448:
31:
3146:
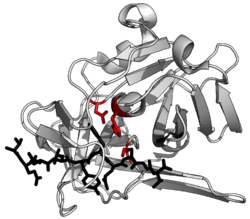
487:(Nu) and attack the substrate. This forms an intermediate where the enzyme is covalently linked to the N-terminal half of the substrate. In a second step, water is activated to hydrolyse this intermediate and complete catalysis. Other enzyme residues (not shown) donate and accept hydrogens and electrostatically stabilise charge build-up along the reaction mechanism.
779:
cycles in the recycling of proteins, and such activity tends to be regulated by nutritional signals in these organisms. The net impact of nutritional regulation of protease activity among the thousands of species present in soil can be observed at the overall microbial community level as proteins
744:
and interfere with the victim's blood clotting cascade. Proteases determine the lifetime of other proteins playing important physiological roles like hormones, antibodies, or other enzymes. This is one of the fastest "switching on" and "switching off" regulatory mechanisms in the physiology of an
571:
link the protease to the substrate protein, releasing the first half of the product. This covalent acyl-enzyme intermediate is then hydrolyzed by activated water to complete catalysis by releasing the second half of the product and regenerating the free
314:, and some superfamilies show divergent evolution to multiple different nucleophiles. Metalloproteases, aspartic, and glutamic proteases utilize their active site residues to activate a water molecule, which then attacks the scissile bond.
620:
Proteases, being themselves proteins, are cleaved by other protease molecules, sometimes of the same variety. This acts as a method of regulation of protease activity. Some proteases are less active after autolysis (e.g.
608:) requires this level of specificity in order to achieve precise cleavage events. This is achieved by proteases having a long binding cleft or tunnel with several pockets that bind to specified residues. For example,
673:). The activity can be a destructive change (abolishing a protein's function or digesting it to its principal components), it can be an activation of a function, or it can be a signal in a signalling pathway.
338:
forms a cyclic chemical structure that cleaves itself at asparagine residues in proteins under the right conditions. Given its fundamentally different mechanism, its inclusion as a peptidase may be debatable.
725:, etc.) play an important role in blood-clotting, as well as lysis of the clots, and the correct action of the immune system. Other proteases are present in leukocytes (
278:
residue that has the cysteine and threonine (proteases) or a water molecule (aspartic, glutamic and metalloproteases) nucleophilic so that it can attack the peptide
2567:
2823:
1827:
1362:
Radzicka A, Wolfenden R (July 1996). "Rates of
Uncatalyzed Peptide Bond Hydrolysis in Neutral Solution and the Transition State Affinities of Proteases".
1058:
found in the seeds of some plants, most notable for humans being soybeans, a major food crop, where they act to discourage predators. Raw soybeans are
649:. These enzymes are involved in a multitude of physiological reactions from simple digestion of food proteins to highly regulated cascades (e.g., the
898:. A variety of proteases are used medically both for their native function (e.g. controlling blood clotting) or for completely artificial functions (
589:, which have to be able to cleave the array of proteins ingested into smaller peptide fragments. Promiscuous proteases typically bind to a single
483:
water, which then hydrolyses the substrate. The bottom panel shows 2-step hydrolysis where a residue within the enzyme is activated to act as a
363:
based on sequence similarity (e.g. the S1 and C3 families within the PA clan). Each family may contain many hundreds of related proteases (e.g.
246:
Proteases were first grouped into 84 families according to their evolutionary relationship in 1993, and classified under four catalytic types:
560:
Aspartic, glutamic, and metallo-proteases activate a water molecule, which performs a nucleophilic attack on the peptide bond to hydrolyze it.
2632:
861:
and protein quality control. Only two ATP-dependent proteases are found in archaea: the membrane associated LonB protease and a soluble
2662:
2175:
Feijoo-Siota L, Villa TG (28 September 2010). "Native and
Biotechnologically Engineered Plant Proteases with Industrial Applications".
1791:
Zelisko A, Jackowski G (October 2004). "Senescence-dependent degradation of Lhcb3 is mediated by a thylakoid membrane-bound protease".
2614:
3176:
2739:
2460:
2447:
2159:
2134:
1636:
834:) have high specificity and only cleave a very restricted set of substrate sequences. They are therefore a common target for
697:
Proteases are used throughout an organism for various metabolic processes. Acid proteases secreted into the stomach (such as
2532:
874:
681:
Plant genomes encode hundreds of proteases, largely of unknown function. Those with known function are largely involved in
2359:
600:
Conversely some proteases are highly specific and only cleave substrates with a certain sequence. Blood clotting (such as
2791:
2421:
661:
pathways, and the invertebrate prophenoloxidase-activating cascade). Proteases can either break specific peptide bonds (
1380:
To assess the relative proficiencies of enzymes that catalyze the hydrolysis of internal and C-terminal peptide bonds
1027:
ligands, attached to lipocalin proteins, have been found to possess tumor protease inhibiting properties. The natural
2865:
1147:
1044:
1032:
1005:
963:
835:
2776:
752:
reactions, which result in rapid and efficient amplification of an organism's response to a physiological signal.
2297:
1847:
3021:
2707:
2622:
585:
such that a wide range of protein substrates are hydrolyzed. This is the case for digestive enzymes such as
2640:
2379:
1152:
1028:
969:
959:
382:
Currently more than 50 clans are known, each indicating an independent evolutionary origin of proteolysis.
2369:
2334:
359:
where P indicates a mixture of nucleophile families). Within each 'clan', proteases are classified into
3136:
2374:
2353:
17:
3122:
3109:
3096:
3083:
3070:
3057:
3044:
3006:
1137:
231:
2316:
803:
3016:
2970:
2913:
2602:
2438:
2393:
1062:
to many animals, including humans, until the protease inhibitors they contain have been denatured.
985:
464:
323:
2918:
2781:
2771:
2597:
2498:
2488:
2414:
407:
2585:
2473:
1652:
Rodriguez J, Gupta N, Smith RD, Pevzner PA (January 2008). "Does trypsin cut before proline?".
1142:
351:
is found in the MEROPS database. In this database, proteases are classified firstly by 'clan' (
306:
as a nucleophile. This is not an evolutionary grouping, however, as the nucleophile types have
2027:"Archaeal membrane-associated proteases: insights on Haloferax volcanii and other haloarchaea"
270:
proteases were not described until 1995 and 2004 respectively. The mechanism used to cleave a
2939:
2858:
2592:
2493:
1759:
682:
125:
In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of
3011:
2766:
2575:
1877:
1700:
1318:
1307:"Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism"
1117:
788:
767:
the peptide bonds in proteins and therefore break the proteins down into their constituent
331:
239:
134:
1768:
1499:
1054:
Other natural protease inhibitors are used as defense mechanisms. Common examples are the
783:
Bacteria contain proteases responsible for general protein quality control (e.g. the AAA+
137:, and different classes of protease can perform the same reaction by completely different
8:
2975:
2725:
2687:
2671:
1394:"New families of carboxyl peptidases: serine-carboxyl peptidases and glutamic peptidases"
1132:
977:
749:
352:
348:
311:
165:
138:
1881:
1704:
1483:
1322:
1255:"Molecular basis of substrate recognition and degradation by human presequence protease"
1185:
902:
for the targeted degradation of pathogenic proteins). Highly specific proteases such as
3166:
2908:
2761:
2702:
2692:
2682:
2407:
2343:
2192:
2102:
2077:
2053:
2026:
1999:
1974:
1868:
Sims GK, Wander MM (2002). "Proteolytic activity under nitrogen or sulfur limitation".
1723:
1688:
1604:
1579:
1552:
1525:
1459:
1434:
1339:
1306:
1279:
1254:
1227:
1200:
1090:
938:
582:
518:
263:
182:
115:
111:
2271:
2246:
2223:
1889:
1689:"A tobacco etch virus protease with increased substrate tolerance at the P1' position"
946:
remedy for digestion and diabetes in the Indian subcontinent. It is also used to make
2756:
2649:
2389:
2276:
2227:
2155:
2130:
2107:
2058:
2004:
1955:
1920:
1808:
1773:
1746:
van der Hoorn RA (2008). "Plant proteases: from phenotypes to molecular mechanisms".
1728:
1669:
1632:
1609:
1557:
1503:
1464:
1415:
1344:
1284:
1232:
1100:
1095:
1080:
1055:
993:
891:
823:
654:
480:
415:
267:
255:
251:
206:
194:
189:
170:
43:
2196:
1008:(which protects the body from inadequate coagulation by blocking protease-triggered
2954:
2949:
2923:
2851:
2806:
2801:
2677:
2357:
2266:
2258:
2219:
2210:
Southan C (July 2001). "A genomic perspective on human proteases as drug targets".
2184:
2097:
2089:
2048:
2038:
1994:
1986:
1947:
1912:
1885:
1839:
1800:
1763:
1755:
1718:
1708:
1661:
1599:
1591:
1547:
1537:
1495:
1454:
1446:
1405:
1371:
1334:
1326:
1274:
1266:
1222:
1212:
880:
795:
563:
Serine, threonine, and cysteine proteases use a nucleophilic residue (usually in a
94:, and spurring the formation of new protein products. They do this by cleaving the
1843:
933:
794:
A secreted bacterial protease may also act as an exotoxin, and be an example of a
3171:
3001:
2985:
2898:
2811:
2796:
2751:
2363:
2338:
1828:"Nitrogen Starvation Promotes Biodegradation of N-Heterocyclic Compounds in Soil"
1713:
1253:
King JV, Liang WG, Scherpelz KP, Schilling AB, Meredith SC, Tang WJ (July 2014).
1186:"Proteolytic enzyme | Description, Types, & Functions | Britannica"
1127:
1105:
1085:
1036:
992:(which protects the body from excessive protease-triggered activation of its own
564:
492:
430:
376:
326:, was described in 2011. Its proteolytic mechanism is unusual since, rather than
307:
287:
259:
247:
218:
213:
201:
158:
2399:
1804:
883:
related to this field were published each year. Proteases are used in industry,
41:) complexed with its peptide substrate in black with catalytic residues in red.(
3150:
3039:
2980:
2832:
2468:
2311:
1048:
911:
895:
850:
722:
686:
522:
514:
360:
119:
34:
2188:
1990:
1938:
Skoreński M, Sieńczyk M (2013). "Viral proteases as targets for drug design".
1270:
3160:
2944:
2903:
2743:
2545:
2348:
2078:"Proteolytic systems of archaea: slicing, dicing, and mincing in the extreme"
2043:
713:) enable us to digest the protein in food. Proteases present in blood serum (
593:
on the substrate and so only have specificity for that residue. For example,
510:
502:
447:
210:
198:
107:
95:
2385:
TopFIND protease database covering cut sites, substrates and protein termini
1542:
1526:"Asparagine peptide lyases: a seventh catalytic type of proteolytic enzymes"
2893:
2451:
2280:
2231:
2111:
2062:
2008:
1959:
1951:
1924:
1812:
1777:
1732:
1673:
1613:
1561:
1507:
1419:
1348:
1288:
1236:
1217:
1059:
1009:
997:
989:
981:
915:
903:
879:
The field of protease research is enormous. Since 2004, approximately 8000
831:
827:
799:
771:. Bacterial and fungal proteases are particularly important to the global
734:
710:
622:
609:
605:
530:
271:
38:
1468:
669:
sequence of a protein, or completely break down a peptide to amino acids (
3117:
3052:
2888:
2697:
2644:
2580:
2540:
2093:
1595:
1122:
1013:
1001:
929:
920:
819:
730:
650:
626:
484:
456:
283:
87:
79:
1410:
1393:
1330:
822:, which needs a protease to cleave this into functional units (e.g. the
597:
is specific for the sequences ...K\... or ...R\... ('\'=cleavage site).
2384:
2262:
1160:- database of protease specificity, substrates, products and inhibitors
1024:
862:
854:
784:
780:
are broken down in response to carbon, nitrogen, or sulfur limitation.
768:
741:
666:
638:
590:
509:
residues. Some detach the terminal amino acids from the protein chain (
506:
472:
452:
335:
327:
275:
235:
99:
91:
30:
1916:
1665:
1450:
1375:
355:) based on structure, mechanism and catalytic residue order (e.g. the
47:
3091:
3065:
2430:
1020:
858:
764:
738:
658:
642:
553:
498:
411:
303:
291:
186:
76:
2375:
Proteolysis Cut Site database - curated expert annotation from users
1023:
proteins, which play a role in cell regulation and differentiation.
918:
in a controlled fashion. Protease-containing plant-solutions called
110:. Proteases are involved in numerous biological pathways, including
3145:
1043:
among them, depend on proteases in their reproductive cycle. Thus,
1040:
943:
907:
884:
806:). Bacterial exotoxic proteases destroy extracellular structures.
776:
760:
726:
714:
702:
601:
568:
542:
423:
372:
368:
299:
279:
174:
83:
1157:
1112:
846:
815:
718:
706:
646:
612:
is specific for the sequence ...ENLYFQ\S... ('\'=cleavage site).
594:
586:
526:
364:
356:
1975:"Improving Viral Protease Inhibitors to Counter Drug Resistance"
3104:
2874:
2520:
2513:
2508:
1163:
1072:
973:
947:
925:
772:
698:
538:
534:
460:
419:
295:
162:
72:
2331:
733:) and play several different roles in metabolic control. Some
685:
regulation. Plant proteases also play a role in regulation of
3078:
2555:
2550:
2483:
2478:
2247:"A genomic analysis of rat proteases and protease inhibitors"
468:
222:
177:
130:
103:
1972:
501:
long protein chains into shorter fragments by splitting the
2326:
1252:
980:(which protects the body from excessive effects of its own
476:
226:
126:
2843:
1651:
849:
use proteases to regulate various cellular processes from
390:
Alternatively, proteases may be classified by the optimal
1305:
Shen Y, Joachimiak A, Rosner MR, Tang WJ (October 2006).
2124:
2024:
1626:
1304:
1201:"Proteases: multifunctional enzymes in life and disease"
748:
By a complex cooperative action, proteases can catalyze
27:
Enzyme that cleaves other proteins into smaller peptides
894:
and are also used extensively in the bread industry in
391:
1686:
1577:
1523:
521:); others attack internal peptide bonds of a protein (
347:
An up-to-date classification of protease evolutionary
3134:
2129:(2nd ed.). London, UK: Elsevier Academic Press.
2076:
Maupin-Furlow JA (December 2018). Robinson NP (ed.).
1973:
Kurt Yilmaz N, Swanstrom R, Schiffer CA (July 2016).
1631:(8th ed.). Philadelphia: Saunders. p. 122.
154:
Proteases can be classified into seven broad groups:
2370:
1524:
Rawlings ND, Barrett AJ, Bateman A (November 2011).
1578:Rawlings ND, Barrett AJ, Bateman A (January 2010).
442:
1019:Natural protease inhibitors include the family of
567:). That residue performs a nucleophilic attack to
342:
129:. Proteases can be found in all forms of life and
2429:
2174:
1937:
1627:Mitchell RS, Kumar V, Abbas AK, Fausto N (2007).
1361:
322:A seventh catalytic type of proteolytic enzymes,
3158:
2244:
1790:
1687:Renicke C, Spadaccini R, Taxis C (2013-06-24).
1573:
1571:
1432:
1248:
1246:
2125:Barrett AJ, Rawlings ND, Woessnerd JF (2004).
1300:
1298:
1198:
2859:
2415:
2075:
2025:Giménez MI, Cerletti M, De Castro RE (2015).
1745:
149:
1568:
1243:
972:. One example of protease inhibitors is the
942:has been in use for thousands of years as a
632:
615:
604:) and viral polyprotein processing (such as
1903:Tong L (December 2002). "Viral proteases".
1295:
2866:
2852:
2422:
2408:
1867:
1166:- Database of protease evolutionary groups
968:The activity of proteases is inhibited by
924:have been in use for hundreds of years in
2392:at the U.S. National Library of Medicine
2354:List of proteases and their specificities
2270:
2101:
2052:
2042:
1998:
1767:
1722:
1712:
1603:
1551:
1541:
1458:
1433:Rawlings ND, Barrett AJ (February 1993).
1409:
1338:
1278:
1226:
1216:
1192:
887:and as a basic biological research tool.
1760:10.1146/annurev.arplant.59.032607.092835
1620:
1364:Journal of the American Chemical Society
1000:(which protects the body from excessive
446:
29:
2209:
1484:"Activity-Based Profiling of Proteases"
1199:López-Otín C, Bond JS (November 2008).
637:Proteases occur in all organisms, from
14:
3159:
2380:Protease cut sites graphical interface
2245:Puente XS, López-Otín C (April 2004).
2149:
1481:
701:) and serine proteases present in the
625:) whilst others are more active (e.g.
556:is achieved by one of two mechanisms:
385:
334:. During this reaction, the catalytic
2847:
2403:
2020:
2018:
1500:10.1146/annurev-biochem-060713-035352
1435:"Evolutionary families of peptidases"
1035:used in antiretroviral therapy. Some
890:Digestive proteases are part of many
1902:
1825:
1519:
1517:
875:Proteases (medical and related uses)
471:in blue. The top panel shows 1-step
2792:Amyloid precursor protein secretase
1530:The Journal of Biological Chemistry
1391:
1205:The Journal of Biological Chemistry
24:
2015:
25:
3188:
2327:International Proteolysis Society
2291:
2152:Proteases in biology and medicine
1514:
1148:Protease inhibitor (pharmacology)
1006:plasminogen activator inhibitor-1
964:Protease inhibitor (pharmacology)
317:
144:
118:(breakdown of old proteins), and
3144:
2777:Proteasome endopeptidase complex
2082:Emerging Topics in Life Sciences
1580:"MEROPS: the peptidase database"
1031:are not to be confused with the
443:Enzymatic function and mechanism
3177:Post-translational modification
2332:MEROPS - the peptidase database
2238:
2203:
2168:
2143:
2127:Handbook of proteolytic enzymes
2118:
2069:
1966:
1931:
1896:
1861:
1832:Soil Biology & Biochemistry
1819:
1784:
1739:
1680:
1645:
343:Based on evolutionary phylogeny
2177:Food and Bioprocess Technology
1769:11858/00-001M-0000-0012-37C7-9
1748:Annual Review of Plant Biology
1482:Sanman, Laura E. (June 2014).
1475:
1426:
1385:
1355:
1178:
789:unfolded or misfolded proteins
576:
13:
1:
2641:Serine type carboxypeptidases
2623:Angiotensin-converting enzyme
2224:10.1016/s1359-6446(01)01793-7
1940:Current Pharmaceutical Design
1890:10.1016/S0929-1393(01)00192-5
1844:10.1016/j.soilbio.2006.01.006
1590:(Database issue): D227–D233.
1488:Annual Review of Biochemistry
1171:
953:
2317:Resources in other libraries
1714:10.1371/journal.pone.0067915
1654:Journal of Proteome Research
1153:Protease inhibitor (biology)
960:Protease inhibitor (biology)
910:are commonly used to cleave
737:are also proteases, such as
548:
294:residue is used to activate
7:
2873:
2344:List of protease inhibitors
1805:10.1016/j.jplph.2004.01.006
1793:Journal of Plant Physiology
1065:
755:
10:
3193:
2349:Protease cutting predictor
2154:. London: Portland Press.
957:
872:
841:
809:
692:
581:Proteolysis can be highly
497:Proteases are involved in
490:
422:. This group includes the
410:. Here, it is released by
394:in which they are active:
150:Based on catalytic residue
133:. They have independently
3030:
3022:Michaelis–Menten kinetics
2994:
2963:
2932:
2881:
2822:
2738:
2718:
2661:
2631:
2613:
2566:
2531:
2459:
2446:
2312:Resources in your library
2189:10.1007/s11947-010-0431-4
2031:Frontiers in Microbiology
1991:10.1016/j.tim.2016.03.010
1271:10.1016/j.str.2014.05.003
1138:Proteases in angiogenesis
976:superfamily. It includes
936:. Vegetarian rennet from
830:). These proteases (e.g.
676:
633:Biodiversity of proteases
616:Degradation and autolysis
475:where the enzyme uses an
414:and causes activation of
282:group. One way to make a
232:Asparagine peptide lyases
2914:Diffusion-limited enzyme
2603:Tripeptidyl peptidase II
2394:Medical Subject Headings
2044:10.3389/fmicb.2015.00039
986:alpha 1-antichymotrypsin
934:kosher and halal Cheeses
451:A comparison of the two
324:asparagine peptide lyase
2772:Threonine endopeptidase
2598:Tripeptidyl peptidase I
2150:Hooper NM, ed. (2002).
1629:Robbins Basic Pathology
1543:10.1074/jbc.M111.260026
1439:The Biochemical Journal
1398:Journal of Biochemistry
1143:Intramembrane proteases
988:(which does likewise),
868:
651:blood-clotting cascade
408:type 1 hypersensitivity
379:within the S1 family).
2762:Aspartic acid protease
2586:Dipeptidyl peptidase-4
1979:Trends in Microbiology
1952:10.2174/13816128130613
1584:Nucleic Acids Research
1392:Oda K (January 2012).
1218:10.1074/jbc.R800035200
488:
135:evolved multiple times
114:of ingested proteins,
52:
3007:Eadie–Hofstee diagram
2940:Allosteric regulation
2593:Tripeptidyl peptidase
763:secrete proteases to
671:unlimited proteolysis
450:
242:(not requiring water)
33:
3017:Lineweaver–Burk plot
2767:Metalloendopeptidase
2672:Metalloexopeptidases
2576:Dipeptidyl peptidase
2212:Drug Discovery Today
2094:10.1042/ETLS20180025
1118:Convergent evolution
1051:therapeutic agents.
814:The genomes of some
665:), depending on the
455:mechanisms used for
332:elimination reaction
308:evolved convergently
240:elimination reaction
139:catalytic mechanisms
2782:HslU—HslV peptidase
2726:Metalloexopeptidase
1882:2002AppSE..19..217S
1705:2013PLoSO...867915R
1536:(44): 38321–38328.
1331:10.1038/nature05143
1323:2006Natur.443..870S
1211:(45): 30433–30437.
1133:The Proteolysis Map
1045:protease inhibitors
1033:protease inhibitors
1029:protease inhibitors
978:alpha 1-antitrypsin
970:protease inhibitors
836:protease inhibitors
818:encode one massive
663:limited proteolysis
467:protein in red and
463:is shown in black,
386:Based on optimal pH
274:involves making an
183:Threonine proteases
102:, a reaction where
98:within proteins by
2976:Enzyme superfamily
2909:Enzyme promiscuity
2362:2011-04-30 at the
2337:2006-11-14 at the
2263:10.1101/gr.1946304
1596:10.1093/nar/gkp971
1056:trypsin inhibitors
939:Withania coagulans
892:laundry detergents
519:carboxypeptidase A
489:
436:alkaline proteases
207:Glutamic proteases
195:Aspartic proteases
171:Cysteine proteases
116:protein catabolism
69:proteolytic enzyme
53:
3132:
3131:
2841:
2840:
2790:Other/ungrouped:
2757:Cysteine protease
2734:
2733:
2652:
2298:Library resources
2161:978-1-85578-147-4
2136:978-0-12-079610-6
1917:10.1021/cr010184f
1911:(12): 4609–4626.
1799:(10): 1157–1170.
1666:10.1021/pr0705035
1638:978-1-4160-2973-1
1451:10.1042/bj2900205
1445:(Pt 1): 205–218.
1411:10.1093/jb/mvr129
1376:10.1021/ja954077c
1370:(26): 6105–6109.
1317:(7113): 870–874.
1047:are developed as
994:complement system
921:vegetarian rennet
824:hepatitis C virus
804:exfoliative toxin
655:complement system
404:Neutral proteases
330:, it performs an
190:secondary alcohol
16:(Redirected from
3184:
3149:
3148:
3140:
3012:Hanes–Woolf plot
2955:Enzyme activator
2950:Enzyme inhibitor
2924:Enzyme catalysis
2868:
2861:
2854:
2845:
2844:
2807:Beta-secretase 2
2802:Beta-secretase 1
2678:Carboxypeptidase
2674:
2650:
2457:
2456:
2424:
2417:
2410:
2401:
2400:
2285:
2284:
2274:
2242:
2236:
2235:
2207:
2201:
2200:
2183:(6): 1066–1088.
2172:
2166:
2165:
2147:
2141:
2140:
2122:
2116:
2115:
2105:
2073:
2067:
2066:
2056:
2046:
2022:
2013:
2012:
2002:
1970:
1964:
1963:
1946:(6): 1126–1153.
1935:
1929:
1928:
1905:Chemical Reviews
1900:
1894:
1893:
1865:
1859:
1858:
1856:
1855:
1846:. Archived from
1838:(8): 2478–2480.
1826:Sims GK (2006).
1823:
1817:
1816:
1788:
1782:
1781:
1771:
1743:
1737:
1736:
1726:
1716:
1684:
1678:
1677:
1649:
1643:
1642:
1624:
1618:
1617:
1607:
1575:
1566:
1565:
1555:
1545:
1521:
1512:
1511:
1479:
1473:
1472:
1462:
1430:
1424:
1423:
1413:
1389:
1383:
1382:
1359:
1353:
1352:
1342:
1302:
1293:
1292:
1282:
1250:
1241:
1240:
1230:
1220:
1196:
1190:
1189:
1182:
796:virulence factor
262:proteases. The
219:Metalloproteases
159:Serine proteases
82:, breaking down
50:
21:
3192:
3191:
3187:
3186:
3185:
3183:
3182:
3181:
3157:
3156:
3155:
3143:
3135:
3133:
3128:
3040:Oxidoreductases
3026:
3002:Enzyme kinetics
2990:
2986:List of enzymes
2959:
2928:
2899:Catalytic triad
2877:
2872:
2842:
2837:
2818:
2812:Gamma secretase
2797:Alpha secretase
2752:Serine protease
2730:
2719:Other/ungrouped
2714:
2670:
2657:
2653:-Transpeptidase
2627:
2609:
2562:
2527:
2442:
2428:
2364:Wayback Machine
2339:Wayback Machine
2323:
2322:
2321:
2306:
2305:
2301:
2294:
2289:
2288:
2251:Genome Research
2243:
2239:
2218:(13): 681–688.
2208:
2204:
2173:
2169:
2162:
2148:
2144:
2137:
2123:
2119:
2074:
2070:
2023:
2016:
1971:
1967:
1936:
1932:
1901:
1897:
1870:Appl. Soil Ecol
1866:
1862:
1853:
1851:
1824:
1820:
1789:
1785:
1744:
1740:
1685:
1681:
1650:
1646:
1639:
1625:
1621:
1576:
1569:
1522:
1515:
1480:
1476:
1431:
1427:
1390:
1386:
1360:
1356:
1303:
1296:
1265:(7): 996–1007.
1251:
1244:
1197:
1193:
1184:
1183:
1179:
1174:
1169:
1128:Catalytic triad
1068:
966:
958:Main articles:
956:
912:fusion proteins
877:
871:
844:
812:
787:) by degrading
758:
695:
679:
635:
618:
579:
565:catalytic triad
551:
515:aminopeptidases
495:
493:Catalytic triad
445:
431:Basic proteases
388:
345:
320:
288:catalytic triad
214:carboxylic acid
202:carboxylic acid
152:
147:
59:(also called a
42:
37:of a protease (
28:
23:
22:
15:
12:
11:
5:
3190:
3180:
3179:
3174:
3169:
3154:
3153:
3130:
3129:
3127:
3126:
3113:
3100:
3087:
3074:
3061:
3048:
3034:
3032:
3028:
3027:
3025:
3024:
3019:
3014:
3009:
3004:
2998:
2996:
2992:
2991:
2989:
2988:
2983:
2978:
2973:
2967:
2965:
2964:Classification
2961:
2960:
2958:
2957:
2952:
2947:
2942:
2936:
2934:
2930:
2929:
2927:
2926:
2921:
2916:
2911:
2906:
2901:
2896:
2891:
2885:
2883:
2879:
2878:
2871:
2870:
2863:
2856:
2848:
2839:
2838:
2836:
2835:
2833:Staphylokinase
2829:
2827:
2820:
2819:
2817:
2816:
2815:
2814:
2809:
2804:
2799:
2787:
2786:
2785:
2784:
2779:
2769:
2764:
2759:
2754:
2748:
2746:
2736:
2735:
2732:
2731:
2729:
2728:
2722:
2720:
2716:
2715:
2713:
2712:
2711:
2710:
2705:
2700:
2695:
2690:
2685:
2675:
2667:
2665:
2659:
2658:
2656:
2655:
2647:
2637:
2635:
2629:
2628:
2626:
2625:
2619:
2617:
2611:
2610:
2608:
2607:
2606:
2605:
2600:
2590:
2589:
2588:
2583:
2572:
2570:
2564:
2563:
2561:
2560:
2559:
2558:
2553:
2548:
2537:
2535:
2529:
2528:
2526:
2525:
2524:
2523:
2518:
2517:
2516:
2511:
2501:
2496:
2491:
2486:
2481:
2476:
2469:Aminopeptidase
2465:
2463:
2454:
2444:
2443:
2427:
2426:
2419:
2412:
2404:
2398:
2397:
2387:
2382:
2377:
2372:
2367:
2351:
2346:
2341:
2329:
2320:
2319:
2314:
2308:
2307:
2296:
2295:
2293:
2292:External links
2290:
2287:
2286:
2257:(4): 609–622.
2237:
2202:
2167:
2160:
2142:
2135:
2117:
2088:(4): 561–580.
2068:
2014:
1985:(7): 547–557.
1965:
1930:
1895:
1860:
1818:
1783:
1738:
1679:
1660:(1): 300–305.
1644:
1637:
1619:
1567:
1513:
1474:
1425:
1384:
1354:
1294:
1242:
1191:
1176:
1175:
1173:
1170:
1168:
1167:
1161:
1155:
1150:
1145:
1140:
1135:
1130:
1125:
1120:
1115:
1110:
1109:
1108:
1103:
1098:
1093:
1088:
1083:
1075:
1069:
1067:
1064:
955:
952:
896:bread improver
873:Main article:
870:
867:
863:20S proteosome
851:cell-signaling
843:
840:
828:picornaviruses
811:
808:
802:(for example,
757:
754:
723:Hageman factor
694:
691:
687:photosynthesis
678:
675:
634:
631:
617:
614:
578:
575:
574:
573:
561:
550:
547:
523:endopeptidases
444:
441:
440:
439:
427:
401:
399:Acid proteases
387:
384:
344:
341:
319:
318:Peptide lyases
316:
244:
243:
238:to perform an
229:
216:
204:
192:
180:
168:
151:
148:
146:
145:Classification
143:
120:cell signaling
35:Ribbon diagram
26:
9:
6:
4:
3:
2:
3189:
3178:
3175:
3173:
3170:
3168:
3165:
3164:
3162:
3152:
3147:
3142:
3141:
3138:
3124:
3120:
3119:
3114:
3111:
3107:
3106:
3101:
3098:
3094:
3093:
3088:
3085:
3081:
3080:
3075:
3072:
3068:
3067:
3062:
3059:
3055:
3054:
3049:
3046:
3042:
3041:
3036:
3035:
3033:
3029:
3023:
3020:
3018:
3015:
3013:
3010:
3008:
3005:
3003:
3000:
2999:
2997:
2993:
2987:
2984:
2982:
2981:Enzyme family
2979:
2977:
2974:
2972:
2969:
2968:
2966:
2962:
2956:
2953:
2951:
2948:
2946:
2945:Cooperativity
2943:
2941:
2938:
2937:
2935:
2931:
2925:
2922:
2920:
2917:
2915:
2912:
2910:
2907:
2905:
2904:Oxyanion hole
2902:
2900:
2897:
2895:
2892:
2890:
2887:
2886:
2884:
2880:
2876:
2869:
2864:
2862:
2857:
2855:
2850:
2849:
2846:
2834:
2831:
2830:
2828:
2825:
2821:
2813:
2810:
2808:
2805:
2803:
2800:
2798:
2795:
2794:
2793:
2789:
2788:
2783:
2780:
2778:
2775:
2774:
2773:
2770:
2768:
2765:
2763:
2760:
2758:
2755:
2753:
2750:
2749:
2747:
2745:
2744:Endopeptidase
2741:
2737:
2727:
2724:
2723:
2721:
2717:
2709:
2706:
2704:
2701:
2699:
2696:
2694:
2691:
2689:
2686:
2684:
2681:
2680:
2679:
2676:
2673:
2669:
2668:
2666:
2664:
2660:
2654:
2648:
2646:
2642:
2639:
2638:
2636:
2634:
2630:
2624:
2621:
2620:
2618:
2616:
2612:
2604:
2601:
2599:
2596:
2595:
2594:
2591:
2587:
2584:
2582:
2579:
2578:
2577:
2574:
2573:
2571:
2569:
2565:
2557:
2554:
2552:
2549:
2547:
2544:
2543:
2542:
2539:
2538:
2536:
2534:
2530:
2522:
2519:
2515:
2512:
2510:
2507:
2506:
2505:
2502:
2500:
2497:
2495:
2492:
2490:
2487:
2485:
2482:
2480:
2477:
2475:
2472:
2471:
2470:
2467:
2466:
2464:
2462:
2458:
2455:
2453:
2449:
2445:
2440:
2436:
2432:
2425:
2420:
2418:
2413:
2411:
2406:
2405:
2402:
2395:
2391:
2388:
2386:
2383:
2381:
2378:
2376:
2373:
2371:
2368:
2365:
2361:
2358:
2355:
2352:
2350:
2347:
2345:
2342:
2340:
2336:
2333:
2330:
2328:
2325:
2324:
2318:
2315:
2313:
2310:
2309:
2304:
2299:
2282:
2278:
2273:
2268:
2264:
2260:
2256:
2252:
2248:
2241:
2233:
2229:
2225:
2221:
2217:
2213:
2206:
2198:
2194:
2190:
2186:
2182:
2178:
2171:
2163:
2157:
2153:
2146:
2138:
2132:
2128:
2121:
2113:
2109:
2104:
2099:
2095:
2091:
2087:
2083:
2079:
2072:
2064:
2060:
2055:
2050:
2045:
2040:
2036:
2032:
2028:
2021:
2019:
2010:
2006:
2001:
1996:
1992:
1988:
1984:
1980:
1976:
1969:
1961:
1957:
1953:
1949:
1945:
1941:
1934:
1926:
1922:
1918:
1914:
1910:
1906:
1899:
1891:
1887:
1883:
1879:
1875:
1871:
1864:
1850:on 2021-04-28
1849:
1845:
1841:
1837:
1833:
1829:
1822:
1814:
1810:
1806:
1802:
1798:
1794:
1787:
1779:
1775:
1770:
1765:
1761:
1757:
1753:
1749:
1742:
1734:
1730:
1725:
1720:
1715:
1710:
1706:
1702:
1699:(6): e67915.
1698:
1694:
1690:
1683:
1675:
1671:
1667:
1663:
1659:
1655:
1648:
1640:
1634:
1630:
1623:
1615:
1611:
1606:
1601:
1597:
1593:
1589:
1585:
1581:
1574:
1572:
1563:
1559:
1554:
1549:
1544:
1539:
1535:
1531:
1527:
1520:
1518:
1509:
1505:
1501:
1497:
1493:
1489:
1485:
1478:
1470:
1466:
1461:
1456:
1452:
1448:
1444:
1440:
1436:
1429:
1421:
1417:
1412:
1407:
1403:
1399:
1395:
1388:
1381:
1377:
1373:
1369:
1365:
1358:
1350:
1346:
1341:
1336:
1332:
1328:
1324:
1320:
1316:
1312:
1308:
1301:
1299:
1290:
1286:
1281:
1276:
1272:
1268:
1264:
1260:
1256:
1249:
1247:
1238:
1234:
1229:
1224:
1219:
1214:
1210:
1206:
1202:
1195:
1187:
1181:
1177:
1165:
1162:
1159:
1156:
1154:
1151:
1149:
1146:
1144:
1141:
1139:
1136:
1134:
1131:
1129:
1126:
1124:
1121:
1119:
1116:
1114:
1111:
1107:
1104:
1102:
1099:
1097:
1094:
1092:
1089:
1087:
1084:
1082:
1079:
1078:
1076:
1074:
1071:
1070:
1063:
1061:
1057:
1052:
1050:
1046:
1042:
1038:
1034:
1030:
1026:
1022:
1017:
1015:
1011:
1007:
1003:
999:
995:
991:
987:
983:
979:
975:
971:
965:
961:
951:
949:
945:
941:
940:
935:
931:
927:
923:
922:
917:
916:affinity tags
913:
909:
905:
901:
897:
893:
888:
886:
882:
876:
866:
864:
860:
856:
852:
848:
839:
837:
833:
829:
825:
821:
817:
807:
805:
801:
798:in bacterial
797:
792:
790:
786:
781:
778:
774:
770:
766:
762:
753:
751:
746:
743:
740:
736:
732:
728:
724:
720:
716:
712:
708:
704:
700:
690:
688:
684:
683:developmental
674:
672:
668:
664:
660:
656:
652:
648:
644:
640:
630:
628:
624:
613:
611:
607:
603:
598:
596:
592:
588:
584:
570:
566:
562:
559:
558:
557:
555:
546:
544:
540:
536:
532:
528:
524:
520:
516:
512:
511:exopeptidases
508:
504:
503:peptide bonds
500:
494:
486:
482:
478:
474:
470:
466:
462:
458:
454:
449:
437:
433:
432:
428:
425:
421:
417:
413:
409:
405:
402:
400:
397:
396:
395:
393:
383:
380:
378:
377:streptogrisin
374:
370:
366:
362:
358:
354:
350:
349:superfamilies
340:
337:
333:
329:
325:
315:
313:
312:superfamilies
310:in different
309:
305:
301:
297:
293:
289:
285:
281:
277:
273:
269:
265:
261:
257:
253:
249:
241:
237:
233:
230:
228:
224:
220:
217:
215:
212:
208:
205:
203:
200:
196:
193:
191:
188:
184:
181:
179:
176:
172:
169:
167:
164:
160:
157:
156:
155:
142:
140:
136:
132:
128:
123:
121:
117:
113:
109:
105:
101:
97:
96:peptide bonds
93:
89:
86:into smaller
85:
81:
78:
74:
70:
66:
62:
58:
49:
45:
40:
36:
32:
19:
3118:Translocases
3115:
3102:
3089:
3076:
3063:
3053:Transferases
3050:
3037:
2894:Binding site
2708:Glutamate II
2503:
2452:Exopeptidase
2434:
2302:
2254:
2250:
2240:
2215:
2211:
2205:
2180:
2176:
2170:
2151:
2145:
2126:
2120:
2085:
2081:
2071:
2034:
2030:
1982:
1978:
1968:
1943:
1939:
1933:
1908:
1904:
1898:
1873:
1869:
1863:
1852:. Retrieved
1848:the original
1835:
1831:
1821:
1796:
1792:
1786:
1751:
1747:
1741:
1696:
1692:
1682:
1657:
1653:
1647:
1628:
1622:
1587:
1583:
1533:
1529:
1491:
1487:
1477:
1442:
1438:
1428:
1404:(1): 13–25.
1401:
1397:
1387:
1379:
1367:
1363:
1357:
1314:
1310:
1262:
1258:
1208:
1204:
1194:
1180:
1053:
1018:
1010:fibrinolysis
998:antithrombin
990:C1-inhibitor
984:proteases),
982:inflammatory
967:
937:
919:
904:TEV protease
899:
889:
878:
845:
832:TEV protease
813:
800:pathogenesis
793:
782:
759:
747:
735:snake venoms
711:chymotrypsin
696:
680:
670:
662:
636:
623:TEV protease
619:
610:TEV protease
606:TEV protease
599:
580:
552:
531:chymotrypsin
496:
435:
429:
406:involved in
403:
398:
389:
381:
346:
321:
272:peptide bond
245:
153:
124:
88:polypeptides
68:
64:
60:
56:
54:
39:TEV protease
2889:Active site
2645:Cathepsin A
2581:Cathepsin C
2541:Dipeptidase
1754:: 191–223.
1494:: 249–273.
1123:Proteolysis
1014:neuroserpin
1002:coagulation
932:for making
930:Middle East
820:polyprotein
769:amino acids
731:cathepsin G
639:prokaryotes
627:trypsinogen
583:promiscuous
577:Specificity
485:nucleophile
457:proteolysis
353:superfamily
284:nucleophile
234:- using an
197:- using an
92:amino acids
80:proteolysis
3161:Categories
3092:Isomerases
3066:Hydrolases
2933:Regulation
2356:(see also
1876:(3): 1–5.
1854:2018-12-29
1172:References
1091:threonine-
1025:Lipophilic
954:Inhibitors
865:complex .
855:metabolism
785:proteasome
745:organism.
742:haemotoxin
667:amino acid
643:eukaryotes
591:amino acid
569:covalently
525:, such as
513:, such as
507:amino acid
505:that link
491:See also:
473:hydrolysis
453:hydrolytic
416:complement
412:mast cells
336:asparagine
328:hydrolysis
290:, where a
276:amino acid
236:asparagine
225:, usually
221:- using a
209:- using a
185:- using a
173:- using a
161:- using a
100:hydrolysis
90:or single
65:proteinase
18:Proteinase
3167:Proteases
2971:EC number
2826:: Unknown
2504:Methionyl
2435:proteases
2431:Hydrolase
2390:Proteases
1259:Structure
1101:glutamic-
1096:aspartic-
1081:cysteine-
1077:Protease
1049:antiviral
1021:lipocalin
944:Ayurvedic
859:secretion
765:hydrolyse
739:pit viper
659:apoptosis
554:Catalysis
549:Catalysis
499:digesting
465:substrate
304:threonine
292:histidine
264:threonine
211:glutamate
199:aspartate
187:threonine
112:digestion
77:catalyzes
61:peptidase
2995:Kinetics
2919:Cofactor
2882:Activity
2499:Glutamyl
2489:Cystinyl
2484:Aspartyl
2360:Archived
2335:Archived
2303:Protease
2281:15060002
2232:11427378
2197:84748291
2112:32953999
2063:25774151
2009:27090931
1960:23016690
1925:12475203
1813:15535125
1778:18257708
1733:23826349
1693:PLOS ONE
1674:18067249
1614:19892822
1562:21832066
1508:24905783
1420:22016395
1349:17051221
1289:24931469
1237:18650443
1106:metallo-
1066:See also
1041:HIV/AIDS
928:and the
908:thrombin
885:medicine
826:and the
777:nitrogen
761:Bacteria
756:Bacteria
727:elastase
715:thrombin
703:duodenum
602:thrombin
543:elastase
481:polarise
424:calpains
373:thrombin
369:elastase
361:families
300:cysteine
286:is by a
280:carbonyl
268:glutamic
256:aspartic
252:cysteine
175:cysteine
84:proteins
71:) is an
57:protease
51:)
3151:Biology
3105:Ligases
2875:Enzymes
2479:Arginyl
2474:Alanine
2103:7497159
2054:4343526
2000:4912444
1878:Bibcode
1724:3691164
1701:Bibcode
1605:2808883
1553:3207474
1469:8439290
1460:1132403
1340:3366509
1319:Bibcode
1280:4128088
1228:2576539
1158:TopFIND
1113:PA clan
1086:serine-
1039:, with
1037:viruses
1012:), and
847:Archaea
842:Archaea
816:viruses
810:Viruses
750:cascade
719:plasmin
707:trypsin
693:Animals
647:viruses
595:trypsin
587:trypsin
527:trypsin
365:trypsin
357:PA clan
260:metallo
166:alcohol
131:viruses
106:breaks
3172:EC 3.4
3137:Portal
3079:Lyases
2824:3.4.99
2740:3.4.21
2663:3.4.17
2633:3.4.16
2615:3.4.15
2568:3.4.14
2533:3.4.13
2494:Leucyl
2461:3.4.11
2448:3.4.11
2396:(MeSH)
2300:about
2279:
2272:383305
2269:
2230:
2195:
2158:
2133:
2110:
2100:
2061:
2051:
2037:: 39.
2007:
1997:
1958:
1923:
1811:
1776:
1731:
1721:
1672:
1635:
1612:
1602:
1560:
1550:
1506:
1467:
1457:
1418:
1347:
1337:
1311:Nature
1287:
1277:
1235:
1225:
1164:MEROPS
1073:Ligase
974:serpin
948:Paneer
926:Europe
881:papers
773:carbon
699:pepsin
677:Plants
653:, the
572:enzyme
539:papain
535:pepsin
461:Enzyme
420:kinins
296:serine
258:, and
248:serine
163:serine
73:enzyme
3031:Types
2742:-25:
2450:-19:
2193:S2CID
1060:toxic
469:water
302:, or
223:metal
178:thiol
127:years
108:bonds
104:water
75:that
67:, or
3123:list
3116:EC7
3110:list
3103:EC6
3097:list
3090:EC5
3084:list
3077:EC4
3071:list
3064:EC3
3058:list
3051:EC2
3045:list
3038:EC1
2441:3.4)
2277:PMID
2228:PMID
2156:ISBN
2131:ISBN
2108:PMID
2059:PMID
2005:PMID
1956:PMID
1921:PMID
1809:PMID
1774:PMID
1729:PMID
1670:PMID
1633:ISBN
1610:PMID
1558:PMID
1504:PMID
1465:PMID
1416:PMID
1345:PMID
1285:PMID
1233:PMID
962:and
914:and
906:and
900:e.g.
869:Uses
775:and
709:and
477:acid
434:(or
418:and
375:and
266:and
227:zinc
48:1LVB
2267:PMC
2259:doi
2220:doi
2185:doi
2098:PMC
2090:doi
2049:PMC
2039:doi
1995:PMC
1987:doi
1948:doi
1913:doi
1909:102
1886:doi
1874:568
1840:doi
1801:doi
1797:161
1764:hdl
1756:doi
1719:PMC
1709:doi
1662:doi
1600:PMC
1592:doi
1548:PMC
1538:doi
1534:286
1496:doi
1455:PMC
1447:doi
1443:290
1406:doi
1402:151
1372:doi
1368:118
1335:PMC
1327:doi
1315:443
1275:PMC
1267:doi
1223:PMC
1213:doi
1209:283
1004:),
996:),
645:to
641:to
629:).
545:).
479:to
44:PDB
3163::
2688:A2
2651:DD
2643::
2439:EC
2433::
2275:.
2265:.
2255:14
2253:.
2249:.
2226:.
2214:.
2191:.
2179:.
2106:.
2096:.
2084:.
2080:.
2057:.
2047:.
2033:.
2029:.
2017:^
2003:.
1993:.
1983:24
1981:.
1977:.
1954:.
1944:19
1942:.
1919:.
1907:.
1884:.
1872:.
1836:38
1834:.
1830:.
1807:.
1795:.
1772:.
1762:.
1752:59
1750:.
1727:.
1717:.
1707:.
1695:.
1691:.
1668:.
1656:.
1608:.
1598:.
1588:38
1586:.
1582:.
1570:^
1556:.
1546:.
1532:.
1528:.
1516:^
1502:.
1492:83
1490:.
1486:.
1463:.
1453:.
1441:.
1437:.
1414:.
1400:.
1396:.
1378:.
1366:.
1343:.
1333:.
1325:.
1313:.
1309:.
1297:^
1283:.
1273:.
1263:22
1261:.
1257:.
1245:^
1231:.
1221:.
1207:.
1203:.
1016:.
950:.
857:,
853:,
838:.
791:.
729:,
721:,
717:,
689:.
657:,
541:,
537:,
533:,
529:,
517:,
459:.
392:pH
371:,
367:,
298:,
254:,
250:,
141:.
122:.
63:,
55:A
46::
3139::
3125:)
3121:(
3112:)
3108:(
3099:)
3095:(
3086:)
3082:(
3073:)
3069:(
3060:)
3056:(
3047:)
3043:(
2867:e
2860:t
2853:v
2703:E
2698:C
2693:B
2683:A
2556:3
2551:2
2546:1
2521:O
2514:2
2509:1
2437:(
2423:e
2416:t
2409:v
2366:)
2283:.
2261::
2234:.
2222::
2216:6
2199:.
2187::
2181:4
2164:.
2139:.
2114:.
2092::
2086:2
2065:.
2041::
2035:6
2011:.
1989::
1962:.
1950::
1927:.
1915::
1892:.
1888::
1880::
1857:.
1842::
1815:.
1803::
1780:.
1766::
1758::
1735:.
1711::
1703::
1697:8
1676:.
1664::
1658:7
1641:.
1616:.
1594::
1564:.
1540::
1510:.
1498::
1471:.
1449::
1422:.
1408::
1374::
1351:.
1329::
1321::
1291:.
1269::
1239:.
1215::
1188:.
705:(
438:)
426:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.