328:(ddNTP) or fluorescently labeled deoxynucleotides (dNTP). With ddNTPs, probes hybridize to the target DNA immediately upstream of SNP nucleotide, and a single, ddNTP complementary to the SNP allele is added to the 3’ end of the probe (the missing 3'-hydroxyl in didioxynucleotide prevents further nucleotides from being added). Each ddNTP is labeled with a different fluorescent signal allowing for the detection of all four alleles in the same reaction. With dNTPs, allele-specific probes have 3’ bases which are complementary to each of the SNP alleles being interrogated. If the target DNA contains an allele complementary to the probe's 3’ base, the target DNA will completely hybridize to the probe, allowing DNA polymerase to extend from the 3’ end of the probe. This is detected by the incorporation of the fluorescently labeled dNTPs onto the end of the probe. If the target DNA does not contain an allele complementary to the probe's 3’ base, the target DNA will produce a mismatch at the 3’ end of the probe and DNA polymerase will not be able to extend from the 3' end of the probe. The benefit of the second approach is that several labeled dNTPs may get incorporated into the growing strand, allowing for increased signal. However, DNA polymerase in some rare cases, can extend from mismatched 3’ probes giving a false positive result.
461:(TGGE) or temperature gradient capillary electrophoresis (TGCE) method is based on the principle that partially denatured DNA is more restricted and travels slower in a porous material such as a gel. This property allows for the separation of DNA by melting temperature. To adapt these methods for SNP detection, two fragments are used; the target DNA which contain the SNP polymorphic site being interrogated and an allele-specific DNA sequence, referred to as the normal DNA fragment. The normal fragment is identical to the target DNA except potentially at the SNP polymorphic site, which is unknown in the target DNA. The fragments are denatured and then reannealed. If the target DNA has the same allele as the normal fragment, homoduplexes will form that will have the same melting temperature. When run on the gel with a temperature gradient, only one band will appear. If the target DNA has a distinct allele, four products will form following the reannealing step; homoduplexes consisting of target DNA, homoduplexes consisting of normal DNA and two heterduplexes of each strand of target DNA hybridized with the normal DNA strand. These four products will have distinct melting temperatures and will appear as four bands in the denaturing gel.
418:
DNA. In the oligonucleotide ligase assay, two probes are designed; an allele-specific probe which hybridizes to the target DNA so that its 3' base is situated directly over the SNP nucleotide and a second probe that hybridizes the template upstream (downstream in the complementary strand) of the SNP polymorphic site providing a 5' end for the ligation reaction. If the allele-specific probe matches the target DNA, it will fully hybridize to the target DNA and ligation can occur. Ligation does not generally occur in the presence of a mismatched 3' base. Ligated or unligated products can be detected by gel electrophoresis, MALDI-TOF mass spectrometry or by capillary electrophoresis for large-scale applications. With appropriate sequences and tags on the oligonucleotides, high-throughput sequence data can be generated from the ligated products and genotypes determined (Curry et al., 2012). The use of large numbers of sample indexes allows high-throughput sequence data on hundreds of SNPs in thousands of samples to be generated in a small portion of a high-throughput sequencing run. This is a massive genotyping by sequencing technology (MGST).
477:
retained in the column. Using PCR, two fragments are generated; target DNA containing the SNP polymorphic site and an allele-specific DNA sequence, referred to as the normal DNA fragment. This normal fragment is identical to the target DNA except potentially at the SNP polymorphic site, which is unknown in the target DNA. The fragments are denatured and then allowed to gradually reanneal. The reannaled products are added to the DHPLC column. If the SNP allele in the target DNA matches the normal DNA fragment, only identical homoduplexes will form during the reannealing step. If the target DNA contains a different SNP allele than the normal DNA fragment, heteroduplexes of the target DNA and normal DNA containing a mismatched polymorphic site will form in addition to homoduplexes. The mismatched heteroduplexes will have a different melting temperature than the homoduplexes and will not be retained in the column as long. This generates a chromatograph pattern that is distinctive from the pattern that would be generated if the target DNA fragment and normal DNA fragments were identical. The eluted DNA is detected by UV absorption.
493:
amplify. You "paint" the amplicon with a double-strand specific dye, included in the PCR mix. The ds-specific dye integrates itself into the PCR product. In essence, the entire amplicon becomes a probe. This opens up new possibilities for discovery. Either you position the primers very close to either side of the SNP in question (small amplicon genotyping, Liew, 2004) or amplify a larger region (100-400bp in length) for scanning purposes. For simple genotyping of an SNP, it is easier to just make the amplicon small to minimize the chances you mistake one SNP for another. The melting temperature (Tm) of the entire amplicon is determined and most homozygotes are sufficiently different (in the better instruments) in Tm to genotype. Heterozygotes are even easier to differentiate because they have heteroduplexes generated (refer to the gel-based explanations) which broadens the melt transition and usually gives two discernible peaks. Amplicon melting using a fluorescently-labeled primer has been described (Gundry et al., 2003) but is less practical than using ds-specific dyes due to the cost of the fluorogenic primer.
442:
between SNP alleles. This method first involves PCR amplification of the target DNA. The double-stranded PCR products are denatured using heat and formaldehyde to produce ssDNA. The ssDNA is applied to a non-denaturing electrophoresis gel and allowed to fold into a tertiary structure. Differences in DNA sequence will alter the tertiary conformation and be detected as a difference in the ssDNA strand mobility (Costabile et al. 2006). This method is widely used because it is technically simple, relatively inexpensive and uses commonly available equipment. However compared to other SNP genotyping methods, the sensitivity of this assay is lower. It has been found that the ssDNA conformation is highly dependent on temperature and it is not generally apparent what the ideal temperature is. Very often the assay will be carried out using several different temperatures. There is also a restriction on the length of fragment because the sensitivity drops when sequences longer than 400 bp are used (Costabile et al. 2006).
225:
the invention is that unexpectedly, oligonucleotides with a mismatched 3'-residue will not function as primers in the PCR under appropriate conditions. As a result, if a given allele is present in the PCR reaction, the primer pair specific to that allele will produce product but not to the alternative allele with a different SNP. The two primer pairs are also designed such that their PCR products are of a significantly different length allowing for easily distinguishable bands by gel electrophoresis or melt temperature analysis. In examining the results, if a genomic sample is homozygous, then the PCR products that result will be from the primer that matches the SNP location and the outer opposite-strand primer, as well from the two outer primers. If the genomic sample is heterozygous, then products will result from the primer of each allele and their respective outer primer counterparts as well as the outer primers.
177:, hundreds of thousands of probes are arrayed on a small chip, allowing for many SNPs to be interrogated simultaneously. Because SNP alleles only differ in one nucleotide and because it is difficult to achieve optimal hybridization conditions for all probes on the array, the target DNA has the potential to hybridize to mismatched probes. This is addressed somewhat by using several redundant probes to interrogate each SNP. Probes are designed to have the SNP site in several different locations as well as containing mismatches to the SNP allele. By comparing the differential amount of hybridization of the target DNA to each of these redundant probes, it is possible to determine specific homozygous and heterozygous alleles. Although oligonucleotide microarrays have a comparatively lower specificity and sensitivity, the scale of SNPs that can be interrogated is a major benefit. The
390:
eliminating the fluorophore's signal. During the PCR amplification step, if the allele-specific probe is perfectly complementary to the SNP allele, it will bind to the target DNA strand and then get degraded by 5’-nuclease activity of the Taq polymerase as it extends the DNA from the PCR primers. The degradation of the probe results in the separation of the fluorophore from the quencher molecule, generating a detectable signal. If the allele-specific probe is not perfectly complementary, it will have lower melting temperature and not bind as efficiently. This prevents the nuclease from acting on the probe (McGuigan & Ralston 2002).
212:(RFLP) is considered to be the simplest and earliest method to detect SNPs. SNP-RFLP makes use of the many different restriction endonucleases and their high affinity to unique and specific restriction sites. By performing a digestion on a genomic sample and determining fragment lengths through a gel assay it is possible to ascertain whether or not the enzymes cut the expected restriction sites. A failure to cut the genomic sample results in an identifiably larger than expected fragment implying that there is a mutation at the point of the restriction site which is rendering it protection from nuclease activity.
309:
This incorporated base is detected and determines the SNP allele (Goelet et al. 1999; Syvanen 2001). Because primer extension is based on the highly accurate DNA polymerase enzyme, the method is generally very reliable. Primer extension is able to genotype most SNPs under very similar reaction conditions making it also highly flexible. The primer extension method is used in a number of assay formats. These formats use a wide range of detection techniques that include
266:
complementary to the SNP nucleotide. If the target DNA contains the desired allele, the
Invader and allele-specific probes will bind to the target DNA forming the tripartite structure. This structure is recognized by cleavase, which will cleave and release the 3’ end of the allele-specific probe. If the SNP nucleotide in the target DNA is not complementary to the allele-specific probe, the correct tripartite structure is not formed and no cleavage occurs. The
343:
based on differential hybridization of probes. Comparatively, APEX methods have greater discriminating power than methods using this differential hybridization, as it is often impossible to obtain the optimal hybridization conditions for the thousands of probes on DNA microarrays (usually this is addressed by having highly redundant probes). However, the same density of probes cannot be achieved in APEX methods, which translates into lower output per run.
451:
158:
96:
146:
isolated state. Attached to one end of the probe is a fluorophore and to the other end a fluorescence quencher. Because of the stem-loop structure of the probe, the fluorophore is close to the quencher, thus preventing the molecule from emitting any fluorescence. The molecule is also engineered such that only the probe sequence is complementary to the genomic DNA that will be used in the assay (Abravaya et al. 2003).
165:
first probe's fluorophore wavelength is detected during the assay then the individual is homozygous to the wild type. If only the second probe's wavelength is detected then the individual is homozygous to the mutant allele. Finally, if both wavelengths are detected, then both molecular beacons must be hybridizing to their complements and thus the individual must contain both alleles and be heterozygous.
238:
497:
composition. Numerous investigators have been able to successfully eliminate the majority of their sequencing through melt-based scanning, allowing accurate locus-based genotyping of large numbers of individuals. Many investigators have found scanning for mutations using high resolution melting as a viable and practical way to study entire genes.
366:(up to 640-plex) and four-color single-base extension on a microarray. The multiplex PCR requires two oligonucleotides per SNP/mutation generating amplicons that contain the tested base pair. The same oligonucleotides are used in the following step as immobilized single-base extension primers on a microarray (Krjutskov et al. 2008).
342:
The flexibility and specificity of primer extension make it amenable to high throughput analysis. Primer extension probes can be arrayed on slides allowing for many SNPs to be genotyped at once. Broadly referred to as arrayed primer extension (APEX), this technology has several benefits over methods
308:
Primer extension is a two step process that first involves the hybridization of a probe to the bases immediately upstream of the SNP nucleotide followed by a ‘mini-sequencing’ reaction, in which DNA polymerase extends the hybridized primer by adding a base that is complementary to the SNP nucleotide.
164:
The unique design of these molecular beacons allows for a simple diagnostic assay to identify SNPs at a given location. If a molecular beacon is designed to match a wild-type allele and another to match a mutant of the allele, the two can be used to identify the genotype of an individual. If only the
86:
Several applications have been developed that interrogate SNPs by hybridizing complementary DNA probes to the SNP site. The challenge of this approach is reducing cross-hybridization between the allele-specific probes. This challenge is generally overcome by manipulating the hybridization stringency
562:
sequence less than 250 bases in a read which limits their ability to sequence whole genomes. However, their ability to generate results in real-time and their potential to be massively scaled up makes them a viable option for sequencing small regions to perform SNP genotyping. Compared to other SNP
480:
DHPLC is easily automated as no labeling or purification of the DNA fragments is needed. The method is also relatively fast and has a high specificity. One major drawback of DHPLC is that the column temperature must be optimized for each target in order to achieve the right degree of denaturation.
417:
DNA ligase catalyzes the ligation of the 3' end of a DNA fragment to the 5' end of a directly adjacent DNA fragment. This mechanism can be used to interrogate a SNP by hybridizing two probes directly over the SNP polymorphic site, whereby ligation can occur if the probes are identical to the target
228:
An alternative strategy is to run multiple qPCR reactions with different primer sets that target each allele separately. Well-designed primers will amplify their target SNP at a much earlier cycle than the other SNPs. This allows more than two alleles to be distinguished, although an individual qPCR
441:
Single-stranded DNA (ssDNA) folds into a tertiary structure. The conformation is sequence dependent and most single base pair mutations will alter the shape of the structure. When applied to a gel, the tertiary shape will determine the mobility of the ssDNA, providing a mechanism to differentiate
353:
assay is an example of a whole-genome genotyping pipeline that is based on primer extension method. In the
Infinium assay, over 100,000 SNPs can be genotyped. The assay uses hapten-labelled nucleotides in a primer extension reaction. The hapten label is recognized by anti-bodies, which in turn are
492:
is the simplest PCR-based method to understand. Basically, the same thermodynamic properties that allowed for the gel techniques to work apply here, and in real-time. A fluorimeter monitors the post-PCR denaturation of the entire dsDNA amplicon. You make primers specific to the site you want to
476:
to interrogate SNPs. The key to DHPLC is the solid phase which has differential affinity for single and double-stranded DNA. In DHPLC, DNA fragments are denatured by heating and then allowed to reanneal. The melting temperature of the reannealed DNA fragments determines the length of time they are
224:
Tetra-primer amplification refractory mutation system PCR, or ARMS-PCR, employs two pairs of primers to amplify two alleles in one PCR reaction. The primers are designed such that the two primer pairs overlap at a SNP location but each match perfectly to only one of the possible SNPs. The basis of
384:
assay is performed concurrently with a PCR reaction and the results can be read in real-time as the PCR reaction proceeds (McGuigan & Ralston 2002). The assay requires forward and reverse PCR primers that will amplify a region that includes the SNP polymorphic site. Allele discrimination is
338:
s iPLEX SNP genotyping method, which uses a MassARRAY mass spectrometer. Extension probes are designed in such a way that 40 different SNP assays can be amplified and analyzed in a PCR cocktail. The extension reaction uses ddNTPs as above, but the detection of the SNP allele is dependent on the
265:
oligonucleotide is a non-matching base that overlaps the SNP nucleotide in the target DNA. The second probe is an allele-specific probe which is complementary to the 5’ end of the target DNA, but also extends past the 3’ side of the SNP nucleotide. The allele-specific probe will contain a base
145:
makes use of a specifically engineered single-stranded oligonucleotide probe. The oligonucleotide is designed such that there are complementary regions at each end and a probe sequence located in between. This design allows the probe to take on a hairpin, or stem-loop, structure in its natural,
295:
assay. The assay has also been adapted in several ways for use in a high-throughput format. In one platform, the allele-specific probes are anchored to microspheres. When cleavage by FEN generates a detectable fluorescent signal, the signal is measured using flow-cytometry. The sensitivity of
270:
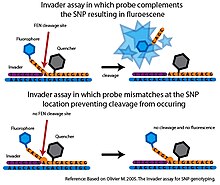
assay is usually coupled with fluorescence resonance energy transfer (FRET) system to detect the cleavage event. In this setup, a quencher molecule is attached to the 3’ end and a fluorophore is attached to the 5’ end of the allele-specific probe. If cleavage occurs, the fluorophore will be
149:
If the probe sequence of the molecular beacon encounters its target genomic DNA during the assay, it will anneal and hybridize. Because of the length of the probe sequence, the hairpin segment of the probe will be denatured in favour of forming a longer, more stable probe-target hybrid. This
215:
The combined factors of the high complexity of most eukaryotic genomes, the requirement for specific endonucleases, the fact that the exact mutation cannot necessarily be resolved in a single experiment, and the slow nature of gel assays make RFLP a poor choice for high throughput analysis.
389:
combined with one or two allele-specific probes that hybridize to the SNP polymorphic site. The probes will have a fluorophore linked to their 5’ end and a quencher molecule linked to their 3’ end. While the probe is intact, the quencher will remain in close proximity to the fluorophore,
77:
project, which aims to provide the minimal set of SNPs needed to genotype the human genome. SNPs can also provide a genetic fingerprint for use in identity testing. The increase of interest in SNPs has been reflected by the furious development of a diverse range of SNP genotyping methods.
496:
Scanning of larger amplicons is based on the same principles as outlined above. However, melting temperature and the overall shape of the melting curve become informative. For amplicons >c.150bp there are often >2 melting peaks, each of which can vary, depending on the DNA template
426:
The characteristic DNA properties of melting temperature and single stranded conformation have been used in several applications to distinguish SNP alleles. These methods very often achieve high specificity but require highly optimized conditions to obtain the best possible results.
278:
assay highly specific. However, in its original format, only one SNP allele could be interrogated per reaction sample and it required a large amount of target DNA to generate a detectable signal in a reasonable time frame. Several developments have extended the original
153:
If on the other hand, the probe sequence encounters a target sequence with as little as one non-complementary nucleotide, the molecular beacon will preferentially stay in its natural hairpin state and no fluorescence will be observed, as the fluorophore remains quenched.
102:
Dynamic allele-specific hybridization (DASH) genotyping takes advantage of the differences in the melting temperature in DNA that results from the instability of mismatched base pairs. The process can be vastly automated and encompasses a few simple principles.
125:
Because DASH genotyping is measuring a quantifiable change in Tm, it is capable of measuring all types of mutations, not just SNPs. Other benefits of DASH include its ability to work with label free probes and its simple design and performance conditions.
284:
229:
reaction is required for each SNP. To achieve high enough specificity, the primer sequence may require placement of an artificial mismatch near its 3'-end, which is an approach generally known as Taq-MAMA.
296:
flow-cytometry, eliminates the need for PCR amplification of the target DNA (Rao et al. 2003). These high-throughput platforms have not progressed beyond the proof-of-principle stage and so far the
257:, a FEN called cleavase is combined with two specific oligonucleotide probes, that together with the target DNA, can form a tripartite structure recognized by cleavase. The first probe, called the
541:
Surveyor nuclease is a mismatch endonuclease enzyme that recognizes all base substitutions and small insertions/deletions (indels), and cleaves the 3′ side of mismatched sites in both DNA strands.
509:
binds different single nucleotide mismatches with different affinities and can be used in capillary electrophoresis to differentiate all six sets of mismatches (Drabovich & Krylov 2006).
405:
assay is limited by the how close the SNPs can be situated. The scale of the assay can be drastically increased by performing many simultaneous reactions in microtitre plates. Generally,
247:(FEN) is an endonuclease that catalyzes structure-specific cleavage. This cleavage is highly sensitive to mismatches and can be used to interrogate SNPs with a high degree of specificity
135:
721:
Bulduk BK, Kiliç HB, Bekircan-Kurt CE, Haliloğlu G, Erdem Özdamar S, Topaloğlu H, Kocaefe YÇ (March 2020). "A Novel
Amplification-Refractory Mutation System-PCR Strategy to Screen
41:, which is the measurement of more general genetic variation. SNPs are one of the most common types of genetic variation. An SNP is a single base pair mutation at a specific
339:
actual mass of the extension product and not on a fluorescent molecule. This method is for low to medium high throughput, and is not intended for whole genome scanning.
409:
is limited to applications that involve interrogating a small number of SNPs since optimal probes and reaction conditions must be designed for each SNP (Syvanen 2001).
17:
118:
is then added in the presence of a molecule that fluoresces when bound to double-stranded DNA. The intensity is then measured as temperature is increased until the
150:
conformational change permits the fluorophore and quencher to be free of their tight proximity due to the hairpin association, allowing the molecule to fluoresce.
990:
Abravaya K, Huff J, Marshall R, Merchant B, Mullen C, Schneider G, Robinson J (April 2003). "Molecular beacons as diagnostic tools: technology and applications".
629:
Howell WM, Jobs M, Gyllensten U, Brookes AJ (January 1999). "Dynamic allele-specific hybridization. A new method for scoring single nucleotide polymorphisms".
505:
DNA mismatch-binding proteins can distinguish single nucleotide mismatches and thus facilitate differential analysis of SNPs. For example, MutS protein from
1070:
Drabovich AP, Krylov SN (March 2006). "Identification of base pairs in single-nucleotide polymorphisms by MutS protein-mediated capillary electrophoresis".
770:"Melt analysis of mismatch amplification mutation assays (Melt-MAMA): a functional study of a cost-effective SNP genotyping assay in bacterial models"
469:
401:
assay can be multiplexed by combining the detection of up to seven SNPs in one reaction. However, since each SNP requires a distinct probe, the
362:
is an arrayed primer extension genotyping method which is able to identify hundreds of SNPs or mutations in parallel using efficient homogeneous
251:
563:
genotyping methods, sequencing is in particular, suited to identifying multiple SNPs in a small region, such as the highly polymorphic
1289:
458:
358:
209:
436:
473:
613:
1204:
McGuigan FE, Ralston SH (September 2002). "Single nucleotide polymorphism detection: allelic discrimination using TaqMan".
917:
Oefner PJ, Underhill PA (1995). "Comparative DNA sequencing by denaturing high-performance liquid chromatography (DHPLC)".
386:
1101:"Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes"
61:. Because SNPs are conserved during evolution, they have been proposed as markers for use in quantitative trait loci (
1151:
564:
181:
Human SNP 5.0 GeneChip performs a genome-wide assay that can genotype over 500,000 human SNPs (Affymetrix 2007)..
115:
50:
34:
1300:
119:
1315:
1233:
Syvänen AC (December 2001). "Accessing genetic variation: genotyping single nucleotide polymorphisms".
555:
550:
324:
Generally, there are two main approaches which use the incorporation of either fluorescently labeled
107:
70:
1295:
1280:
672:
Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. (April 1989).
536:
1325:
937:
1171:"Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons"
674:"Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS)"
489:
8:
1330:
66:
261:
oligonucleotide is complementary to the 3’ end of the target DNA. The last base of the
1258:
1053:
1028:
1015:
968:
938:"Rapid identification of chloroplast haplotypes using High Resolution Melting analysis"
891:
866:
796:
769:
750:
654:
524:
421:
325:
110:
with a biotinylated primer. In the second step, the amplified product is attached to a
1143:
786:
698:
673:
1250:
1221:
1217:
1192:
1157:
1147:
1122:
1087:
1058:
1007:
960:
956:
896:
836:
801:
768:
Birdsell DN, Pearson T, Price EP, Hornstra HM, Nera RD, Stone N, et al. (2012).
754:
742:
703:
646:
609:
244:
1262:
1019:
658:
1242:
1213:
1182:
1139:
1112:
1079:
1048:
1040:
999:
972:
952:
886:
878:
828:
791:
781:
734:
693:
685:
638:
142:
106:
In the first step, a genomic segment is amplified and attached to a bead through a
58:
42:
1284:
1187:
1170:
882:
832:
287:(SISAR) allows both SNP alleles to be interrogated in a single reaction. SISAR
1099:
Gundry CN, Vandersteen JG, Reed GH, Pryor RJ, Chen J, Wittwer CT (March 2003).
559:
291:
assay also requires less target DNA, improving the sensitivity of the original
194:
1169:
Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C (July 2004).
1309:
819:
Li B, Kadura I, Fu DJ, Watson DE (February 2004). "Genotyping with TaqMAMA".
363:
1134:
Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, et al. (2006).
1029:"Molecular approaches in the diagnosis of primary immunodeficiency diseases"
689:
1320:
1254:
1225:
1196:
1161:
1126:
1091:
1062:
1011:
964:
900:
840:
805:
746:
738:
111:
1003:
707:
650:
450:
1044:
190:
178:
157:
134:
38:
1083:
1246:
1117:
1100:
310:
300:
system has not been used in any large scale SNP genotyping projects.
271:
separated from the quencher molecule generating a detectable signal.
201:
have been employed to generate high-fidelity SNP genotyping methods.
198:
174:
936:
Dang XD, Kelleher CT, Howard-Williams E, Meade CV (September 2012).
464:
422:
Other post-amplification methods based on physical properties of DNA
114:
column and washed with NaOH to remove the unbiotinylated strand. An
95:
333:
314:
54:
122:
can be determined. A SNP will result in a lower than expected Tm.
57:
of many human diseases and are becoming of particular interest in
720:
642:
484:
397:
assay is based on PCR, it is relatively simple to implement. The
46:
935:
237:
518:
376:
274:
Only minimal cleavage occurs with mismatched probes making the
74:
671:
318:
283:
assay. By carrying out secondary FEN cleavage reactions, the
989:
767:
628:
445:
204:
1138:. Methods in Enzymology. Vol. 410. pp. 359–376.
1098:
430:
1277:
374:
Taq DNA polymerase's 5’-nuclease activity is used in the
62:
354:
coupled to a detectable signal (Gunderson et al. 2006).
90:
500:
1168:
1133:
1026:
37:(SNPs) between members of a species. It is a form of
523:
SNPlex is a proprietary genotyping platform sold by
1296:Genotyping by Oligonucleotide Ligation Assay (OLA)
1027:Costabile M, Quach A, Ferrante A (December 2006).
53:is > 1%). SNPs are found to be involved in the
470:Denaturing high performance liquid chromatography
465:Denaturing high performance liquid chromatography
412:
1307:
818:
1203:
1069:
916:
725:Pathogenic Variants in Patient Repositories".
485:High-resolution melting of the entire amplicon
81:
1290:Delaware Valley Personalized Medicine Project
860:
858:
856:
854:
852:
850:
603:
599:
285:Serial Invasive Signal Amplification Reaction
912:
910:
597:
595:
593:
591:
589:
587:
585:
583:
581:
579:
33:is the measurement of genetic variations of
929:
73:. The use of SNPs is being extended in the
992:Clinical Chemistry and Laboratory Medicine
847:
530:
1186:
1116:
1052:
907:
890:
795:
785:
697:
622:
576:
18:Single-nucleotide polymorphism genotyping
1292:Uses SNPs to help make medicine personal
727:Genetic Testing and Molecular Biomarkers
459:temperature gradient gel electrophoresis
449:
446:Temperature gradient gel electrophoresis
236:
210:Restriction fragment length polymorphism
205:Restriction fragment length polymorphism
156:
94:
1232:
864:
606:Molecular analysis and genome discovery
437:Single_strand_conformation_polymorphism
431:Single strand conformation polymorphism
184:
175:high-density oligonucleotide SNP arrays
14:
1308:
867:"The Invader assay for SNP genotyping"
608:. London: John Wiley & Sons Ltd.
91:Dynamic allele-specific hybridization
501:Use of DNA mismatch-binding proteins
232:
219:
129:
303:
189:A broad range of enzymes including
24:
982:
168:
133:
25:
1342:
1271:
787:10.1371/journal.pone.0032866.s002
27:Measurement of genetic variations
1218:10.1097/00041444-200209000-00003
957:10.1111/j.1755-0998.2012.03164.x
565:Major Histocompatibility Complex
490:High Resolution Melting analysis
331:A different approach is used by
380:assay for SNP genotyping. The
369:
116:allele-specific oligonucleotide
35:single nucleotide polymorphisms
812:
761:
714:
665:
413:Oligonucleotide Ligation Assay
13:
1:
1144:10.1016/S0076-6879(06)10017-8
570:
544:
1278:International HapMap Project
1188:10.1373/clinchem.2004.032136
883:10.1016/j.mrfmmm.2004.08.016
604:Harbron S, Rapley R (2004).
472:(DHPLC) uses reversed-phase
45:, usually consisting of two
7:
945:Molecular Ecology Resources
833:10.1016/j.ygeno.2003.08.005
311:MALDI-TOF Mass spectrometry
82:Hybridization-based methods
10:
1347:
556:Next-generation sequencing
548:
534:
516:
434:
551:SNV calling from NGS data
512:
1235:Nature Reviews. Genetics
120:melting temperature (Tm)
1301:Why SNP test your mice?
1136:Whole-genome genotyping
865:Olivier M (June 2005).
537:Surveyor_nuclease_assay
531:Surveyor nuclease assay
739:10.1089/gtmb.2019.0079
678:Nucleic Acids Research
567:region of the genome.
454:
241:
161:
141:SNP detection through
138:
99:
1004:10.1515/CCLM.2003.070
690:10.1093/nar/17.7.2503
558:technologies such as
549:Further information:
453:
240:
160:
137:
98:
51:rare allele frequency
1206:Psychiatric Genetics
1072:Analytical Chemistry
631:Nature Biotechnology
185:Enzyme-based methods
67:association studies
1283:2014-04-16 at the
1175:Clinical Chemistry
1105:Clinical Chemistry
1045:10.1002/humu.20412
525:Applied Biosystems
455:
326:dideoxynucleotides
242:
162:
139:
100:
65:) analysis and in
1316:Molecular biology
1084:10.1021/ac0520386
1039:(12): 1163–1173.
871:Mutation Research
615:978-0-471-49919-0
507:Thermus aquaticus
245:Flap endonuclease
233:Flap endonuclease
220:PCR-based methods
143:molecular beacons
130:Molecular beacons
16:(Redirected from
1338:
1266:
1247:10.1038/35103535
1229:
1200:
1190:
1181:(7): 1156–1164.
1165:
1130:
1120:
1118:10.1373/49.3.396
1095:
1078:(6): 2035–2038.
1066:
1056:
1023:
977:
976:
942:
933:
927:
926:
914:
905:
904:
894:
877:(1–2): 103–110.
862:
845:
844:
816:
810:
809:
799:
789:
765:
759:
758:
718:
712:
711:
701:
684:(7): 2503–2516.
669:
663:
662:
626:
620:
619:
601:
304:Primer extension
59:pharmacogenetics
21:
1346:
1345:
1341:
1340:
1339:
1337:
1336:
1335:
1306:
1305:
1285:Wayback Machine
1274:
1269:
1241:(12): 930–942.
1154:
985:
983:Further reading
980:
940:
934:
930:
915:
908:
863:
848:
817:
813:
766:
762:
719:
715:
670:
666:
627:
623:
616:
602:
577:
573:
553:
547:
539:
533:
521:
515:
503:
487:
467:
448:
439:
433:
424:
415:
385:achieved using
372:
349:Incorporated's
321:-like methods.
306:
235:
222:
207:
187:
171:
169:SNP microarrays
132:
93:
84:
71:microsatellites
28:
23:
22:
15:
12:
11:
5:
1344:
1334:
1333:
1328:
1323:
1318:
1304:
1303:
1298:
1293:
1287:
1273:
1272:External links
1270:
1268:
1267:
1230:
1212:(3): 133–136.
1201:
1166:
1152:
1131:
1111:(3): 396–406.
1096:
1067:
1033:Human Mutation
1024:
998:(4): 468–474.
986:
984:
981:
979:
978:
951:(5): 894–908.
928:
919:Am J Hum Genet
906:
846:
827:(2): 311–320.
811:
760:
733:(3): 165–170.
713:
664:
621:
614:
574:
572:
569:
560:pyrosequencing
546:
543:
535:Main article:
532:
529:
517:Main article:
514:
511:
502:
499:
486:
483:
466:
463:
447:
444:
435:Main article:
432:
429:
423:
420:
414:
411:
371:
368:
305:
302:
234:
231:
221:
218:
206:
203:
195:DNA polymerase
186:
183:
170:
167:
131:
128:
92:
89:
83:
80:
31:SNP genotyping
26:
9:
6:
4:
3:
2:
1343:
1332:
1329:
1327:
1326:Biotechnology
1324:
1322:
1319:
1317:
1314:
1313:
1311:
1302:
1299:
1297:
1294:
1291:
1288:
1286:
1282:
1279:
1276:
1275:
1264:
1260:
1256:
1252:
1248:
1244:
1240:
1236:
1231:
1227:
1223:
1219:
1215:
1211:
1207:
1202:
1198:
1194:
1189:
1184:
1180:
1176:
1172:
1167:
1163:
1159:
1155:
1153:9780121828158
1149:
1145:
1141:
1137:
1132:
1128:
1124:
1119:
1114:
1110:
1106:
1102:
1097:
1093:
1089:
1085:
1081:
1077:
1073:
1068:
1064:
1060:
1055:
1050:
1046:
1042:
1038:
1034:
1030:
1025:
1021:
1017:
1013:
1009:
1005:
1001:
997:
993:
988:
987:
974:
970:
966:
962:
958:
954:
950:
946:
939:
932:
924:
920:
913:
911:
902:
898:
893:
888:
884:
880:
876:
872:
868:
861:
859:
857:
855:
853:
851:
842:
838:
834:
830:
826:
822:
815:
807:
803:
798:
793:
788:
783:
780:(3): e32866.
779:
775:
771:
764:
756:
752:
748:
744:
740:
736:
732:
728:
724:
717:
709:
705:
700:
695:
691:
687:
683:
679:
675:
668:
660:
656:
652:
648:
644:
640:
636:
632:
625:
617:
611:
607:
600:
598:
596:
594:
592:
590:
588:
586:
584:
582:
580:
575:
568:
566:
561:
557:
552:
542:
538:
528:
526:
520:
510:
508:
498:
494:
491:
482:
478:
475:
471:
462:
460:
452:
443:
438:
428:
419:
410:
408:
404:
400:
396:
391:
388:
383:
379:
378:
367:
365:
364:multiplex PCR
361:
360:
355:
352:
348:
344:
340:
337:
335:
329:
327:
322:
320:
316:
312:
301:
299:
294:
290:
286:
282:
277:
272:
269:
264:
260:
256:
254:
250:In the basic
248:
246:
239:
230:
226:
217:
213:
211:
202:
200:
196:
192:
182:
180:
176:
166:
159:
155:
151:
147:
144:
136:
127:
123:
121:
117:
113:
109:
104:
97:
88:
79:
76:
72:
68:
64:
60:
56:
52:
48:
44:
40:
36:
32:
19:
1238:
1234:
1209:
1205:
1178:
1174:
1135:
1108:
1104:
1075:
1071:
1036:
1032:
995:
991:
948:
944:
931:
922:
918:
874:
870:
824:
820:
814:
777:
773:
763:
730:
726:
722:
716:
681:
677:
667:
643:10.1038/5270
637:(1): 87–88.
634:
630:
624:
605:
554:
540:
522:
506:
504:
495:
488:
479:
468:
456:
440:
425:
416:
406:
402:
398:
394:
392:
381:
375:
373:
370:5’- nuclease
357:
356:
350:
346:
345:
341:
332:
330:
323:
307:
297:
292:
288:
280:
275:
273:
267:
262:
258:
252:
249:
243:
227:
223:
214:
208:
188:
172:
163:
152:
148:
140:
124:
112:streptavidin
108:PCR reaction
105:
101:
87:conditions.
85:
69:in place of
30:
29:
49:(where the
1331:Gene tests
1310:Categories
571:References
545:Sequencing
393:Since the
191:DNA ligase
179:Affymetrix
39:genotyping
925:: 103–10.
755:212693790
199:nucleases
1281:Archived
1263:15411761
1255:11733746
1226:12218656
1197:15229148
1162:16938560
1127:12600951
1092:16536443
1063:16960849
1020:36220311
1012:12747588
965:22783911
901:15829241
841:14706460
821:Genomics
806:22438886
774:PLOS ONE
747:32167396
659:37367067
351:Infinium
347:Illumina
334:Sequenom
315:Sequenom
55:etiology
1054:7165860
973:8384592
892:2771639
797:3306377
708:2785681
651:9920276
298:Invader
293:Invader
289:Invader
281:Invader
276:Invader
268:Invader
263:Invader
259:Invader
253:Invader
47:alleles
1261:
1253:
1224:
1195:
1160:
1150:
1125:
1090:
1061:
1051:
1018:
1010:
971:
963:
899:
889:
839:
804:
794:
753:
745:
723:MT-TL1
706:
699:317639
696:
657:
649:
612:
519:SNPlex
513:SNPlex
407:TaqMan
403:TaqMan
399:TaqMan
395:TaqMan
382:TaqMan
377:TaqMan
359:APEX-2
317:) and
75:HapMap
1259:S2CID
1016:S2CID
969:S2CID
941:(PDF)
751:S2CID
655:S2CID
319:ELISA
313:(see
255:assay
43:locus
1251:PMID
1222:PMID
1193:PMID
1158:PMID
1148:ISBN
1123:PMID
1088:PMID
1059:PMID
1008:PMID
961:PMID
897:PMID
837:PMID
802:PMID
743:PMID
704:PMID
647:PMID
610:ISBN
474:HPLC
457:The
387:FRET
197:and
1321:DNA
1243:doi
1214:doi
1183:doi
1140:doi
1113:doi
1080:doi
1049:PMC
1041:doi
1000:doi
953:doi
887:PMC
879:doi
875:573
829:doi
792:PMC
782:doi
735:doi
694:PMC
686:doi
639:doi
173:In
63:QTL
1312::
1257:.
1249:.
1237:.
1220:.
1210:12
1208:.
1191:.
1179:50
1177:.
1173:.
1156:.
1146:.
1121:.
1109:49
1107:.
1103:.
1086:.
1076:78
1074:.
1057:.
1047:.
1037:27
1035:.
1031:.
1014:.
1006:.
996:41
994:.
967:.
959:.
949:12
947:.
943:.
923:57
921:.
909:^
895:.
885:.
873:.
869:.
849:^
835:.
825:83
823:.
800:.
790:.
776:.
772:.
749:.
741:.
731:24
729:.
702:.
692:.
682:17
680:.
676:.
653:.
645:.
635:17
633:.
578:^
527:.
193:,
1265:.
1245::
1239:2
1228:.
1216::
1199:.
1185::
1164:.
1142::
1129:.
1115::
1094:.
1082::
1065:.
1043::
1022:.
1002::
975:.
955::
903:.
881::
843:.
831::
808:.
784::
778:7
757:.
737::
710:.
688::
661:.
641::
618:.
336:'
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.