030:
6024:
6023:
5987:
5981:
5980:
5969:
5963:
5962:
5952:
5943:(7): 4255–4266.
5928:
5922:
5921:
5911:
5887:
5881:
5880:
5862:
5838:
5832:
5831:
5821:
5781:
5775:
5774:
5764:
5754:
5730:
5724:
5723:
5713:
5673:
5667:
5666:
5656:
5619:
5613:
5612:
5602:
5569:
5563:
5562:
5526:
5520:
5519:
5508:
5502:
5501:
5491:
5467:
5461:
5460:
5416:
5410:
5409:
5399:
5389:
5357:
5351:
5350:
5332:
5308:
5302:
5301:
5273:
5267:
5266:
5256:
5227:G-Quadruplex DNA
5222:
5216:
5215:
5205:
5173:
5167:
5166:
5149:(10): 997–1013.
5140:
5131:
5125:
5124:
5084:
5078:
5077:
5067:
5027:
5021:
5020:
5000:
4994:
4993:
4974:10.1038/330495a0
4949:
4943:
4942:
4932:
4915:(14): 4598–608.
4900:
4894:
4893:
4865:
4859:
4858:
4848:
4816:
4810:
4809:
4777:
4771:
4770:
4760:
4750:
4718:
4712:
4711:
4701:
4661:
4655:
4654:
4617:
4611:
4610:
4600:
4583:(22): 7698–713.
4568:
4562:
4561:
4551:
4519:
4513:
4512:
4495:(49): 16341–50.
4483:
4477:
4476:
4466:
4425:
4419:
4418:
4408:
4376:
4370:
4369:
4347:
4338:
4332:
4331:
4302:
4293:
4292:
4282:
4259:The FEBS Journal
4250:
4244:
4243:
4237:
4233:
4231:
4223:
4213:
4179:
4173:
4172:
4146:
4137:
4131:
4130:
4120:
4088:
4082:
4081:
4071:
4048:The EMBO Journal
4039:
4033:
4032:
4022:
3990:
3984:
3983:
3973:
3941:
3932:
3931:
3921:
3889:
3880:
3879:
3869:
3837:
3828:
3827:
3817:
3785:
3772:
3771:
3754:(3): 1109–1152.
3748:Chemical Reviews
3743:
3737:
3736:
3726:
3694:
3685:
3684:
3674:
3657:(9): 2863–2874.
3642:
3633:
3632:
3622:
3605:(6): 1293–1300.
3590:
3577:
3576:
3566:
3556:
3524:
3505:
3504:
3498:
3490:
3436:
3427:
3426:
3386:
3380:
3379:
3369:
3337:
3331:
3330:
3320:
3280:
3274:
3273:
3263:
3232:Nature Chemistry
3223:
3217:
3216:
3206:
3196:
3172:
3166:
3165:
3155:
3145:
3121:
3115:
3114:
3104:
3094:
3070:
3059:
3058:
3038:
3032:
3031:
3021:
2989:
2980:
2979:
2977:
2953:
2947:
2946:
2936:
2927:(46): 38117–20.
2912:
2906:
2905:
2895:
2885:
2853:
2847:
2846:
2836:
2804:
2798:
2797:
2787:
2755:
2749:
2748:
2738:
2706:
2697:
2696:
2686:
2654:
2648:
2647:
2637:
2605:
2599:
2598:
2588:
2578:
2546:
2540:
2539:
2529:
2497:
2491:
2490:
2480:
2470:
2446:
2440:
2439:
2429:
2419:
2389:
2383:
2382:
2346:
2340:
2339:
2329:
2319:
2287:
2281:
2280:
2270:
2238:
2232:
2231:
2221:
2197:
2191:
2190:
2180:
2148:
2142:
2141:
2113:
2107:
2106:
2096:
2072:
2066:
2065:
2055:
2045:
2013:
2007:
2006:
1996:
1964:
1958:
1957:
1947:
1915:
1909:
1908:
1872:
1866:
1865:
1855:
1822:
1816:
1815:
1805:
1774:Nature Chemistry
1765:
1759:
1758:
1722:
1716:
1715:
1705:
1695:
1663:
1657:
1656:
1636:
1630:
1629:
1619:
1587:
1581:
1580:
1570:
1538:
1532:
1531:
1521:
1489:
1483:
1482:
1472:
1440:
1434:
1433:
1405:
1399:
1398:
1369:
1363:
1362:
1342:
1329:
1328:
1318:
1286:
1273:
1272:
1253:10.1038/334364a0
1228:
1217:
1216:
1197:10.1038/342825a0
1172:
1163:
1162:
1134:
1123:
1114:
1113:
1103:
1071:
1065:
1064:
1054:
1044:
1012:
969:oligonucleotides
835:(FMR1) gene and
712:electron density
576:Promoter Regions
518:thymidine kinase
480:oxidative stress
147:
7285:
7284:
7280:
7279:
7278:
7276:
7275:
7274:
7250:
7249:
7220:
7200:Wayback Machine
7160:
7155:
7150:
7130:(23): 7515–22.
7111:
7030:Genome Research
6983:(18): 11593–8.
6940:(19): 5402–15.
6778:(38): 10563–5.
6765:
6763:Further reading
6760:
6748:
6747:
6738:
6737:
6717:
6695:
6691:
6644:
6640:
6619:(12): 2602–12.
6609:
6605:
6574:
6570:
6525:
6521:
6489:
6485:
6440:
6436:
6405:(12): 2178–85.
6391:
6387:
6332:
6328:
6289:(7409): 111–5.
6275:
6271:
6225:
6221:
6180:(47): E4530–9.
6165:
6161:
6115:
6111:
6066:
6062:
6031:
6027:
5988:
5984:
5971:
5970:
5966:
5929:
5925:
5888:
5884:
5839:
5835:
5782:
5778:
5731:
5727:
5674:
5670:
5620:
5616:
5570:
5566:
5537:(11): 1615–21.
5527:
5523:
5510:
5509:
5505:
5482:(14): 1653–68.
5468:
5464:
5417:
5413:
5358:
5354:
5309:
5305:
5274:
5270:
5245:
5223:
5219:
5174:
5170:
5138:
5132:
5128:
5085:
5081:
5028:
5024:
5001:
4997:
4960:(6147): 495–7.
4950:
4946:
4901:
4897:
4866:
4862:
4817:
4813:
4778:
4774:
4733:(18): 11593–8.
4719:
4715:
4662:
4658:
4618:
4614:
4569:
4565:
4534:(33): 10220–8.
4520:
4516:
4484:
4480:
4426:
4422:
4391:(18): 6070–80.
4377:
4373:
4345:
4339:
4335:
4303:
4296:
4265:(17): 3459–69.
4251:
4247:
4235:
4234:
4225:
4224:
4202:
4180:
4176:
4144:
4138:
4134:
4089:
4085:
4040:
4036:
3991:
3987:
3942:
3935:
3890:
3883:
3838:
3831:
3786:
3775:
3744:
3740:
3695:
3688:
3643:
3636:
3591:
3580:
3525:
3508:
3492:
3491:
3463:
3437:
3430:
3407:10.1038/ng.3662
3395:Nature Genetics
3387:
3383:
3338:
3334:
3281:
3277:
3224:
3220:
3187:(4): e1003468.
3177:"The G4 genome"
3173:
3169:
3122:
3118:
3071:
3062:
3039:
3035:
2990:
2983:
2954:
2950:
2913:
2909:
2854:
2850:
2813:Genome Research
2805:
2801:
2756:
2752:
2721:(25): 7854–60.
2707:
2700:
2655:
2651:
2606:
2602:
2561:(18): 11593–8.
2547:
2543:
2498:
2494:
2447:
2443:
2398:
2390:
2386:
2363:10.1038/nsmb982
2347:
2343:
2288:
2284:
2253:(14): 6229–37.
2239:
2235:
2198:
2194:
2149:
2145:
2124:(24): 7957–63.
2114:
2110:
2073:
2069:
2028:(1): e0146174.
2014:
2010:
1965:
1961:
1930:(19): 5402–15.
1916:
1912:
1873:
1869:
1823:
1819:
1766:
1762:
1723:
1719:
1664:
1660:
1637:
1633:
1602:(18): 8627–37.
1588:
1584:
1539:
1535:
1490:
1486:
1461:10.1038/nrg3296
1441:
1437:
1406:
1402:
1370:
1366:
1343:
1332:
1295:Genome Research
1287:
1276:
1239:(6180): 364–6.
1229:
1220:
1183:(6251): 825–9.
1173:
1166:
1151:
1132:
1124:
1117:
1072:
1068:
1027:(7): e1000861.
1013:
1009:
1005:
946:
938:heterochromatin
934:DNA methylation
900:
896:
892:
888:
884:
880:
876:
872:
864:
860:
820:
795:
786:
778:
753:nucleotide base
750:
746:
742:
738:
734:
730:
726:
720:
660:
647:porphyrin rings
643:
611:promoter region
578:
558:
553:
526:
497:ChIP-sequencing
467:
446:ChIP-sequencing
438:ChIP-sequencing
429:
395:
323:proto-oncogenes
286:
233:
175:
155:
139:
95:
17:
12:
11:
5:
7283:
7273:
7272:
7267:
7262:
7248:
7247:
7242:
7237:
7232:
7227:
7219:
7216:
7215:
7214:
7208:
7203:
7190:
7184:
7179:
7173:
7167:
7159:
7156:
7154:
7153:External links
7151:
7149:
7148:
7119:
7117:on 2007-09-30.
7109:
7094:
7076:(22): 7800–5.
7064:
7021:
6968:
6925:
6882:
6854:(9): 2908–16.
6839:
6811:(8): 1250–63.
6796:
6766:
6764:
6761:
6759:
6758:
6749:|journal=
6715:
6689:
6638:
6603:
6584:(8): 1675–80.
6568:
6519:
6500:(2): 564–568.
6483:
6434:
6385:
6346:(11): 4221–6.
6326:
6269:
6219:
6159:
6109:
6060:
6041:(6): 397–400.
6025:
5998:(4): 817–822.
5982:
5964:
5923:
5882:
5853:(4): 489–499.
5833:
5776:
5745:(14): 9860–6.
5725:
5668:
5614:
5564:
5521:
5503:
5462:
5411:
5352:
5303:
5284:(4): 324–331.
5268:
5243:
5217:
5188:(2): 537–542.
5168:
5126:
5079:
5036:Bioinformatics
5022:
4995:
4944:
4895:
4860:
4811:
4792:(5): 2723–31.
4772:
4713:
4656:
4612:
4563:
4514:
4478:
4420:
4371:
4333:
4314:(16): 5671–9.
4294:
4245:
4236:|journal=
4201:978-0471142706
4200:
4174:
4132:
4103:(3): 832–845.
4083:
4034:
3985:
3933:
3881:
3829:
3773:
3738:
3709:(2): 430–438.
3686:
3634:
3578:
3506:
3461:
3428:
3381:
3332:
3275:
3218:
3167:
3116:
3060:
3033:
3004:(1): 172–183.
2981:
2948:
2907:
2848:
2799:
2770:(9): 2908–16.
2750:
2698:
2649:
2600:
2541:
2512:(5): 1167–72.
2492:
2441:
2396:
2384:
2357:(10): 847–54.
2341:
2302:(15): 8572–7.
2282:
2233:
2192:
2143:
2108:
2067:
2008:
1959:
1910:
1867:
1838:(3): 301–310.
1817:
1760:
1733:(6): 899–908.
1717:
1678:(12): 2013–8.
1658:
1631:
1582:
1533:
1484:
1455:(11): 770–80.
1435:
1400:
1364:
1330:
1301:(5): 644–655.
1274:
1218:
1164:
1149:
1115:
1066:
1006:
1004:
1001:
989:aromatic rings
945:
942:
898:
894:
890:
886:
882:
878:
874:
870:
862:
858:
819:
816:
794:
791:
785:
782:
777:
774:
748:
744:
740:
736:
732:
728:
724:
719:
716:
659:
656:
642:
639:
577:
574:
557:
554:
552:
549:
525:
522:
510:carcinogenesis
466:
463:
428:
425:
394:
391:
387:non-coding DNA
375:downregulation
294:proto-oncogene
285:
282:
232:
229:
191:thermodynamics
174:
171:
154:
151:
94:
91:
75:intramolecular
44:guanine tetrad
15:
9:
6:
4:
3:
2:
7282:
7271:
7268:
7266:
7263:
7261:
7258:
7257:
7255:
7245:
7243:
7240:
7238:
7235:
7233:
7231:
7228:
7225:
7222:
7221:
7212:
7209:
7207:
7204:
7201:
7197:
7194:
7191:
7188:
7185:
7183:
7180:
7177:
7174:
7171:
7168:
7165:
7162:
7161:
7145:
7141:
7137:
7133:
7129:
7125:
7120:
7116:
7112:
7106:
7102:
7101:
7095:
7091:
7087:
7083:
7079:
7075:
7071:
7065:
7061:
7057:
7052:
7047:
7043:
7039:
7036:(5): 644–55.
7035:
7031:
7027:
7022:
7018:
7014:
7009:
7004:
6999:
6994:
6990:
6986:
6982:
6978:
6974:
6969:
6965:
6961:
6956:
6951:
6947:
6943:
6939:
6935:
6931:
6926:
6922:
6918:
6913:
6908:
6904:
6900:
6897:(9): 2901–7.
6896:
6892:
6888:
6883:
6879:
6875:
6870:
6865:
6861:
6857:
6853:
6849:
6845:
6840:
6836:
6832:
6827:
6822:
6818:
6814:
6810:
6806:
6802:
6797:
6793:
6789:
6785:
6781:
6777:
6773:
6768:
6767:
6754:
6742:
6734:
6730:
6726:
6722:
6718:
6716:9783110470734
6712:
6708:
6704:
6700:
6693:
6685:
6681:
6676:
6671:
6666:
6661:
6658:(8): 4653–9.
6657:
6653:
6649:
6642:
6634:
6630:
6626:
6622:
6618:
6614:
6607:
6599:
6595:
6591:
6587:
6583:
6579:
6572:
6564:
6560:
6555:
6550:
6546:
6542:
6538:
6534:
6530:
6523:
6515:
6511:
6507:
6503:
6499:
6495:
6487:
6479:
6475:
6470:
6465:
6461:
6457:
6453:
6449:
6445:
6438:
6430:
6426:
6421:
6416:
6412:
6408:
6404:
6400:
6396:
6389:
6381:
6377:
6372:
6367:
6362:
6357:
6353:
6349:
6345:
6341:
6337:
6330:
6322:
6318:
6313:
6308:
6304:
6300:
6296:
6292:
6288:
6284:
6280:
6273:
6265:
6261:
6256:
6251:
6247:
6243:
6239:
6235:
6231:
6223:
6215:
6211:
6206:
6201:
6196:
6191:
6187:
6183:
6179:
6175:
6171:
6163:
6155:
6151:
6146:
6141:
6137:
6133:
6130:(2): 415–28.
6129:
6125:
6121:
6113:
6105:
6101:
6096:
6091:
6087:
6083:
6080:(5): 515–23.
6079:
6075:
6071:
6064:
6056:
6052:
6048:
6044:
6040:
6036:
6029:
6021:
6017:
6013:
6009:
6005:
6001:
5997:
5993:
5986:
5978:
5974:
5968:
5960:
5956:
5951:
5946:
5942:
5938:
5934:
5927:
5919:
5915:
5910:
5905:
5901:
5897:
5893:
5886:
5878:
5874:
5870:
5866:
5861:
5856:
5852:
5848:
5844:
5837:
5829:
5825:
5820:
5815:
5811:
5807:
5803:
5799:
5795:
5791:
5787:
5780:
5772:
5768:
5763:
5758:
5753:
5748:
5744:
5740:
5736:
5729:
5721:
5717:
5712:
5707:
5703:
5699:
5695:
5691:
5687:
5683:
5679:
5672:
5664:
5660:
5655:
5650:
5646:
5642:
5639:(3): 345–53.
5638:
5634:
5630:
5626:
5618:
5610:
5606:
5601:
5596:
5592:
5588:
5584:
5580:
5576:
5568:
5560:
5556:
5552:
5548:
5544:
5540:
5536:
5532:
5525:
5517:
5513:
5507:
5499:
5495:
5490:
5485:
5481:
5477:
5473:
5466:
5458:
5454:
5450:
5446:
5442:
5438:
5434:
5430:
5426:
5422:
5415:
5407:
5403:
5398:
5393:
5388:
5383:
5379:
5375:
5371:
5367:
5363:
5356:
5348:
5344:
5340:
5336:
5331:
5326:
5322:
5318:
5314:
5307:
5299:
5295:
5291:
5287:
5283:
5279:
5272:
5264:
5260:
5255:
5250:
5246:
5244:9781588299505
5240:
5236:
5232:
5228:
5221:
5213:
5209:
5204:
5199:
5195:
5191:
5187:
5183:
5179:
5172:
5164:
5160:
5156:
5152:
5148:
5144:
5137:
5130:
5122:
5118:
5114:
5110:
5106:
5102:
5098:
5094:
5090:
5083:
5075:
5071:
5066:
5061:
5057:
5053:
5049:
5045:
5041:
5037:
5033:
5026:
5018:
5014:
5011:(4): 207–15.
5010:
5006:
4999:
4991:
4987:
4983:
4979:
4975:
4971:
4967:
4963:
4959:
4955:
4948:
4940:
4936:
4931:
4926:
4922:
4918:
4914:
4910:
4906:
4899:
4891:
4887:
4883:
4879:
4875:
4871:
4864:
4856:
4852:
4847:
4842:
4838:
4834:
4831:(9): 2901–7.
4830:
4826:
4822:
4815:
4807:
4803:
4799:
4795:
4791:
4787:
4783:
4776:
4768:
4764:
4759:
4754:
4749:
4744:
4740:
4736:
4732:
4728:
4724:
4717:
4709:
4705:
4700:
4695:
4691:
4687:
4683:
4679:
4675:
4671:
4667:
4660:
4652:
4648:
4644:
4640:
4636:
4632:
4628:
4624:
4616:
4608:
4604:
4599:
4594:
4590:
4586:
4582:
4578:
4574:
4567:
4559:
4555:
4550:
4545:
4541:
4537:
4533:
4529:
4525:
4518:
4510:
4506:
4502:
4498:
4494:
4490:
4482:
4474:
4470:
4465:
4460:
4456:
4452:
4448:
4444:
4440:
4436:
4432:
4424:
4416:
4412:
4407:
4402:
4398:
4394:
4390:
4386:
4382:
4375:
4367:
4363:
4359:
4355:
4351:
4344:
4337:
4329:
4325:
4321:
4317:
4313:
4309:
4301:
4299:
4290:
4286:
4281:
4276:
4272:
4268:
4264:
4260:
4256:
4249:
4241:
4229:
4221:
4217:
4212:
4207:
4203:
4197:
4193:
4189:
4185:
4178:
4170:
4166:
4162:
4158:
4154:
4150:
4143:
4136:
4128:
4124:
4119:
4114:
4110:
4106:
4102:
4098:
4094:
4087:
4079:
4075:
4070:
4065:
4061:
4057:
4053:
4049:
4045:
4038:
4030:
4026:
4021:
4016:
4012:
4008:
4004:
4000:
3996:
3989:
3981:
3977:
3972:
3967:
3963:
3959:
3955:
3951:
3947:
3940:
3938:
3929:
3925:
3920:
3915:
3911:
3907:
3903:
3899:
3895:
3888:
3886:
3877:
3873:
3868:
3863:
3859:
3855:
3851:
3847:
3843:
3836:
3834:
3825:
3821:
3816:
3811:
3807:
3803:
3799:
3795:
3791:
3784:
3782:
3780:
3778:
3769:
3765:
3761:
3757:
3753:
3749:
3742:
3734:
3730:
3725:
3720:
3716:
3712:
3708:
3704:
3700:
3693:
3691:
3682:
3678:
3673:
3668:
3664:
3660:
3656:
3652:
3648:
3641:
3639:
3630:
3626:
3621:
3616:
3612:
3608:
3604:
3600:
3596:
3589:
3587:
3585:
3583:
3574:
3570:
3565:
3560:
3555:
3550:
3546:
3542:
3538:
3534:
3530:
3523:
3521:
3519:
3517:
3515:
3513:
3511:
3502:
3496:
3488:
3484:
3480:
3476:
3472:
3468:
3464:
3458:
3454:
3450:
3446:
3442:
3435:
3433:
3424:
3420:
3416:
3412:
3408:
3404:
3400:
3396:
3392:
3385:
3377:
3373:
3368:
3363:
3359:
3355:
3351:
3347:
3343:
3336:
3328:
3324:
3319:
3314:
3310:
3306:
3302:
3298:
3294:
3290:
3286:
3279:
3271:
3267:
3262:
3257:
3253:
3249:
3245:
3241:
3237:
3233:
3229:
3222:
3214:
3210:
3205:
3200:
3195:
3190:
3186:
3182:
3181:PLOS Genetics
3178:
3171:
3163:
3159:
3154:
3149:
3144:
3139:
3135:
3131:
3127:
3120:
3112:
3108:
3103:
3098:
3093:
3088:
3084:
3080:
3076:
3069:
3067:
3065:
3056:
3052:
3049:(44): 28–30.
3048:
3044:
3037:
3029:
3025:
3020:
3015:
3011:
3007:
3003:
2999:
2995:
2988:
2986:
2976:
2971:
2967:
2963:
2959:
2952:
2944:
2940:
2935:
2930:
2926:
2922:
2918:
2911:
2903:
2899:
2894:
2889:
2884:
2879:
2875:
2871:
2868:(1): e30189.
2867:
2863:
2859:
2852:
2844:
2840:
2835:
2830:
2826:
2822:
2819:(5): 644–55.
2818:
2814:
2810:
2803:
2795:
2791:
2786:
2781:
2777:
2773:
2769:
2765:
2761:
2754:
2746:
2742:
2737:
2732:
2728:
2724:
2720:
2716:
2712:
2705:
2703:
2694:
2690:
2685:
2680:
2676:
2672:
2669:(4): 1096–8.
2668:
2664:
2660:
2653:
2645:
2641:
2636:
2631:
2627:
2623:
2620:(2): 406–13.
2619:
2615:
2611:
2604:
2596:
2592:
2587:
2582:
2577:
2572:
2568:
2564:
2560:
2556:
2552:
2545:
2537:
2533:
2528:
2523:
2519:
2515:
2511:
2507:
2503:
2496:
2488:
2484:
2479:
2474:
2469:
2464:
2460:
2456:
2452:
2445:
2437:
2433:
2428:
2423:
2418:
2413:
2409:
2405:
2401:
2399:
2388:
2380:
2376:
2372:
2368:
2364:
2360:
2356:
2352:
2345:
2337:
2333:
2328:
2323:
2318:
2313:
2309:
2305:
2301:
2297:
2293:
2286:
2278:
2274:
2269:
2264:
2260:
2256:
2252:
2248:
2244:
2237:
2229:
2225:
2220:
2215:
2212:(4): 1984–9.
2211:
2207:
2203:
2196:
2188:
2184:
2179:
2174:
2170:
2166:
2163:(1): 327–38.
2162:
2158:
2154:
2147:
2139:
2135:
2131:
2127:
2123:
2119:
2112:
2104:
2100:
2095:
2090:
2086:
2082:
2078:
2071:
2063:
2059:
2054:
2049:
2044:
2039:
2035:
2031:
2027:
2023:
2019:
2012:
2004:
2000:
1995:
1990:
1986:
1982:
1978:
1974:
1970:
1963:
1955:
1951:
1946:
1941:
1937:
1933:
1929:
1925:
1921:
1914:
1906:
1902:
1898:
1894:
1890:
1886:
1882:
1878:
1871:
1863:
1859:
1854:
1849:
1845:
1841:
1837:
1833:
1829:
1821:
1813:
1809:
1804:
1799:
1795:
1791:
1787:
1783:
1779:
1775:
1771:
1764:
1756:
1752:
1748:
1744:
1740:
1736:
1732:
1728:
1721:
1713:
1709:
1704:
1699:
1694:
1689:
1685:
1681:
1677:
1673:
1669:
1662:
1654:
1650:
1647:(44): 28–30.
1646:
1642:
1635:
1627:
1623:
1618:
1613:
1609:
1605:
1601:
1597:
1593:
1586:
1578:
1574:
1569:
1564:
1560:
1556:
1552:
1548:
1544:
1537:
1529:
1525:
1520:
1515:
1511:
1507:
1503:
1499:
1495:
1488:
1480:
1476:
1471:
1466:
1462:
1458:
1454:
1450:
1446:
1439:
1431:
1427:
1423:
1419:
1416:(4): 136–42.
1415:
1411:
1404:
1396:
1392:
1388:
1384:
1380:
1376:
1368:
1360:
1356:
1353:(22): 12–17.
1352:
1348:
1341:
1339:
1337:
1335:
1326:
1322:
1317:
1312:
1308:
1304:
1300:
1296:
1292:
1285:
1283:
1281:
1279:
1270:
1266:
1262:
1258:
1254:
1250:
1246:
1242:
1238:
1234:
1227:
1225:
1223:
1214:
1210:
1206:
1202:
1198:
1194:
1190:
1186:
1182:
1178:
1171:
1169:
1160:
1156:
1152:
1146:
1142:
1138:
1131:
1130:
1122:
1120:
1111:
1107:
1102:
1097:
1093:
1092:10.3791/55496
1089:
1085:
1081:
1077:
1070:
1062:
1058:
1053:
1048:
1043:
1038:
1034:
1030:
1026:
1022:
1018:
1011:
1007:
1000:
996:
994:
990:
986:
981:
979:
975:
970:
965:
962:
958:
956:
951:
941:
939:
935:
931:
927:
923:
919:
915:
910:
908:
904:
868:
856:
852:
849:
845:
840:
838:
834:
830:
826:
815:
813:
809:
805:
801:
790:
781:
773:
769:
766:
762:
758:
754:
715:
713:
707:
705:
700:
698:
693:
692:nucleic acids
688:
682:
680:
676:
671:
669:
665:
655:
651:
648:
638:
634:
631:
626:
622:
620:
615:
612:
606:
602:
600:
594:
591:
587:
583:
573:
571:
567:
563:
548:
545:
540:
536:
532:
521:
519:
515:
511:
505:
504:
500:
498:
493:
489:
485:
481:
477:
473:
462:
460:
456:
452:
447:
443:
439:
434:
424:
422:
421:
416:
412:
408:
404:
400:
390:
388:
384:
383:transcription
380:
376:
372:
368:
364:
360:
356:
353:
349:
345:
343:
339:
335:
331:
327:
324:
320:
316:
313:
310:
306:
302:
298:
295:
291:
281:
279:
275:
271:
268:
264:
261:
257:
253:
249:
248:
243:
242:
237:
228:
225:
219:
216:
210:
208:
204:
200:
196:
192:
188:
184:
180:
170:
167:
163:
161:
146:
142:
135:
131:
129:
124:
120:
116:
112:
108:
104:
100:
90:
88:
84:
80:
76:
73:, and may be
72:
68:
64:
60:
56:
53:, especially
52:
47:
45:
41:
37:
33:
30:
21:
7260:G-quadruplex
7127:
7123:
7115:the original
7099:
7073:
7069:
7033:
7029:
6980:
6976:
6937:
6933:
6894:
6890:
6851:
6847:
6808:
6804:
6775:
6771:
6698:
6692:
6655:
6651:
6641:
6616:
6612:
6606:
6581:
6578:Biochemistry
6577:
6571:
6536:
6532:
6522:
6497:
6493:
6486:
6451:
6448:Biochemistry
6447:
6437:
6402:
6398:
6388:
6343:
6339:
6329:
6286:
6282:
6272:
6237:
6233:
6222:
6177:
6173:
6162:
6127:
6123:
6112:
6077:
6073:
6063:
6038:
6034:
6028:
5995:
5991:
5985:
5976:
5967:
5940:
5936:
5926:
5899:
5895:
5885:
5850:
5846:
5836:
5793:
5789:
5779:
5742:
5738:
5728:
5685:
5681:
5671:
5636:
5632:
5617:
5582:
5578:
5567:
5534:
5530:
5524:
5515:
5506:
5479:
5476:FEBS Letters
5475:
5465:
5424:
5421:Nano Letters
5420:
5414:
5369:
5365:
5355:
5323:(1): 74–78.
5320:
5317:FEBS Letters
5316:
5306:
5281:
5277:
5271:
5226:
5220:
5185:
5181:
5171:
5146:
5142:
5129:
5096:
5092:
5082:
5039:
5035:
5025:
5008:
5004:
4998:
4957:
4953:
4947:
4912:
4908:
4898:
4876:(9): 65–95.
4873:
4869:
4863:
4828:
4824:
4814:
4789:
4785:
4775:
4730:
4726:
4716:
4673:
4669:
4659:
4626:
4623:Biochemistry
4622:
4615:
4580:
4576:
4566:
4531:
4527:
4517:
4492:
4489:Biochemistry
4488:
4481:
4441:(1): 11541.
4438:
4434:
4423:
4388:
4384:
4374:
4349:
4336:
4311:
4307:
4262:
4258:
4248:
4183:
4177:
4152:
4148:
4135:
4100:
4096:
4086:
4051:
4047:
4037:
4002:
3998:
3988:
3953:
3949:
3901:
3897:
3849:
3845:
3797:
3793:
3751:
3747:
3741:
3706:
3702:
3654:
3650:
3602:
3598:
3536:
3532:
3444:
3398:
3394:
3384:
3349:
3345:
3335:
3292:
3288:
3278:
3238:(3): 182–6.
3235:
3231:
3221:
3184:
3180:
3170:
3133:
3129:
3119:
3082:
3078:
3046:
3042:
3036:
3001:
2997:
2965:
2961:
2951:
2924:
2920:
2910:
2865:
2861:
2851:
2816:
2812:
2802:
2767:
2763:
2753:
2718:
2715:Biochemistry
2714:
2666:
2662:
2652:
2617:
2613:
2603:
2558:
2554:
2544:
2509:
2505:
2495:
2461:(14): 7381.
2458:
2454:
2444:
2407:
2403:
2395:
2387:
2354:
2350:
2344:
2299:
2295:
2285:
2250:
2246:
2236:
2209:
2205:
2195:
2160:
2156:
2146:
2121:
2117:
2111:
2087:(25): 22–9.
2084:
2080:
2070:
2025:
2021:
2011:
1976:
1972:
1962:
1927:
1923:
1913:
1883:(4): 621–8.
1880:
1876:
1870:
1835:
1831:
1820:
1777:
1773:
1763:
1730:
1726:
1720:
1675:
1671:
1661:
1644:
1640:
1634:
1599:
1595:
1585:
1550:
1546:
1536:
1501:
1497:
1487:
1452:
1448:
1438:
1413:
1409:
1403:
1378:
1374:
1367:
1350:
1346:
1298:
1294:
1236:
1232:
1180:
1176:
1128:
1083:
1079:
1069:
1024:
1020:
1010:
997:
982:
966:
959:
947:
911:
841:
821:
796:
787:
779:
770:
721:
708:
703:
701:
683:
679:telomestatin
672:
661:
652:
644:
641:Therapeutics
635:
627:
623:
619:angiogenesis
616:
607:
603:
595:
589:
579:
570:cell's death
559:
531:8-oxoguanine
527:
506:
502:
501:
468:
430:
418:
396:
371:upregulation
366:
358:
346:
287:
278:telomestatin
245:
239:
234:
220:
211:
176:
168:
164:
156:
127:
122:
114:
110:
96:
87:antiparallel
48:
29:G-quadruplex
28:
26:
7166:{NAB group}
5099:: 115–118.
3800:: 207–219.
3445:The Nucleus
1979:(1): 3–10.
855:presynaptic
755:(including
363:prokaryotes
252:vertebrates
79:bimolecular
7254:Categories
5977:Gene Cards
2968:(8): 253.
1003:References
918:elongation
761:algorithms
675:porphyrins
562:eukaryotic
514:epigenetic
484:tautomeric
270:telomerase
258:, TEM and
224:telomerase
203:chromosome
119:Aaron Klug
7193:GRS_UTRdb
6805:Biochimie
6751:ignored (
6741:cite book
5625:Rossor MN
5531:Neurology
5457:203638480
5093:Biochimie
5056:1367-4803
4676:: 11385.
4366:214472968
4238:ignored (
4228:cite book
3495:cite book
3487:220631202
3471:1940-6029
3352:: 161–7.
978:SCID mice
907:ribosomes
903:nucleolin
851:cytoplasm
827:(ALS) or
765:eumetazoa
673:Cationic
582:telomeric
556:Telomeres
433:AP sites.
420:Neisseria
381:blocking
319:ubiquitin
236:Telomeric
215:Telomeres
183:chromatin
107:telomeric
55:potassium
40:Hoogsteen
7196:Archived
7176:Greglist
7144:24148836
7090:19435350
7060:16651665
7017:12195017
6964:17012276
6921:15914666
6878:15914667
6835:18331848
6792:21858307
6733:44981950
6725:29394031
6684:24371143
6633:24814531
6598:19173611
6563:23471001
6514:19099510
6478:21028906
6429:21971427
6380:22371589
6321:22859208
6264:24154603
6214:24170860
6154:24139042
6104:25188012
6020:31455523
5959:19097999
5918:14570712
5869:11719189
5828:24598541
5771:23423380
5720:23264878
5688:: 1016.
5663:23434116
5609:22840558
5559:45904851
5551:12058088
5498:25979174
5449:31577148
5406:25225404
5298:17967702
5263:20012416
5163:28755976
5121:47005625
5113:29885355
5074:30721922
5017:20656986
4939:18614607
4855:15914666
4806:22280460
4767:12195017
4708:26077929
4651:53093959
4643:30369235
4607:17984069
4558:17672459
4509:16331995
4473:28912501
4415:16239639
4328:21774525
4289:20670278
4220:22956454
4169:26840940
4127:19934257
4078:14633989
4029:18809583
3980:27994014
3928:31226203
3876:28936535
3824:31993111
3768:11848927
3733:11139613
3681:19385599
3629:24485507
3573:32404420
3479:32681486
3423:20967177
3415:27618450
3376:27239262
3327:29899445
3270:23422559
3213:23637633
3162:31575660
3111:28717007
3028:19033359
2962:Crystals
2943:16150737
2902:22272300
2862:PLOS ONE
2843:16651665
2794:15914667
2745:16784237
2693:16433524
2644:17169996
2595:12195017
2487:34299001
2436:29674319
2371:16142245
2336:11438689
2277:21441540
2228:26810457
2187:19858105
2138:16771510
2103:24584093
2062:26727593
2022:PLOS ONE
2003:22652626
1954:17012276
1905:43536134
1897:11405224
1862:22306580
1812:21107376
1755:37343642
1712:13947099
1626:26350216
1577:27185890
1528:17962308
1479:23032257
1430:10740289
1395:18767830
1325:16651665
1159:26860303
1110:28362374
1061:20676380
922:dendrite
848:neuronal
800:nanopore
697:π-π bond
668:proteins
586:oncogene
566:helicase
393:Function
379:promoter
317:, human
312:β-globin
305:promoter
301:nuclease
290:telomere
241:in vitro
153:Topology
148:)
111:in vitro
83:parallel
7051:1457047
6985:Bibcode
6955:1636468
6912:1140077
6869:1140081
6826:2585026
6675:3931028
6554:3627599
6469:2991413
6420:3242664
6371:3306674
6348:Bibcode
6312:4221572
6291:Bibcode
6255:4090945
6205:3839752
6182:Bibcode
6145:4098943
6095:4165481
6055:1301913
6012:1878973
5877:8203054
5819:4046618
5798:Bibcode
5762:3617286
5711:3527825
5690:Bibcode
5654:3591848
5600:3617405
5429:Bibcode
5397:4209999
5374:Bibcode
5347:1306129
5339:9755862
5278:Methods
5254:2797547
5212:9862977
5065:6748775
4990:4360764
4982:2825028
4962:Bibcode
4930:2504309
4890:7574496
4846:1140077
4735:Bibcode
4699:4468576
4678:Bibcode
4598:2190695
4549:2566970
4464:5599563
4443:Bibcode
4406:1266068
4350:bioRxiv
4280:2971675
4211:3463244
4118:2817463
4020:2593380
3971:5335514
3919:6895268
3867:5636683
3815:6974700
3672:2757002
3620:5576885
3564:7260947
3541:Bibcode
3367:4872678
3318:6261253
3297:Bibcode
3261:3622242
3240:Bibcode
3204:3630100
3153:6879327
3102:5602382
3019:2615625
2893:3260238
2870:Bibcode
2834:1457047
2785:1140081
2736:2195898
2684:2556172
2635:1802602
2563:Bibcode
2536:9469822
2478:8306923
2427:6005428
2379:6079323
2304:Bibcode
2268:3152327
2178:2800236
2053:4699641
2030:Bibcode
1994:3701776
1973:Methods
1945:1636468
1853:3433707
1803:3119466
1782:Bibcode
1747:3690664
1680:Bibcode
1617:4605312
1568:4987949
1519:2238983
1470:3725559
1316:1457047
1269:4351855
1261:3393228
1241:Bibcode
1213:4357161
1205:2601741
1185:Bibcode
1101:5409278
1086:(121).
1052:2908698
1029:Bibcode
985:ligands
955:C9orf72
950:ligands
853:and at
844:C9orf72
757:guanine
664:ligands
490:enzyme
472:Guanine
367:E. coli
359:in vivo
309:chicken
274:cancers
247:in vivo
207:hairpin
187:histone
160:meiosis
128:in vivo
123:in vivo
115:in vivo
99:guanine
93:History
36:guanine
7142:
7107:
7088:
7058:
7048:
7015:
7008:129314
7005:
6962:
6952:
6919:
6909:
6876:
6866:
6833:
6823:
6790:
6731:
6723:
6713:
6682:
6672:
6631:
6596:
6561:
6551:
6512:
6476:
6466:
6427:
6417:
6378:
6368:
6319:
6309:
6283:Nature
6262:
6252:
6212:
6202:
6152:
6142:
6124:Neuron
6102:
6092:
6053:
6018:
6010:
5957:
5916:
5875:
5867:
5826:
5816:
5790:Nature
5769:
5759:
5718:
5708:
5661:
5651:
5607:
5597:
5557:
5549:
5496:
5455:
5447:
5404:
5394:
5345:
5337:
5296:
5261:
5251:
5241:
5210:
5203:148212
5200:
5161:
5119:
5111:
5072:
5062:
5054:
5015:
4988:
4980:
4954:Nature
4937:
4927:
4888:
4853:
4843:
4804:
4765:
4758:129314
4755:
4706:
4696:
4649:
4641:
4605:
4595:
4556:
4546:
4507:
4471:
4461:
4413:
4403:
4364:
4326:
4287:
4277:
4218:
4208:
4198:
4167:
4125:
4115:
4076:
4069:291836
4066:
4027:
4017:
3978:
3968:
3926:
3916:
3874:
3864:
3822:
3812:
3766:
3731:
3721:
3679:
3669:
3627:
3617:
3571:
3561:
3485:
3477:
3469:
3459:
3421:
3413:
3374:
3364:
3325:
3315:
3289:Nature
3268:
3258:
3211:
3201:
3160:
3150:
3109:
3099:
3026:
3016:
2941:
2900:
2890:
2841:
2831:
2792:
2782:
2743:
2733:
2691:
2681:
2642:
2632:
2593:
2586:129314
2583:
2534:
2527:147388
2524:
2485:
2475:
2434:
2424:
2377:
2369:
2334:
2324:
2275:
2265:
2226:
2185:
2175:
2136:
2101:
2060:
2050:
2001:
1991:
1952:
1942:
1903:
1895:
1860:
1850:
1810:
1800:
1753:
1745:
1710:
1703:221115
1700:
1624:
1614:
1575:
1565:
1526:
1516:
1477:
1467:
1428:
1393:
1323:
1313:
1267:
1259:
1233:Nature
1211:
1203:
1177:Nature
1157:
1147:
1108:
1098:
1059:
1049:
926:autism
867:intron
704:c-Myc.
630:kinase
551:Cancer
355:genome
348:Genome
267:enzyme
179:genome
69:, and
51:cation
7187:GRSDB
6729:S2CID
6016:S2CID
5873:S2CID
5555:S2CID
5453:S2CID
5343:S2CID
5139:(PDF)
5117:S2CID
4986:S2CID
4647:S2CID
4362:S2CID
4346:(PDF)
4145:(PDF)
3956:(6).
3724:29662
3483:S2CID
3419:S2CID
2375:S2CID
2327:37477
1901:S2CID
1751:S2CID
1265:S2CID
1209:S2CID
1133:(PDF)
687:BCL-2
590:c-Myc
411:DHX36
352:human
342:N-ras
338:H-ras
330:bcl-2
326:c-kit
297:c-myc
260:X-ray
7140:PMID
7105:ISBN
7086:PMID
7056:PMID
7013:PMID
6960:PMID
6917:PMID
6874:PMID
6831:PMID
6788:PMID
6753:help
6721:PMID
6711:ISBN
6680:PMID
6629:PMID
6594:PMID
6559:PMID
6510:PMID
6474:PMID
6425:PMID
6376:PMID
6317:PMID
6260:PMID
6210:PMID
6150:PMID
6100:PMID
6051:PMID
6008:PMID
5992:Cell
5955:PMID
5914:PMID
5865:PMID
5847:Cell
5824:PMID
5767:PMID
5716:PMID
5659:PMID
5605:PMID
5547:PMID
5494:PMID
5445:PMID
5402:PMID
5335:PMID
5294:PMID
5259:PMID
5239:ISBN
5208:PMID
5159:PMID
5109:PMID
5070:PMID
5052:ISSN
5013:PMID
4978:PMID
4935:PMID
4886:PMID
4851:PMID
4802:PMID
4763:PMID
4704:PMID
4639:PMID
4603:PMID
4554:PMID
4505:PMID
4469:PMID
4411:PMID
4324:PMID
4285:PMID
4240:help
4216:PMID
4196:ISBN
4165:PMID
4123:PMID
4074:PMID
4025:PMID
3976:PMID
3924:PMID
3872:PMID
3820:PMID
3764:PMID
3729:PMID
3677:PMID
3625:PMID
3569:PMID
3501:link
3475:PMID
3467:ISSN
3457:ISBN
3411:PMID
3372:PMID
3323:PMID
3266:PMID
3209:PMID
3158:PMID
3107:PMID
3024:PMID
2939:PMID
2898:PMID
2839:PMID
2790:PMID
2741:PMID
2689:PMID
2640:PMID
2591:PMID
2532:PMID
2483:PMID
2432:PMID
2367:PMID
2332:PMID
2273:PMID
2224:PMID
2183:PMID
2134:PMID
2099:PMID
2058:PMID
1999:PMID
1950:PMID
1893:PMID
1858:PMID
1808:PMID
1743:PMID
1727:Cell
1708:PMID
1622:PMID
1573:PMID
1524:PMID
1475:PMID
1426:PMID
1391:PMID
1321:PMID
1257:PMID
1201:PMID
1155:PMID
1145:ISBN
1106:PMID
1057:PMID
930:exon
810:and
539:OGG1
535:OGG1
492:OGG1
405:and
340:and
334:VEGF
315:gene
145:2HY9
117:” -
7270:RNA
7265:DNA
7132:doi
7078:doi
7074:131
7046:PMC
7038:doi
7003:PMC
6993:doi
6950:PMC
6942:doi
6907:PMC
6899:doi
6864:PMC
6856:doi
6821:PMC
6813:doi
6780:doi
6703:doi
6670:PMC
6660:doi
6656:289
6621:doi
6586:doi
6549:PMC
6541:doi
6502:doi
6464:PMC
6456:doi
6415:PMC
6407:doi
6366:PMC
6356:doi
6344:109
6307:PMC
6299:doi
6287:488
6250:PMC
6242:doi
6200:PMC
6190:doi
6178:110
6140:PMC
6132:doi
6090:PMC
6082:doi
6043:doi
6000:doi
5945:doi
5941:284
5904:doi
5855:doi
5851:107
5814:PMC
5806:doi
5794:507
5757:PMC
5747:doi
5743:288
5706:PMC
5698:doi
5649:PMC
5641:doi
5595:PMC
5587:doi
5539:doi
5484:doi
5480:589
5437:doi
5392:PMC
5382:doi
5370:111
5325:doi
5321:435
5286:doi
5249:PMC
5231:doi
5198:PMC
5190:doi
5151:doi
5101:doi
5097:151
5060:PMC
5044:doi
4970:doi
4958:330
4925:PMC
4917:doi
4878:doi
4841:PMC
4833:doi
4794:doi
4790:134
4753:PMC
4743:doi
4694:PMC
4686:doi
4631:doi
4593:PMC
4585:doi
4544:PMC
4536:doi
4532:129
4497:doi
4459:PMC
4451:doi
4401:PMC
4393:doi
4354:doi
4316:doi
4275:PMC
4267:doi
4263:277
4206:PMC
4188:doi
4157:doi
4113:PMC
4105:doi
4064:PMC
4056:doi
4015:PMC
4007:doi
3966:PMC
3958:doi
3914:PMC
3906:doi
3862:PMC
3854:doi
3810:PMC
3802:doi
3756:doi
3719:PMC
3711:doi
3667:PMC
3659:doi
3615:PMC
3607:doi
3559:PMC
3549:doi
3537:117
3449:doi
3403:doi
3362:PMC
3354:doi
3313:PMC
3305:doi
3293:558
3256:PMC
3248:doi
3199:PMC
3189:doi
3148:PMC
3138:doi
3134:294
3097:PMC
3087:doi
3083:292
3051:doi
3014:PMC
3006:doi
2970:doi
2929:doi
2925:280
2888:PMC
2878:doi
2829:PMC
2821:doi
2780:PMC
2772:doi
2731:PMC
2723:doi
2679:PMC
2671:doi
2667:128
2630:PMC
2622:doi
2581:PMC
2571:doi
2522:PMC
2514:doi
2473:PMC
2463:doi
2422:PMC
2412:doi
2408:293
2359:doi
2322:PMC
2312:doi
2263:PMC
2255:doi
2214:doi
2173:PMC
2165:doi
2126:doi
2122:128
2089:doi
2048:PMC
2038:doi
1989:PMC
1981:doi
1940:PMC
1932:doi
1885:doi
1881:382
1848:PMC
1840:doi
1798:PMC
1790:doi
1735:doi
1698:PMC
1688:doi
1649:doi
1612:PMC
1604:doi
1563:PMC
1555:doi
1514:PMC
1506:doi
1465:PMC
1457:doi
1418:doi
1383:doi
1355:doi
1311:PMC
1303:doi
1249:doi
1237:334
1193:doi
1181:342
1137:doi
1096:PMC
1088:doi
1084:121
1047:PMC
1037:doi
745:1-7
737:1-7
729:1-7
723:d(G
403:WRN
373:or
256:NMR
193:of
141:PDB
103:DNA
85:or
71:PNA
67:LNA
63:RNA
59:DNA
7256::
7138:.
7128:21
7126:.
7084:.
7072:.
7054:.
7044:.
7034:16
7032:.
7028:.
7011:.
7001:.
6991:.
6981:99
6979:.
6975:.
6958:.
6948:.
6938:34
6936:.
6932:.
6915:.
6905:.
6895:33
6893:.
6889:.
6872:.
6862:.
6852:33
6850:.
6846:.
6829:.
6819:.
6809:90
6807:.
6803:.
6786:.
6776:47
6774:.
6745::
6743:}}
6739:{{
6727:.
6719:.
6709:.
6678:.
6668:.
6654:.
6650:.
6627:.
6617:24
6615:.
6592:.
6582:48
6580:.
6557:.
6547:.
6537:41
6535:.
6531:.
6508:.
6498:52
6496:.
6472:.
6462:.
6452:49
6450:.
6446:.
6423:.
6413:.
6403:19
6401:.
6397:.
6374:.
6364:.
6354:.
6342:.
6338:.
6315:.
6305:.
6297:.
6285:.
6281:.
6258:.
6248:.
6236:.
6232:.
6208:.
6198:.
6188:.
6176:.
6172:.
6148:.
6138:.
6128:80
6126:.
6122:.
6098:.
6088:.
6078:27
6076:.
6072:.
6049:.
6037:.
6014:.
6006:.
5996:66
5994:.
5975:.
5953:.
5939:.
5935:.
5912:.
5900:12
5898:.
5894:.
5871:.
5863:.
5849:.
5845:.
5822:.
5812:.
5804:.
5792:.
5788:.
5765:.
5755:.
5741:.
5737:.
5714:.
5704:.
5696:.
5684:.
5680:.
5657:.
5647:.
5637:92
5635:.
5631:.
5603:.
5593:.
5583:33
5581:.
5577:.
5553:.
5545:.
5535:58
5533:.
5514:.
5492:.
5478:.
5474:.
5451:.
5443:.
5435:.
5425:19
5423:.
5400:.
5390:.
5380:.
5368:.
5364:.
5341:.
5333:.
5319:.
5315:.
5292:.
5282:43
5280:.
5257:.
5247:.
5237:.
5206:.
5196:.
5186:27
5184:.
5180:.
5157:.
5147:35
5145:.
5141:.
5115:.
5107:.
5095:.
5091:.
5068:.
5058:.
5050:.
5040:35
5038:.
5034:.
5007:.
4984:.
4976:.
4968:.
4956:.
4933:.
4923:.
4913:36
4911:.
4907:.
4884:.
4874:64
4872:.
4849:.
4839:.
4829:33
4827:.
4823:.
4800:.
4788:.
4784:.
4761:.
4751:.
4741:.
4731:99
4729:.
4725:.
4702:.
4692:.
4684:.
4672:.
4668:.
4645:.
4637:.
4627:57
4625:.
4601:.
4591:.
4581:35
4579:.
4575:.
4552:.
4542:.
4530:.
4526:.
4503:.
4493:44
4491:.
4467:.
4457:.
4449:.
4437:.
4433:.
4409:.
4399:.
4389:33
4387:.
4383:.
4360:.
4352:.
4348:.
4322:.
4312:54
4310:.
4297:^
4283:.
4273:.
4261:.
4257:.
4232::
4230:}}
4226:{{
4214:.
4204:.
4194:.
4163:.
4153:59
4151:.
4147:.
4121:.
4111:.
4101:38
4099:.
4095:.
4072:.
4062:.
4052:22
4050:.
4046:.
4023:.
4013:.
4003:28
4001:.
3997:.
3974:.
3964:.
3954:37
3952:.
3948:.
3936:^
3922:.
3912:.
3902:47
3900:.
3896:.
3884:^
3870:.
3860:.
3850:15
3848:.
3844:.
3832:^
3818:.
3808:.
3798:18
3796:.
3792:.
3776:^
3762:.
3752:98
3750:.
3727:.
3717:.
3707:29
3705:.
3701:.
3689:^
3675:.
3665:.
3655:52
3653:.
3649:.
3637:^
3623:.
3613:.
3603:35
3601:.
3597:.
3581:^
3567:.
3557:.
3547:.
3535:.
3531:.
3509:^
3497:}}
3493:{{
3481:.
3473:.
3465:.
3455:.
3443:.
3431:^
3417:.
3409:.
3399:48
3397:.
3393:.
3370:.
3360:.
3350:14
3348:.
3344:.
3321:.
3311:.
3303:.
3291:.
3287:.
3264:.
3254:.
3246:.
3234:.
3230:.
3207:.
3197:.
3183:.
3179:.
3156:.
3146:.
3132:.
3128:.
3105:.
3095:.
3081:.
3077:.
3063:^
3047:87
3045:.
3022:.
3012:.
3002:37
3000:.
2996:.
2984:^
2964:.
2960:.
2937:.
2923:.
2919:.
2896:.
2886:.
2876:.
2864:.
2860:.
2837:.
2827:.
2817:16
2815:.
2811:.
2788:.
2778:.
2768:33
2766:.
2762:.
2739:.
2729:.
2719:45
2717:.
2713:.
2701:^
2687:.
2677:.
2665:.
2661:.
2638:.
2628:.
2618:35
2616:.
2612:.
2589:.
2579:.
2569:.
2559:99
2557:.
2553:.
2530:.
2520:.
2510:26
2508:.
2504:.
2481:.
2471:.
2459:22
2457:.
2453:.
2430:.
2420:.
2406:.
2402:.
2373:.
2365:.
2355:12
2353:.
2330:.
2320:.
2310:.
2300:98
2298:.
2294:.
2271:.
2261:.
2251:39
2249:.
2245:.
2222:.
2210:88
2208:.
2204:.
2181:.
2171:.
2161:38
2159:.
2155:.
2132:.
2120:.
2097:.
2085:25
2083:.
2079:.
2056:.
2046:.
2036:.
2026:11
2024:.
2020:.
1997:.
1987:.
1977:57
1975:.
1971:.
1948:.
1938:.
1928:34
1926:.
1922:.
1899:.
1891:.
1879:.
1856:.
1846:.
1834:.
1830:.
1806:.
1796:.
1788:.
1776:.
1772:.
1749:.
1741:.
1731:51
1729:.
1706:.
1696:.
1686:.
1676:48
1674:.
1670:.
1645:87
1643:.
1620:.
1610:.
1600:43
1598:.
1594:.
1571:.
1561:.
1551:44
1549:.
1545:.
1522:.
1512:.
1502:36
1500:.
1496:.
1473:.
1463:.
1453:13
1451:.
1447:.
1424:.
1414:21
1412:.
1389:.
1379:51
1377:.
1351:85
1349:.
1333:^
1319:.
1309:.
1299:16
1297:.
1293:.
1277:^
1263:.
1255:.
1247:.
1235:.
1221:^
1207:.
1199:.
1191:.
1179:.
1167:^
1153:.
1143:.
1118:^
1104:.
1094:.
1082:.
1078:.
1055:.
1045:.
1035:.
1023:.
1019:.
839:.
749:3+
741:3+
733:3+
725:3+
681:.
572:.
344:.
336:,
332:,
328:,
280:.
143::
77:,
65:,
61:,
7146:.
7134::
7092:.
7080::
7062:.
7040::
7019:.
6995::
6987::
6966:.
6944::
6923:.
6901::
6880:.
6858::
6837:.
6815::
6794:.
6782::
6755:)
6735:.
6705::
6686:.
6662::
6635:.
6623::
6600:.
6588::
6565:.
6543::
6516:.
6504::
6480:.
6458::
6431:.
6409::
6382:.
6358::
6350::
6323:.
6301::
6293::
6266:.
6244::
6238:5
6216:.
6192::
6184::
6156:.
6134::
6106:.
6084::
6057:.
6045::
6039:1
6022:.
6002::
5979:.
5961:.
5947::
5920:.
5906::
5879:.
5857::
5830:.
5808::
5800::
5773:.
5749::
5722:.
5700::
5692::
5686:2
5665:.
5643::
5611:.
5589::
5561:.
5541::
5500:.
5486::
5459:.
5439::
5431::
5408:.
5384::
5376::
5349:.
5327::
5300:.
5288::
5265:.
5233::
5214:.
5192::
5165:.
5153::
5123:.
5103::
5076:.
5046::
5019:.
5009:7
4992:.
4972::
4964::
4941:.
4919::
4892:.
4880::
4857:.
4835::
4808:.
4796::
4769:.
4745::
4737::
4710:.
4688::
4680::
4674:5
4653:.
4633::
4609:.
4587::
4560:.
4538::
4511:.
4499::
4475:.
4453::
4445::
4439:7
4417:.
4395::
4368:.
4356::
4330:.
4318::
4291:.
4269::
4242:)
4222:.
4190::
4171:.
4159::
4129:.
4107::
4080:.
4058::
4031:.
4009::
3982:.
3960::
3930:.
3908::
3878:.
3856::
3826:.
3804::
3770:.
3758::
3735:.
3713::
3683:.
3661::
3631:.
3609::
3575:.
3551::
3543::
3503:)
3489:.
3451::
3425:.
3405::
3378:.
3356::
3329:.
3307::
3299::
3272:.
3250::
3242::
3236:5
3215:.
3191::
3185:9
3164:.
3140::
3113:.
3089::
3057:.
3053::
3030:.
3008::
2978:.
2972::
2966:7
2945:.
2931::
2904:.
2880::
2872::
2866:7
2845:.
2823::
2796:.
2774::
2747:.
2725::
2695:.
2673::
2646:.
2624::
2597:.
2573::
2565::
2538:.
2516::
2489:.
2465::
2438:.
2414::
2397:n
2381:.
2361::
2338:.
2314::
2306::
2279:.
2257::
2230:.
2216::
2189:.
2167::
2140:.
2128::
2105:.
2091::
2064:.
2040::
2032::
2005:.
1983::
1956:.
1934::
1907:.
1887::
1864:.
1842::
1836:8
1814:.
1792::
1784::
1778:2
1757:.
1737::
1714:.
1690::
1682::
1655:.
1651::
1628:.
1606::
1579:.
1557::
1530:.
1508::
1481:.
1459::
1432:.
1420::
1397:.
1385::
1361:.
1357::
1327:.
1305::
1271:.
1251::
1243::
1215:.
1195::
1187::
1161:.
1139::
1112:.
1090::
1063:.
1039::
1031::
1025:6
899:2
897:C
895:4
891:2
889:C
887:4
883:2
881:C
879:4
875:2
873:C
871:4
863:2
861:C
859:4
747:G
743:N
739:G
735:N
731:G
727:N
138:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.