20:
28:
34:
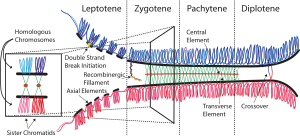
Homologous chromosomes (light blue) align and synapse together via transverse filaments (black lines) and longitudinal filaments (dark blue). Recombination nodules (gray ellipsoids) on the central region may help in completing recombination. Chromatin (red loops) is attached to its sexual leg and
193:
does not appear to require synaptonemal complex formation. Research has shown that not only does the SC form after genetic recombination but mutant yeast cells unable to assemble a synaptonemal complex can still engage in the exchange of genetic information. However, in other organisms like the
137:
show only "partial synapsis" as they usually form only a short SC in the XY pair. The SC shows very little structural variability among eukaryotic organisms despite some significant protein differences. In many organisms the SC carries one or several "recombination nodules" associated with its
113:
The synaptonemal complex was described by
Montrose J. Moses in 1956 in primary spermatocytes of crayfish and by D. Fawcett in spermatocytes of pigeon, cat and man. As seen with the electron microscope, the synaptonemal complex is formed by two "lateral elements", mainly formed by SYCP3 and
114:
secondarily by SYCP2, a "central element" that contains at least two additional proteins and the amino terminal region of SYCP1, and a "central region" spanned between the two lateral elements, that contains the "transverse filaments" composed mainly by the protein SYCP1.
516:- Synaptonemal complex by 3D-Structured Illumination, photograph by Dr. Chung-Ju Rachel Wang, University of California Berkeley, Department of Molecular and Cell Biology, Berkeley, CA, USA, second place winner of the 2009 Olympus Bioscapes Digital Imaging Competition.
99:. Previous to the pachytene stage, during leptonema, the lateral elements begin to form and they initiate and complete their pairing during the zygotene stage. After pachynema ends, the SC usually becomes disassembled and can no longer be identified.
39:
Top: Set of tomato SCs. Chromatin "sheaths" visible around each SC. Bottom: Two tomato SCs with the chromatin removed, allowing kinetochores ("ball-like" structures) at centromeres to be revealed.
133:
and may be used to probe the presence of pairing abnormalities in individuals carrying chromosomal abnormalities, either in number or in the chromosomal structure. The sex chromosomes in
87:
The synaptonemal complex is a tripartite structure consisting of two parallel lateral regions and a central element. This "tripartite structure" is seen during the
102:
In humans, three specific components of the synaptonemal complex have been characterized: SC protein-1 (SYCP1), SC protein-2 (SYCP2), and SC protein-3 (
117:
The SCs can be seen with the light microscope using silver staining or with immunofluorescence techniques that label the proteins SYCP3 or SYCP2.
302:"Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair"
169:
It is now evident that the synaptonemal complex is not required for genetic recombination in some organisms. For instance, in
271:
161:
In cell development the synaptonemal complex disappears during the late prophase of meiosis I. It is formed during zygotene.
138:
central space. These nodules are thought to correspond to mature genetic recombination events or "crossovers". In male mice,
150:
are likely repaired by crossover recombination in SCs. The finding of an interaction between a SC structural component and
75:. It is currently thought that the SC functions primarily as a scaffold to allow interacting chromatids to complete their
472:"Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway"
423:"Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex"
147:
110:
is on chromosome 1p13; the SYCP2 gene is on chromosome 20q13.33; and the gene for SYCP3 is on chromosome 12q.
540:
215:
Page SL, Hawley RS (2004-10-08). "The genetics and molecular biology of the synaptonemal complex".
151:
386:
Zickler D, Kleckner N (1999-12-01). "Meiotic chromosomes: integrating structure and function".
300:
Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, et al. (February 2009).
228:
190:
76:
68:
8:
471:
364:
447:
422:
328:
301:
514:
493:
452:
403:
368:
333:
277:
267:
250:
Yang F, Wang PJ (2009). "The
Mammalian synaptonemal complex: a scaffold and beyond".
232:
56:
198:
nematode, formation of chiasmata require the formation of the synaptonemal complex.
483:
442:
434:
399:
395:
360:
323:
313:
259:
224:
318:
519:
534:
372:
96:
488:
497:
456:
407:
337:
281:
236:
134:
23:
Schematic of the synaptonemal complex at different stages during
Prophase I
438:
178:
130:
185:
27:
19:
263:
139:
88:
72:
170:
126:
92:
64:
55:
structure that forms between homologous chromosomes (two pairs of
173:
143:
60:
52:
155:
103:
470:
Chi J, Mahé F, Loidl J, Logsdon J, Dunthorn M (March 2014).
299:
420:
107:
351:
Moses, Montrose J. (1968-12-01). "Synaptinemal complex".
421:
Lukaszewicz A, Howard-Till RA, Loidl J (November 2013).
125:
Formation of the SC usually reflects the pairing or "
469:
532:
385:
217:Annual Review of Cell and Developmental Biology
158:also suggests a role for the SC in DNA repair.
146:in SCs. This indicates that exogenously caused
525:Meiosis in Mice without a Synaptonemal Complex
35:toe, extending from both sister chromatids.
120:
214:
164:
487:
446:
327:
317:
249:
229:10.1146/annurev.cellbio.19.111301.155141
26:
18:
533:
95:, both in males and in females during
350:
295:
293:
291:
71:during prophase I during meiosis in
365:10.1146/annurev.ge.02.120168.002051
13:
14:
552:
508:
288:
476:Molecular Biology and Evolution
463:
414:
400:10.1146/annurev.genet.33.1.603
379:
344:
243:
208:
82:
1:
201:
319:10.1371/journal.pgen.1000393
7:
91:stage of the first meiotic
10:
557:
63:and is thought to mediate
388:Annual Review of Genetics
353:Annual Review of Genetics
121:Assembly and disassembly
165:Necessity in eukaryotes
427:Nucleic Acids Research
152:recombinational repair
40:
24:
523:Kounetsova A. et al,
489:10.1093/molbev/mst258
30:
22:
45:synaptonemal complex
541:Molecular genetics
439:10.1093/nar/gkt703
144:meiotic crossovers
41:
25:
273:978-3-8055-8967-3
264:10.1159/000166620
191:genetic crossover
140:gamma irradiation
57:sister chromatids
16:Protein structure
548:
502:
501:
491:
467:
461:
460:
450:
433:(20): 9296–309.
418:
412:
411:
383:
377:
376:
348:
342:
341:
331:
321:
297:
286:
285:
247:
241:
240:
212:
129:" of homologous
556:
555:
551:
550:
549:
547:
546:
545:
531:
530:
527:PLOS ONE (2011)
511:
506:
505:
468:
464:
419:
415:
384:
380:
349:
345:
312:(2): e1000393.
298:
289:
274:
252:Genome Dynamics
248:
244:
213:
209:
204:
167:
123:
85:
17:
12:
11:
5:
554:
544:
543:
529:
528:
521:
517:
510:
509:External links
507:
504:
503:
462:
413:
394:(1): 603–754.
378:
359:(1): 363–412.
343:
287:
272:
242:
206:
205:
203:
200:
166:
163:
122:
119:
84:
81:
15:
9:
6:
4:
3:
2:
553:
542:
539:
538:
536:
526:
522:
520:
518:
515:
513:
512:
499:
495:
490:
485:
482:(3): 660–72.
481:
477:
473:
466:
458:
454:
449:
444:
440:
436:
432:
428:
424:
417:
409:
405:
401:
397:
393:
389:
382:
374:
370:
366:
362:
358:
354:
347:
339:
335:
330:
325:
320:
315:
311:
307:
306:PLOS Genetics
303:
296:
294:
292:
283:
279:
275:
269:
265:
261:
257:
253:
246:
238:
234:
230:
226:
223:(1): 525–58.
222:
218:
211:
207:
199:
197:
192:
189:
187:
182:
180:
175:
172:
162:
159:
157:
153:
149:
145:
141:
136:
132:
128:
118:
115:
111:
109:
106:). The SYCP1
105:
100:
98:
97:gametogenesis
94:
90:
80:
78:
74:
70:
69:recombination
66:
62:
58:
54:
50:
46:
38:
33:
29:
21:
524:
479:
475:
465:
430:
426:
416:
391:
387:
381:
356:
352:
346:
309:
305:
255:
251:
245:
220:
216:
210:
195:
184:
177:
168:
160:
135:male mammals
124:
116:
112:
101:
86:
79:activities.
48:
44:
42:
36:
31:
188:tetraurelia
181:thermophila
179:Tetrahymena
148:DNA damages
131:chromosomes
83:Composition
202:References
196:C. elegans
186:Paramecium
142:increases
73:eukaryotes
373:0066-4197
258:: 69–80.
171:protozoan
89:pachytene
77:crossover
59:) during
535:Category
498:24336924
457:23935123
408:10690419
338:19247432
282:18948708
237:15473851
176:such as
174:ciliates
154:protein
127:synapsis
93:prophase
65:synapsis
448:3814389
329:2640461
61:meiosis
53:protein
51:) is a
496:
455:
445:
406:
371:
336:
326:
280:
270:
235:
156:RAD51
104:SYCP3
494:PMID
453:PMID
404:PMID
369:ISSN
334:PMID
278:PMID
268:ISBN
233:PMID
183:and
108:gene
67:and
43:The
484:doi
443:PMC
435:doi
396:doi
361:doi
324:PMC
314:doi
260:doi
225:doi
537::
492:.
480:31
478:.
474:.
451:.
441:.
431:41
429:.
425:.
402:.
392:33
390:.
367:.
355:.
332:.
322:.
308:.
304:.
290:^
276:.
266:.
254:.
231:.
221:20
219:.
49:SC
500:.
486::
459:.
437::
410:.
398::
375:.
363::
357:2
340:.
316::
310:5
284:.
262::
256:5
239:.
227::
47:(
37:B
32:A
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.