308:
585:
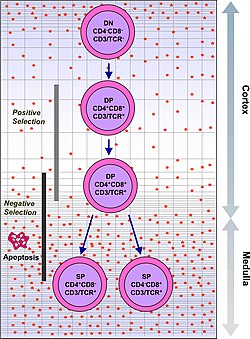
is the medulla, once cells are at the single positive stage. In order to remove thymocytes reactive to peripheral organs, the transcription factors Aire and Fezf2 drive the expression of multiple peripheral antigens, such as insulin, resulting in deletion of cells specific for those antigens. This allows single positive thymocytes to be exposed to a more complex set of self-antigens than is present in the cortex, and therefore more efficiently deletes those T cells which are autoreactive.
500:
644:
Natural Killer lymphocytes (NK cells).), myeloid cells, and dendritic cells. However, the thymus is not a source of B, NKC, or myeloid development (this statement is not true for all B-cells or NKC). The development of these cells in the thymus reflects the multi-potent nature of hematopoietic progenitors that seed the thymus. Mature B-cells and other APCs can also be found in the medulla which contribute to negative selection processes.
543:
T cell receptor) and then upregulation of CD4 only. Thymocytes that start receiving signal again are those that recognise MHC class II, and they become CD4+ T cells. Thymocytes that do not start receiving signal again are those that recognize MHC class I, and they downregulate CD4 and upregulate CD8, to become CD8+ T cells. Both of these thymocytes types are known as single positive thymocytes.
429:
421:
385:
Lymphoid
Progenitors (ELP) are proposed to settle the thymus and are likely the precursors of at least some thymocytes. ELPs are Lineage-CD44+CD25-CD117+ and thus closely resemble ETPs, the earliest progenitors in the thymus. Precursors enter the thymus at the cortico-medullary junction. Molecules known to be important for thymus entry include
542:
The double positive thymocytes undergo lineage commitment, maturing into a CD8+ T cell (recognising MHC class I) or a CD4+ T cell (recognising MHC class II). Lineage commitment occurs at the late stage of positive selection and works by downregulation of both CD4 and CD8 (reducing the signal from the
487:
In the β-selection stage similarly to the mature TCR, pre-TCR also forms an immunological synapse. Although the pre-TCR and the peptide-bound MHC interaction is not essential for T cell development, it plays a critical role in encouraging the preferential proliferation of cells whose pre-TCR can bind
618:
results in that T cells are taught to avoid reacting with donor antigens instead, and may still react with many self-antigens in the body. Autoimmune disease is a frequent complication after thymus transplantation, found in 42% of subjects over 1 year post-transplantation. However, this is partially
568:
is that by random chance, some arrangements of gene fragments will create a T cell receptor capable of binding self-peptides presented on MHC class I or MHC class II. If T cells bearing these T cell receptors were to enter the periphery, they would be capable of activating an immune response against
584:
Negative selection can occur at the double positive stage in the cortex. However the repertoire of peptides in the cortex is limited to those expressed by epithelial cells, and double positive cells are poor at undergoing negative selection. Therefore, the most important site for negative selection
384:
Thymocytes are ultimately derived from bone marrow hematopoietic progenitor cells which reach the thymus through the circulation. The number of progenitors that enter the thymus each day is thought to be extremely small. Therefore, which progenitors colonize the thymus is unknown. Currently Early
643:
As well as classical αβ T cells (their development of which is outlined above), a number of other T lineages develop in the thymus, including γδ T cells and
Natural Killer T (NKT)cells. Additionally, other non-T hematopoietic lineages can develop in the thymus, including B lymphocytes (B cells),
352:
Negative selection is the active induction of apoptosis in thymocytes with a high affinity for self peptides or MHC. This eliminates cells which would direct immune responses towards self-proteins in the periphery. Negative selection is not 100% effective, some autoreactive T cells escape thymic
66:
class I or II molecules with at least a weak affinity. This eliminates (by a process called "death by neglect") those T cells which would be non-functional due to an inability to bind MHC. Negative selection destroys thymocytes with a high affinity for self peptides or MHC. This eliminates cells
83:
Thymocytes are classified into a number of distinct maturational stages based on the expression of cell surface markers. The earliest thymocyte stage is the double negative stage (negative for both CD4 and CD8), which more recently has been better described as
Lineage-negative, and which can be
471:
TCR rearrangement occurs in two steps. First the TCRβ chain is rearranged at the DN3 stage of T cell development. The TCRβ chain is paired with the pre-Tα to generate the pre-TCR. The cellular disadvantage in the rearrangement process is that many of the combinations of the
455:
is made up of a series of alternative gene fragments. In order to create a functional T cell receptor, the double negative thymocytes use a series of DNA-interacting enzymes to clip the DNA and bring separate gene fragments together. The outcome of this process is that each
484:. This process is referred to as the beta-selection checkpoint. Successful beta-selection requires that TCRβ is produced, TCRβ is capable of pairing with pre-Tα to generate the pre-TCR, and that the pre-TCR can interact on the cell surface with the TCR signalling proteins.
53:
describes the process which turns thymocytes into mature T cells according to either negative or positive selection. This selection process is vitally important in shaping the population of thymocytes into a peripheral pool of T cells that are able to respond to foreign
403:
Following thymus entry, progenitors proliferate to generate the ETP population. This step is followed by the generation of DN2 thymocytes which migrate from the cortico-medullary junction toward the thymus capsule. DN3 thymocytes are generated at the subcapsular zone.
534:
Double positive thymocytes that have a T cell receptor capable of binding MHC class I or class II (even with a weak affinity) receive signalling through the T cell receptor. Thymocytes that have a T cell receptor incapable of binding MHC class I or class II undergo
588:
Single positive thymocytes remain in the medulla for 1–2 weeks, surveying self-antigens to test for autoreactivity. During this time they undergo final maturational changes, and then exit the thymus using S1P and CCR7. Upon entry to the peripheral
407:
In addition to proliferation, differentiation and T lineage commitment occurs within the DN thymocyte population. Commitment, or loss of alternative lineage potentials (such as myeloid, B, and NK lineage potentials), is dependent on
519:. The next major stage of thymocyte development is positive selection, to keep only those thymocytes which have a T cell receptor capable of binding MHC. The T cell receptor requires CD8 as a coreceptor to bind to MHC class I, and
797:
67:
which would direct immune responses towards self-proteins in the periphery. Negative selection is not 100% effective, and some autoreactive T cells escape and are released into the circulation. Additional mechanisms of
573:. Negative selection is the process evolved to reduce this risk. During negative selection, all thymocytes with a high affinity for binding self peptides presented on MHC class I or class II are induced to upregulate
318:
rearranged by the thymocyte must retain the structural properties allowing it to be presented on the surface of the thymocyte with pre-TCRα. This eliminates thymocytes with gross defects introduced into the
480:, only cells that have successfully rearranged the beta chain to produce a functional pre-TCR are allowed to develop beyond the DN3 stage. Cells that fail to produce a functional pre-TCR are eliminated by
84:
divided into four substages. The next major stage is the double positive stage (positive for both CD4 and CD8). The final stage in maturation is the single positive stage (positive for either CD4 or CD8).
659:
1025:
Li, Xiaolong; Mizsei, Réka; Tan, Kemin; Mallis, Robert J.; Duke-Cohan, Jonathan S.; Akitsu, Aoi; Tetteh, Paul W.; Dubey, Abhinav; Hwang, Wonmuk; Wagner, Gerhard; Lang, Matthew J. (2021-01-08).
557:
Success in positive selection allows the thymocyte to undergo a number of maturational changes during the transition to a single positive T cell. The single positive T cells upregulate the
347:
class I or II molecules with at least a weak affinity. This eliminates (by a process called "death by neglect") those T cells which would be non-functional due to an inability to bind MHC.
794:
767:
Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol. 2007 Feb 15;178(4):2008-17.
561:
receptor CCR7, causing migration from the cortex to the medulla. At this stage the key maturation process involves negative selection, the elimination of autoreactive thymocytes.
577:, a protein which drives apoptosis. Cells which do not have a high affinity for self-antigens survive negative selection. At this stage, some cells are also selected to become
539:. Some thymocytes are able to rescue failed positive selection by receptor editing (rearrangement of the other T cell receptor allele to produce a new T cell receptor).
448:. The purpose of thymocyte development is to produce mature T cells with a diverse array of functional T cell receptors, through the process of TCR gene rearrangement.
1454:
596:
Negative selection is not 100% effective, some autoreactive T cells escape thymic censorship, and are released into the circulation. Additional mechanisms of
778:
C. Clare
Blackburn & Nancy R. Manley "Developing a new paradigm for thymus organogenesis" Nature Reviews Immunology April 2004 278-289 Retrieved 10/4/12
1738:
491:
Following β-selection thymocytes generate CD4+CD8+ double positive cells, which then undergo TCRα rearrangement, resulting in completely assembled TCR.
460:
has a different sequence, due to different choice of gene fragments and the errors introduced during the cutting and joining process (see section on
966:
Mizsei, Réka; Li, Xiaolong; Chen, Wan-Na; Szabo, Monika; Wang, Jia-huai; Wagner, Gerhard; Reinherz, Ellis L.; Mallis, Robert J. (January 2021).
1149:"Review of 54 patients with complete DiGeorge anomaly enrolled in protocols for thymus transplantation: outcome of 44 consecutive transplants"
1089:
Anderson, M.S. et al. (2002) Projection of an
Immunological Self-Shadow Within the Thymus by the Aire Protein. Science 298 (5597), 1395-1401
1098:
Takaba, H. et al. (2015) Fezf2 Orchestrates a Thymic
Programs of Self-Antigen Expression for Immune Tolerance. Cell 163, 975 - 987
968:"A general chemical crosslinking strategy for structural analyses of weakly interacting proteins applied to preTCR-pMHC complexes"
1731:
1368:
1127:
1119:
687:
664:
468:
is that each T cell is capable of recognizing a different peptide, providing a defense against rapidly evolving pathogens.
1587:
1724:
2070:
1623:
516:
445:
344:
333:
63:
2065:
2008:
1546:
1668:
1638:
1618:
464:
for more information on TCR rearrangement). The evolutionary advantage in having a large number of unique
1656:
1651:
1491:
412:, and is complete by the DN3 stage. Following T lineage commitment, DN3 thymocytes undergo β-selection.
1894:
1361:
1294:
Passos, Geraldo A.; Speck-Hernandez, Cesar A.; Assis, Amanda F.; Mendes-da-Cruz, Daniella A. (2018).
1716:
874:"Thymocyte Development in the Absence of Pre-T Cell Receptor Extracellular Immunoglobulin Domains"
779:
677:
503:
A figure depicting the process of T cell / thymocyte positive and negative selection in the thymus
2074:
1832:
1695:
815:"Developing T cells form an immunological synapse for passage through the β-selection checkpoint"
219:
36:
356:
Additional mechanisms of tolerance active in the periphery exist to silence these cells such as
2096:
1814:
1750:
1538:
929:"Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. 1990"
631:
Thymocytes that gain oncogenic mutations allowing uncontrolled proliferation can become thymic
615:
2043:
1899:
1877:
1577:
1554:
1530:
1198:"Inhibition of Notch signaling biases rat thymocyte development towards the NK cell lineage"
747:
Schwarz BA, Bhandoola A. Trafficking from the bone marrow to the thymus: a prerequisite for
476:
gene fragments are non-functional. To eliminate thymocytes which have made a non-functional
2167:
2060:
1993:
1904:
1828:
1601:
1418:
1354:
1038:
885:
873:
597:
365:
68:
8:
2120:
2055:
2038:
1872:
1790:
1610:
1449:
1247:"Human natural killer cell committed thymocytes and their relation to the T cell lineage"
461:
1042:
927:
Koller, Beverly H.; Marrack, Philippa; Kappler, John W.; Smithies, Oliver (2010-05-01).
889:
1940:
1582:
1569:
1328:
1295:
1271:
1246:
1227:
1173:
1148:
1107:
Thymus
Transplantation Book Thymus Gland Pathology, pages 255-267, Springer Milan 2008
1067:
1026:
1002:
967:
849:
814:
789:
Sleckman BP, Lymphocyte antigen receptor gene assembly: multiple layers of regulation.
756:
708:
605:
578:
451:
Unlike most genes, which have a stable sequence in each cell which expresses them, the
390:
361:
768:
1988:
1754:
1633:
1514:
1433:
1413:
1333:
1315:
1276:
1219:
1178:
1123:
1115:
1072:
1054:
1027:"Pre–T cell receptors topologically sample self-ligands during thymocyte β-selection"
1007:
989:
948:
940:
909:
901:
854:
836:
730:
683:
620:
552:
307:
2108:
2048:
2020:
2015:
1983:
1970:
1960:
1506:
1482:
1323:
1307:
1266:
1258:
1231:
1209:
1168:
1160:
1131:
1108:
1062:
1046:
997:
979:
893:
844:
826:
720:
24:
2003:
1498:
1164:
1135:
897:
801:
725:
678:
Mitchell, Richard
Sheppard; Kumar, Vinay; Abbas, Abul K.; Fausto, Nelson (2007).
565:
512:
508:
477:
473:
465:
457:
452:
437:
409:
340:
320:
315:
1196:
van den Brandt J, Voss K, Schott M, Hünig T, Wolfe MS, Reichardt HM (May 2004).
222:
reside in bone marrow. They produce precursors of T lymphocytes, which seed the
2030:
1975:
1916:
1836:
1809:
523:
as a coreceptor to bind MHC class II. At this stage thymocytes upregulate both
1112:
984:
2161:
1998:
1887:
1705:
1687:
1525:
1487:
1408:
1319:
1058:
993:
944:
905:
840:
440:(TCR), which is a surface protein able to recognize short protein sequences (
238:
227:
1050:
813:
Allam, Amr H.; Charnley, Mirren; Pham, Kim; Russell, Sarah M. (2021-03-01).
1965:
1950:
1945:
1882:
1780:
1520:
1428:
1423:
1337:
1262:
1223:
1182:
1076:
1011:
952:
928:
858:
748:
734:
609:
570:
369:
72:
1280:
1214:
1197:
913:
831:
1700:
1403:
1398:
590:
40:
1293:
436:
The ability of T cells to recognize foreign antigens is mediated by the
332:, thymocytes will have to interact with several cell surface molecules,
2088:
1841:
1747:
1377:
386:
1311:
581:, usually cells which have an intermediate affinity for self-peptide.
511:
which is capable of assembling on the surface. However, many of these
2146:
1851:
1746:
1673:
1628:
632:
558:
536:
481:
233:
Early, double negative thymocytes express (and can be identified by)
226:(thus becoming thymocytes) and differentiate under influence of the
2136:
1921:
1909:
1867:
1821:
1785:
1442:
1393:
499:
441:
55:
872:
Irving, Bryan A.; Alt, Frederick W.; Killeen, Nigel (1998-05-08).
2141:
1955:
1804:
1775:
1661:
574:
59:
709:"The fourth way? Harnessing aggressive tendencies in the thymus"
314:
In order to pass the β-selection checkpoint, the β chain of the
193:
TCR-alpha rearrangement, positive selection, negative selection
2113:
2101:
1846:
1797:
1464:
1385:
623:(absence of thymus), increases the risk of autoimmune disease.
601:
593:, the cells are considered mature T cells, and not thymocytes.
357:
223:
32:
28:
1346:
1245:
Sánchez MJ, Spits H, Lanier LL, Phillips JH (December 1993).
1195:
600:
active in the periphery exist to silence these cells such as
121:
44:
1147:
Markert ML, Devlin BH, Alexieff MJ, et al. (May 2007).
926:
494:
619:
explained by that the indication itself, that is, complete
428:
397:
393:
246:
117:
113:
1244:
515:
will still be non-functional, due to an inability to bind
62:. Positive selection selects cells which are able to bind
564:
The key disadvantage in a gene rearrangement process for
528:
524:
520:
420:
250:
242:
234:
184:
180:
812:
253:
is expressed. Expression of both CD4 and CD8 makes them
706:
608:. If these peripheral tolerance mechanisms also fail,
424:
Histology of the thymus showing the cortex and medulla
1146:
109:
Double negative 1 or ETP (Early T lineage
Progenitor)
353:censorship, and are released into the circulation.
1024:
965:
71:exist to silence these cells, but if these fail,
2159:
871:
130:Proliferation, Loss of B and myeloid potentials
1296:"Update on Aire and thymic negative selection"
702:
700:
257:, and matures into either CD4+ or CD8+ cells.
1732:
1362:
1189:
707:Baldwin TA, Hogquist KA, Jameson SC (2004).
507:Thymocytes which pass β-selection express a
31:, before it undergoes transformation into a
1140:
697:
670:
260:
58:but remain tolerant towards the body's own
1739:
1725:
1369:
1355:
245:. Still during the double negative stage,
1327:
1270:
1213:
1172:
1066:
1001:
983:
848:
830:
724:
495:Positive selection and lineage commitment
498:
427:
419:
339:Positive selection selects cells with a
336:, to ensure reactivity and specificity.
1609:
638:
158:TCR-beta rearrangement, beta selection
78:
2160:
1720:
1350:
546:
1287:
1588:Mucosal associated invariant T cell
13:
531:, becoming double positive cells.
379:
14:
2179:
284:autoreactive (negative selection)
1624:Lymphokine-activated killer cell
306:
1376:
1238:
1101:
1092:
1083:
1018:
972:Journal of Biological Chemistry
959:
920:
279:functional (positive selection)
2066:Immunoglobulin class switching
865:
806:
783:
772:
761:
741:
653:
415:
1:
647:
218:In humans, circulating CD34+
43:and reach the thymus via the
35:. Thymocytes are produced as
1669:Type 3 innate lymphoid cells
1657:Type 2 innate lymphoid cells
1652:Type 1 innate lymphoid cells
1639:Uterine natural killer cells
1619:Cytokine-induced killer cell
1165:10.1182/blood-2006-10-048652
1136:10.1007/978-88-470-0828-1_30
898:10.1126/science.280.5365.905
726:10.4049/jimmunol.173.11.6515
665:Dorland's Medical Dictionary
213:
7:
432:Minute structure of thymus.
274:functional (beta selection)
10:
2184:
1895:Polyclonal B cell response
682:. Philadelphia: Saunders.
550:
87:
2129:
2087:
2029:
1930:
1860:
1768:
1761:
1686:
1647:
1600:
1568:
1472:
1463:
1384:
1113:10.1007/978-88-470-0828-1
985:10.1016/j.jbc.2021.100255
626:
444:) that are presented on
261:Events during maturation
220:hematopoietic stem cells
138:Lineage-CD44+CD25+CD117+
98:Defining surface markers
1696:Hematopoietic stem cell
1455:Lymphoplasmacytoid cell
1051:10.1126/science.abe0918
819:Journal of Cell Biology
680:Robbins Basic Pathology
323:by gene rearrangement.
2009:Tolerance in pregnancy
1751:adaptive immune system
1263:10.1084/jem.178.6.1857
616:Thymus transplantation
504:
433:
425:
368:mechanisms also fail,
2044:Somatic hypermutation
1878:Polyclonal antibodies
1873:Monoclonal antibodies
1602:Innate lymphoid cells
1578:Natural killer T cell
1215:10.1002/eji.200324735
933:Journal of Immunology
832:10.1083/jcb.201908108
502:
431:
423:
249:expression stops and
2061:Junctional diversity
1829:Antigen presentation
639:Alternative lineages
598:peripheral tolerance
366:peripheral tolerance
201:CD4+CD8- or CD4-CD8+
79:Stages of maturation
69:peripheral tolerance
2056:V(D)J recombination
2039:Affinity maturation
1791:Antigenic variation
1570:Innate-like T cells
1450:Transitional B cell
1043:2021Sci...371..181L
890:1998Sci...280..905I
569:self, resulting in
462:V(D)J recombination
391:chemokine receptors
330:positively-selected
207:Negative selection
104:Significant events
800:2008-01-27 at the
606:regulatory T cells
579:regulatory T cells
547:Negative selection
505:
434:
426:
362:regulatory T cells
166:Lineage-CD44-CD25-
152:Lineage-CD44-CD25+
2155:
2154:
2083:
2082:
1833:professional APCs
1714:
1713:
1682:
1681:
1596:
1595:
1312:10.1111/imm.12831
1128:978-88-470-0828-1
1120:978-88-470-0827-4
1037:(6525): 181–185.
884:(5365): 905–908.
825:(3): e201908108.
689:978-1-4160-2973-1
676:Figure 12-13 in:
621:DiGeorge syndrome
553:Central tolerance
377:
376:
230:and its ligands.
211:
210:
163:Double negative 4
149:Double negative 3
135:Double negative 2
2175:
2049:Clonal selection
2021:Immune privilege
2016:Immunodeficiency
1971:Cross-reactivity
1961:Hypersensitivity
1766:
1765:
1741:
1734:
1727:
1718:
1717:
1634:Adaptive NK cell
1607:
1606:
1470:
1469:
1371:
1364:
1357:
1348:
1347:
1342:
1341:
1331:
1291:
1285:
1284:
1274:
1242:
1236:
1235:
1217:
1193:
1187:
1186:
1176:
1144:
1138:
1105:
1099:
1096:
1090:
1087:
1081:
1080:
1070:
1022:
1016:
1015:
1005:
987:
963:
957:
956:
939:(9): 4592–4595.
924:
918:
917:
869:
863:
862:
852:
834:
810:
804:
793:32:153-8, 2005.
787:
781:
776:
770:
765:
759:
745:
739:
738:
728:
704:
695:
693:
674:
668:
657:
604:, deletion, and
566:T cell receptors
513:T cell receptors
466:T cell receptors
389:(CD62P) and the
360:, deletion, and
310:
265:
264:
92:
91:
2183:
2182:
2178:
2177:
2176:
2174:
2173:
2172:
2158:
2157:
2156:
2151:
2125:
2079:
2025:
2004:Clonal deletion
1932:
1926:
1856:
1757:
1745:
1715:
1710:
1678:
1643:
1592:
1564:
1558:
1550:
1542:
1534:
1510:
1502:
1495:
1459:
1437:
1380:
1375:
1345:
1292:
1288:
1243:
1239:
1202:Eur. J. Immunol
1194:
1190:
1159:(10): 4539–47.
1145:
1141:
1106:
1102:
1097:
1093:
1088:
1084:
1023:
1019:
964:
960:
925:
921:
870:
866:
811:
807:
802:Wayback Machine
788:
784:
777:
773:
766:
762:
746:
742:
719:(11): 6515–20.
705:
698:
690:
675:
671:
658:
654:
650:
641:
629:
555:
549:
509:T cell receptor
497:
478:T cell receptor
474:T cell receptor
458:T cell receptor
453:T cell receptor
438:T cell receptor
418:
410:Notch signaling
382:
380:Thymus settling
341:T cell receptor
328:In order to be
321:T cell receptor
316:T cell receptor
301:cortex/medulla
263:
255:double positive
216:
198:Single positive
177:Double positive
90:
81:
27:present in the
17:
12:
11:
5:
2181:
2171:
2170:
2153:
2152:
2150:
2149:
2144:
2139:
2133:
2131:
2127:
2126:
2124:
2123:
2118:
2117:
2116:
2106:
2105:
2104:
2093:
2091:
2085:
2084:
2081:
2080:
2078:
2077:
2068:
2063:
2058:
2053:
2052:
2051:
2046:
2035:
2033:
2031:Immunogenetics
2027:
2026:
2024:
2023:
2018:
2013:
2012:
2011:
2006:
2001:
1996:
1991:
1979:
1978:
1976:Co-stimulation
1973:
1968:
1963:
1958:
1953:
1948:
1943:
1936:
1934:
1928:
1927:
1925:
1924:
1919:
1917:Immune complex
1913:
1912:
1907:
1902:
1897:
1892:
1891:
1890:
1885:
1880:
1875:
1864:
1862:
1858:
1857:
1855:
1854:
1849:
1844:
1839:
1837:Dendritic cell
1825:
1824:
1819:
1818:
1817:
1815:Conformational
1812:
1801:
1800:
1795:
1794:
1793:
1788:
1783:
1772:
1770:
1763:
1759:
1758:
1744:
1743:
1736:
1729:
1721:
1712:
1711:
1709:
1708:
1703:
1698:
1692:
1690:
1684:
1683:
1680:
1679:
1677:
1676:
1671:
1666:
1665:
1664:
1654:
1648:
1645:
1644:
1642:
1641:
1636:
1631:
1626:
1621:
1615:
1613:
1604:
1598:
1597:
1594:
1593:
1591:
1590:
1585:
1580:
1574:
1572:
1566:
1565:
1563:
1562:
1561:
1560:
1556:
1552:
1548:
1544:
1540:
1536:
1532:
1523:
1518:
1508:
1500:
1493:
1485:
1479:
1473:
1467:
1461:
1460:
1458:
1457:
1452:
1447:
1446:
1445:
1435:
1431:
1426:
1421:
1416:
1411:
1406:
1401:
1396:
1390:
1388:
1382:
1381:
1374:
1373:
1366:
1359:
1351:
1344:
1343:
1286:
1257:(6): 1857–66.
1237:
1208:(5): 1405–13.
1188:
1139:
1100:
1091:
1082:
1017:
958:
919:
864:
805:
782:
771:
760:
755:209:47, 2006.
740:
696:
688:
669:
651:
649:
646:
640:
637:
628:
625:
551:Main article:
548:
545:
496:
493:
417:
414:
381:
378:
375:
374:
349:
325:
311:
303:
302:
299:
296:
293:
287:
286:
281:
276:
271:
262:
259:
215:
212:
209:
208:
205:
202:
199:
195:
194:
191:
188:
178:
174:
173:
170:
167:
164:
160:
159:
156:
153:
150:
146:
145:
142:
139:
136:
132:
131:
128:
125:
110:
106:
105:
102:
99:
96:
89:
86:
80:
77:
15:
9:
6:
4:
3:
2:
2180:
2169:
2166:
2165:
2163:
2148:
2145:
2143:
2140:
2138:
2135:
2134:
2132:
2128:
2122:
2119:
2115:
2112:
2111:
2110:
2107:
2103:
2100:
2099:
2098:
2095:
2094:
2092:
2090:
2086:
2076:
2072:
2069:
2067:
2064:
2062:
2059:
2057:
2054:
2050:
2047:
2045:
2042:
2041:
2040:
2037:
2036:
2034:
2032:
2028:
2022:
2019:
2017:
2014:
2010:
2007:
2005:
2002:
2000:
1999:Clonal anergy
1997:
1995:
1992:
1990:
1987:
1986:
1985:
1981:
1980:
1977:
1974:
1972:
1969:
1967:
1964:
1962:
1959:
1957:
1954:
1952:
1949:
1947:
1944:
1942:
1938:
1937:
1935:
1929:
1923:
1920:
1918:
1915:
1914:
1911:
1908:
1906:
1903:
1901:
1898:
1896:
1893:
1889:
1888:Microantibody
1886:
1884:
1881:
1879:
1876:
1874:
1871:
1870:
1869:
1866:
1865:
1863:
1859:
1853:
1850:
1848:
1845:
1843:
1840:
1838:
1834:
1830:
1827:
1826:
1823:
1820:
1816:
1813:
1811:
1808:
1807:
1806:
1803:
1802:
1799:
1796:
1792:
1789:
1787:
1784:
1782:
1779:
1778:
1777:
1774:
1773:
1771:
1767:
1764:
1760:
1756:
1752:
1749:
1742:
1737:
1735:
1730:
1728:
1723:
1722:
1719:
1707:
1706:Prolymphocyte
1704:
1702:
1699:
1697:
1694:
1693:
1691:
1689:
1688:Lymphopoiesis
1685:
1675:
1672:
1670:
1667:
1663:
1660:
1659:
1658:
1655:
1653:
1650:
1649:
1646:
1640:
1637:
1635:
1632:
1630:
1627:
1625:
1622:
1620:
1617:
1616:
1614:
1612:
1608:
1605:
1603:
1599:
1589:
1586:
1584:
1581:
1579:
1576:
1575:
1573:
1571:
1567:
1559:
1553:
1551:
1545:
1543:
1537:
1535:
1529:
1528:
1527:
1526:Memory T cell
1524:
1522:
1519:
1516:
1512:
1504:
1496:
1489:
1486:
1484:
1483:Cytotoxic CD8
1480:
1478:
1475:
1474:
1471:
1468:
1466:
1462:
1456:
1453:
1451:
1448:
1444:
1441:
1440:
1439:
1432:
1430:
1427:
1425:
1422:
1420:
1419:Marginal zone
1417:
1415:
1412:
1410:
1407:
1405:
1402:
1400:
1397:
1395:
1392:
1391:
1389:
1387:
1383:
1379:
1372:
1367:
1365:
1360:
1358:
1353:
1352:
1349:
1339:
1335:
1330:
1325:
1321:
1317:
1313:
1309:
1305:
1301:
1297:
1290:
1282:
1278:
1273:
1268:
1264:
1260:
1256:
1252:
1248:
1241:
1233:
1229:
1225:
1221:
1216:
1211:
1207:
1203:
1199:
1192:
1184:
1180:
1175:
1170:
1166:
1162:
1158:
1154:
1150:
1143:
1137:
1133:
1129:
1125:
1121:
1117:
1114:
1110:
1104:
1095:
1086:
1078:
1074:
1069:
1064:
1060:
1056:
1052:
1048:
1044:
1040:
1036:
1032:
1028:
1021:
1013:
1009:
1004:
999:
995:
991:
986:
981:
977:
973:
969:
962:
954:
950:
946:
942:
938:
934:
930:
923:
915:
911:
907:
903:
899:
895:
891:
887:
883:
879:
875:
868:
860:
856:
851:
846:
842:
838:
833:
828:
824:
820:
816:
809:
803:
799:
796:
792:
786:
780:
775:
769:
764:
758:
754:
750:
744:
736:
732:
727:
722:
718:
714:
710:
703:
701:
691:
685:
681:
673:
667:
666:
661:
656:
652:
645:
636:
634:
624:
622:
617:
613:
611:
607:
603:
599:
594:
592:
586:
582:
580:
576:
572:
567:
562:
560:
554:
544:
540:
538:
532:
530:
526:
522:
518:
514:
510:
501:
492:
489:
485:
483:
479:
475:
469:
467:
463:
459:
454:
449:
447:
443:
439:
430:
422:
413:
411:
405:
401:
399:
395:
392:
388:
373:
371:
367:
363:
359:
354:
350:
348:
346:
343:able to bind
342:
337:
335:
331:
326:
324:
322:
317:
312:
309:
305:
304:
300:
297:
294:
292:
289:
288:
285:
282:
280:
277:
275:
272:
270:
267:
266:
258:
256:
252:
248:
244:
240:
236:
231:
229:
228:Notch protein
225:
221:
206:
203:
200:
197:
196:
192:
189:
186:
182:
179:
176:
175:
171:
168:
165:
162:
161:
157:
154:
151:
148:
147:
143:
140:
137:
134:
133:
129:
126:
123:
119:
115:
111:
108:
107:
103:
100:
97:
94:
93:
85:
76:
74:
70:
65:
61:
57:
52:
48:
46:
42:
38:
34:
30:
26:
22:
1966:Inflammation
1951:Alloimmunity
1946:Autoimmunity
1931:Immunity vs.
1883:Autoantibody
1781:Superantigen
1476:
1306:(1): 10–20.
1303:
1299:
1289:
1254:
1250:
1240:
1205:
1201:
1191:
1156:
1152:
1142:
1103:
1094:
1085:
1034:
1030:
1020:
975:
971:
961:
936:
932:
922:
881:
877:
867:
822:
818:
808:
790:
785:
774:
763:
752:
749:thymopoiesis
743:
716:
712:
694:8th edition.
679:
672:
663:
655:
642:
630:
614:
610:autoimmunity
595:
587:
583:
571:autoimmunity
563:
556:
541:
533:
506:
490:
486:
470:
450:
435:
406:
402:
383:
370:autoimmunity
355:
351:
338:
329:
327:
313:
290:
283:
278:
273:
268:
254:
232:
217:
82:
73:autoimmunity
51:Thymopoiesis
50:
49:
20:
18:
2168:Lymphocytes
2089:Lymphocytes
1748:Lymphocytic
1701:Lymphoblast
1399:Plasmablast
1378:Lymphocytes
1251:J. Exp. Med
791:Immunol Res
753:Immunol Rev
660:"thymocyte"
612:may arise.
591:bloodstream
416:β-selection
372:may arise.
364:. If these
75:may arise.
41:bone marrow
25:immune cell
16:Immune cell
2130:Substances
1994:Peripheral
1982:Inaction:
1861:Antibodies
1842:Macrophage
1755:complement
1515:Regulatory
1488:Helper CD4
1414:Follicular
1300:Immunology
978:: 100255.
648:References
488:self-MHC.
387:P-selectin
37:stem cells
2147:Cytolysin
2137:Cytokines
1984:Tolerance
1933:tolerance
1852:Immunogen
1674:LTi cells
1629:Null cell
1477:Thymocyte
1320:1365-2567
1130:(Online)
1059:0036-8075
994:0021-9258
945:1550-6606
906:0036-8075
841:0021-9525
795:full text
757:full text
713:J Immunol
633:lymphomas
559:chemokine
537:apoptosis
482:apoptosis
291:location:
214:In humans
56:pathogens
21:thymocyte
2162:Category
2097:Cellular
1941:Immunity
1939:Action:
1922:Paratope
1910:Idiotype
1900:Allotype
1868:Antibody
1822:Mimotope
1786:Allergen
1769:Antigens
1762:Lymphoid
1662:Nuocytes
1611:NK cells
1443:B10 cell
1338:28871661
1224:15114674
1183:17284531
1122:(Print)
1077:33335016
1012:33837736
953:20410496
859:33464309
798:Archived
735:15557139
442:peptides
112:Lineage-
101:Location
60:antigens
2142:Opsonin
2121:NK cell
2109:Humoral
1989:Central
1956:Allergy
1905:Isotype
1805:Epitope
1776:Antigen
1465:T cells
1394:B1 cell
1386:B cells
1329:5721245
1281:7504051
1272:2191276
1232:6270092
1174:1885498
1068:8011828
1039:Bibcode
1031:Science
1003:7948749
914:9572735
886:Bibcode
878:Science
850:7814350
575:BCL2L11
204:medulla
88:In mice
39:in the
2114:B cell
2102:T cell
1847:B cell
1810:Linear
1798:Hapten
1409:Memory
1404:Plasma
1336:
1326:
1318:
1279:
1269:
1230:
1222:
1181:
1171:
1126:
1118:
1075:
1065:
1057:
1010:
1000:
992:
951:
943:
912:
904:
857:
847:
839:
733:
686:
627:Cancer
602:anergy
358:anergy
298:cortex
295:cortex
224:thymus
190:cortex
169:cortex
155:cortex
141:cortex
127:cortex
33:T cell
29:thymus
23:is an
1521:Naïve
1429:Pre-B
1424:Naïve
1228:S2CID
1153:Blood
269:type:
122:CD117
95:Stage
45:blood
1753:and
1481:αβ (
1438:cell
1334:PMID
1316:ISSN
1277:PMID
1220:PMID
1179:PMID
1124:ISBN
1116:ISBN
1073:PMID
1055:ISSN
1008:PMID
990:ISSN
949:PMID
941:ISSN
910:PMID
902:ISSN
855:PMID
837:ISSN
731:PMID
684:ISBN
527:and
398:CCR9
396:and
394:CCR7
247:CD34
241:and
118:CD25
114:CD44
2075:HLA
2071:MHC
1436:reg
1324:PMC
1308:doi
1304:153
1267:PMC
1259:doi
1255:178
1210:doi
1169:PMC
1161:doi
1157:109
1132:doi
1109:doi
1063:PMC
1047:doi
1035:371
998:PMC
980:doi
976:296
937:184
894:doi
882:280
845:PMC
827:doi
823:220
721:doi
717:173
662:at
529:CD8
525:CD4
521:CD4
517:MHC
446:MHC
345:MHC
334:MHC
251:CD1
243:CD7
239:CD5
235:CD2
185:CD8
181:CD4
64:MHC
2164::
1835::
1583:γδ
1557:VM
1549:RM
1541:EM
1533:CM
1513:/
1511:17
1505:/
1497:/
1494:FH
1490:/
1332:.
1322:.
1314:.
1302:.
1298:.
1275:.
1265:.
1253:.
1249:.
1226:.
1218:.
1206:34
1204:.
1200:.
1177:.
1167:.
1155:.
1151:.
1071:.
1061:.
1053:.
1045:.
1033:.
1029:.
1006:.
996:.
988:.
974:.
970:.
947:.
935:.
931:.
908:.
900:.
892:.
880:.
876:.
853:.
843:.
835:.
821:.
817:.
751:.
729:.
715:.
711:.
699:^
635:.
400:.
237:,
172:-
144:-
47:.
19:A
2073:/
1831:/
1740:e
1733:t
1726:v
1555:T
1547:T
1539:T
1531:T
1517:)
1509:h
1507:T
1503:3
1501:h
1499:T
1492:T
1434:B
1370:e
1363:t
1356:v
1340:.
1310::
1283:.
1261::
1234:.
1212::
1185:.
1163::
1134::
1111::
1079:.
1049::
1041::
1014:.
982::
955:.
916:.
896::
888::
861:.
829::
737:.
723::
692:.
187:+
183:+
124:+
120:-
116:+
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.