464:
142:
707:
differ from prokaryotes in distribution of overlap types: while unidirectional (i.e., same-strand) overlaps are most common in prokaryotes, opposite or antiparallel-strand overlaps are more common in eukaryotes. Among the opposite-strand overlaps, convergent orientation is most common. Most studies of eukaryotic gene overlap have found that overlapping genes are extensively subject to genomic reorganization even in closely related species, and thus the presence of an overlap is not always well-conserved. Overlap with older or less taxonomically restricted genes is also a common feature of genes likely to have originated
387:
488:
246:(HBV), whose DNA genome contains numerous overlapping genes, showed the mean number of synonymous nucleotide substitutions per site in overlapping coding regions was significantly lower than that of non-overlapping regions. The same study showed that it was possible for some of these overlapping regions and their proteins to diverge significantly from the original when there's weak selection against amino acid change. The
770:
may be inappropriate for the detection of overlapping genes as they are reliant on already curated genes while overlapping genes are generally overlooked contain atypical sequence composition. Genome annotation standards are also often biased against feature overlaps, such as genes entirely contained
706:
Compared to prokaryotic genomes, eukaryotic genomes are often poorly annotated and thus identifying genuine overlaps is relatively challenging. However, examples of validated gene overlaps have been documented in a variety of eukaryotic organisms, including mammals such as mice and humans. Eukaryotes
119:
regions of the genome. It is believed that most overlapping genes, or genes whose expressible nucleotide sequences partially overlap with each other, evolved in part due to this mechanism, suggesting that each overlap is composed of one ancestral gene and one novel gene. Subsequently, overprinting is
680:
co-regulated. In prokaryotic genomes, unidirectional overlaps are most common, possibly due to the tendency of adjacent prokaryotic genes to share orientation. Among unidirectional overlaps, long overlaps are more commonly read with a one-nucleotide offset in reading frame (i.e., phase 1) and short
539:
longer than the measured length of its genome. Analysis of the fully sequenced 5386 nucleotide genome showed that the virus possessed extensive overlap between coding regions, revealing that some genes (like genes D and E) were translated from the same DNA sequences but in different reading frames.
719:
The precise functions of overlapping genes seems to vary across the domains of life but several experiments have shown that they are important for virus lifecycles through proper protein expression and stoichiometry as well as playing a role in proper protein folding. A version of
128:
organization of viruses, likely to greatly increase the number of potential expressible genes from a small set of viral genetic information. It is likely that overprinting is responsible for the generation of numerous novel proteins by viruses over the course of their
775:
markedly penalizes overlaps between predicted ORFs. However, rapid advancement of genome-scale protein and RNA measurement tools along with increasingly advanced prediction algorithms have revealed an avalanche of overlapping genes and ORFs within numerous genomes.
454:
than older members, but the older members are also more disordered than other proteins, presumably as a way of alleviating the increased evolutionary constraints posed by overlap. Overlaps are more likely to originate in proteins that already have high disorder.
800:
is also used to identify genomic regions containing overlapping transcripts. It has been utilized to identify 180,000 alternate ORFs within previously annotated coding regions found in humans. Newly discovered ORFs such as these are verified using a variety of
757:
to deliver large human genes such as CFTR81. Therefore, it is suggested that overlapping genes evolved as a means to overcome these physical constraints, increasing genetic diversity by utilizing only the existing sequence rather than increasing genome length.
70:, gene overlap is almost always defined as mRNA transcript overlap. Specifically, a gene overlap in eukaryotes is defined when at least one nucleotide is shared between the boundaries of the primary mRNA transcripts of two or more genes, such that a DNA base
619:
and less restrictive genome sizes. The lower mutation rate of DNA viruses facilitates greater genomic novelty and evolutionary exploration within a structurally constrained genome and may be the primary driver of the evolution of overlapping genes.
591:
geometry. However, other studies dispute this conclusion and argue that the distribution of overlaps in viral genomes is more likely to reflect overprinting as the evolutionary origin of overlapping viral genes. Overprinting is a common source of
560:
was shown to express a novel protein that induces lysis of E. coli by inhibiting biosynthesis of its cell wall, suggesting that de novo protein creation through the process of overprinting can be a significant factor in the evolution of
284:
are encoded by overlapping genes that form a 549 nt coding region, and p19 is shown to be under positive selection while p22 is under purifying selection. Additional examples are mentioned in studies involving overlapping genes of the
1378:
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (April 1981). "Sequence and organization of the human mitochondrial genome".
241:
change is favored. Overlapping genes are reasoned to evolve under strict constraints as a single nucleotide substitution is able to alter the structure and function of the two proteins simultaneously. A study on the
667:
genomes typically find that around one third of bacterial genes are overlapped, though usually only by a few base pairs. Most studies of overlap in bacterial genomes find evidence that overlap serves a function in
233:
Studies on overlapping genes suggest that their evolution can be summarized in two possible models. In one model, the two proteins encoded by their respective overlapping genes evolve under similar
225:
occurs when the shared sequences use different reading frames. This can occur in "phase 1" or "phase 2", depending on whether the reading frames are offset by 1 or 2 nucleotides. Because a
1800:"Sequence analysis of Potato leafroll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products"
215:
occurs when the shared sequences use the same reading frame. This is also known as "phase 0". Unidirectional genes with phase 0 overlap are not considered distinct genes, but rather as
734:
size limitations. Dramatic viability loss was observed in viruses with genomes engineered to be longer than the wild-type genome. Increasing the single-stranded DNA genome length of
615:
or in separate capsids, are more likely to contain an overlapping sequence than non-segmented viruses. RNA viruses have fewer overlapping genes than DNA viruses which possess lower
35:
partially overlaps with the expressible nucleotide sequence of another gene. In this way, a nucleotide sequence may make a contribution to the function of one or more
1575:
Rogozin IB, Spiridonov AN, Sorokin AV, Wolf YI, Jordan I, Tatusov RL, Koonin EV (May 2002). "Purifying and directional selection in overlapping prokaryotic genes".
611:
contain overlapping coding sequences. Segmented viruses in particular, or viruses with their genome split into separate pieces and packaged either all in the same
168:
end of another gene on the same strand. This arrangement can be symbolized with the notation → → where arrows indicate the reading frame from start to end.
2365:
Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes JC, Hutchison CA, Slocombe PM, Smith M (1977). "Nucleotide sequence of bacteriophage ΦX174 DNA".
426:, who identified a candidate gene that may have arisen by this mechanism. Some de novo genes originating in this way may not remain overlapping, but
467:
Overlapping genes in the bacteriophage ΦX174 genome. There are 11 genes in this genome (A, A*, B-H, J, K). Genes B, K, E overlap with genes A, C, D.
264:
The second model suggests that the two proteins and their respective overlap genes evolve under opposite selection pressures: one frame experiences
2607:"Finally, a Role Befitting A star : Strongly Conserved, Unessential Microvirus A* Proteins Ensure the Product Fidelity of Packaging Reactions"
572:
4367:"Viral Proteins Originated De Novo by Overprinting Can Be Identified by Codon Usage: Application to the "Gene Nursery" of Deltaretroviruses"
3220:
Cock PJ, Whitworth DE (19 March 2007). "Evolution of Gene
Overlaps: Relative Reading Frame Bias in Prokaryotic Two-Component System Genes".
2663:
1321:"Viral Proteins Originated De Novo by Overprinting Can Be Identified by Codon Usage: Application to the "Gene Nursery" of Deltaretroviruses"
4186:"Targeted replacement of full-length CFTR in human airway stem cells by CRISPR-Cas9 for pan-mutation correction in the endogenous locus"
261:
compared to the overlap regions that were highly conserved among different HBV strains, which are absolutely essential for the process.
254:
and the pre-S1 region of a surface protein of HBV, for example, had a percentage of conserved amino acids of 30% and 40%, respectively.
4623:
Prensner JR, Enache OM, Luria V, Krug K, Clauser KR, Dempster JM, Karger A, Wang L, Stumbraite K, Wang VM, Botta G (28 January 2021).
3648:"Overlapping genes of Aedes aegypti: evolutionary implications from comparison with orthologs of Anopheles gambiae and other insects"
3371:
Fellner L, Simon S, Scherling C, Witting M, Schober S, Polte C, Schmitt-Kopplin P, Keim DA, Scherer S, Neuhaus K (18 December 2015).
4184:
Vaidyanathan S, Baik R, Chen L, Bravo DT, Suarez CJ, Abazari SM, Salahudeen AA, Dudek AM, Teran CA, Davis TH, Lee CM (March 2021).
2020:
Saha D, Podder S, Panda A, Ghosh TC (May 2016). "Overlapping genes: A significant genomic correlate of prokaryotic growth rates".
4525:"Decision letter: Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins"
269:
4300:"Overlapping genes and the proteins they encode differ significantly in their sequence composition from non-overlapping genes"
74:
at any point of the overlapping region would affect the transcripts of all genes involved. This definition includes 5′ and 3′
2175:
1290:
297:. This phenomenon of overlapping genes experiencing different selection pressures is suggested to be a consequence of a high
813:. Attempts at proof-by-synthesis are also performed to show beyond doubt the absence of any undiscovered overlapping genes.
1745:"Conserved and non-conserved regions in the Sendai virus genome: Evolution of a gene possessing overlapping reading frames"
378:
of the overlapping genes. Gene overlaps introduce novel evolutionary constraints on the sequences of the overlap regions.
1678:"Positive Selection or Free to Vary? Assessing the Functional Significance of Sequence Change Using Molecular Dynamics"
639:. In some cases overprinted proteins do have well-defined, but novel, three-dimensional structures; one example is the
463:
2193:"Birth of a unique enzyme from an alternative reading frame of the preexisted, internally repetitious coding sequence"
780:
methods have been essential in discovering numerous overlapping genes and include a combination of techniques such as
66:
transcripts, and is defined when these coding sequences share a nucleotide on either the same or opposite strands. In
50:. The current definition of an overlapping gene varies significantly between eukaryotes, prokaryotes, and viruses. In
3027:"Overlapping Genes Produce Proteins with Unusual Sequence Properties and Offer Insight into De Novo Protein Creation"
1533:
Normark S., Bergstrom S., Edlund T., Grundstrom T., Jaurin B., Lindberg F.P., Olsson O. (1983). "Overlapping genes".
451:
281:
3432:"New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation"
3373:"Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting"
623:
Studies of overprinted viral genes suggest that their protein products tend to be accessory proteins which are not
402:
associated with blood cancers. This region contains numerous overlapping genes, several of which likely originated
120:
also believed to be a source of novel proteins, as de novo proteins coded by these novel genes usually lack remote
145:
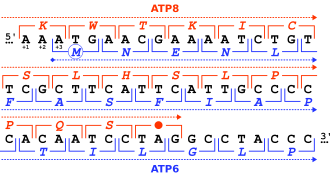
Tandem out-of-phase overlap of the human mitochondrial genes ATP8 (+1 frame, in red) and ATP6 (+3 frame, in blue)
4680:
Cao X, Khitun A, Luo Y, Na Z, Phoodokmai T, Sappakhaw K, Olatunji E, Uttamapinant C, Slavoff SA (5 March 2020).
1847:
Stamenković GG, Ćirković VS, Šiljić MM, Blagojević JV, Knežević AM, Joksić ID, Stanojević MP (24 October 2016).
552:
of the original A protein but possessing a different function It was concluded that other undiscovered sites of
108:
while still preserving the function of the original gene. Overprinting has been hypothesized as a mechanism for
4571:"Faculty Opinions recommendation of Pervasive functional translation of noncanonical human open reading frames"
742:, believed to be the result of the strict physical constraints imposed by the finite capsid volume. Studies on
556:
could be hidden through the genome due to overlapping genes. An identified de novo gene of another overlapping
727:
has also been created where all gene overlaps were removed proving they were not necessary for replication.
101:
640:
492:
395:
277:
1435:
Fukuda Y, Nakayama Y, Tomita M (December 2003). "On dynamics of overlapping genes in bacterial genomes".
599:
The proportion of viruses with overlapping coding sequences within their genomes varies. Double-stranded
545:
149:
Genes may overlap in a variety of ways and can be classified by their positions relative to each other.
753:
showed that viral packaging is constrained by genetic cargo size limits, requiring the use of multiple
750:
302:
3815:"Genome Modularization Reveals Overlapped Gene Topology Is Necessary for Efficient Viral Reproduction"
301:
of nucleotide substitution with different effects on the two frames; the substitutions may be majorly
88:
refers to a type of overlap in which all or part of the sequence of one gene is read in an alternate
4609:
4555:
4509:
4463:
4298:
Pavesi A, Vianelli A, Chirico N, Bao Y, Blinkova O, Belshaw R, Firth A, Karlin D (19 October 2018).
4479:"Faculty Opinions recommendation of The RAST Server: rapid annotations using subsystems technology"
673:
496:
368:
141:
689:. Robustly validated examples of long overlaps in bacterial genomes are rare; in the well-studied
3869:
3322:"Large gene overlaps in prokaryotic genomes: result of functional constraints or mispredictions?"
2970:"New insights into the evolutionary features of viral overlapping genes by discriminant analysis"
1093:
Normark S, Bergström S, Edlund T, Grundström T, Jaurin B, Lindberg FP, Olsson O (December 1983).
754:
306:
4625:"Noncanonical open reading frames encode functional proteins essential for cancer cell survival"
418:
by mutations to introduce novel ORFs in alternate reading frames; he described the mechanism as
743:
677:
372:
216:
3074:
Abroi A (1 December 2015). "A protein domain-based view of the virosphere–host relationship".
685:
are more common for convergent genes; however, putative long overlaps have very high rates of
4596:
4542:
4496:
4450:
290:
265:
234:
32:
583:
toward small genome sizes mediated by the physical constraints of packaging the genome in a
411:
4752:
4693:
4378:
4311:
3969:
3909:"A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast"
3718:
3659:
3551:
3384:
3229:
2555:
2492:
2432:
2374:
2204:
2071:
1974:
1860:
1689:
1626:
1388:
1332:
1215:
1156:
810:
781:
553:
427:
8:
1094:
806:
286:
237:. The proteins and the overlap region are highly conserved when strong selection against
75:
4756:
4682:"Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24"
4382:
4315:
3973:
3722:
3663:
3555:
3388:
3233:
2559:
2496:
2436:
2378:
2208:
2075:
1978:
1864:
1693:
1630:
1546:
1392:
1336:
1219:
1160:
1110:
317:
Overlapping genes are particularly common in rapidly evolving genomes, such as those of
229:
is three nucleotides long, an offset of three nucleotides is an in-phase, phase 0 frame.
4783:
4740:
4716:
4681:
4657:
4584:
4409:
4366:
4342:
4299:
4275:
4223:
4210:
4185:
4161:
4109:
3870:"Translational Coupling Controls Expression and Function of the DrrAB Drug Efflux Pump"
3850:
3790:
3765:
3741:
3706:
3682:
3647:
3574:
3539:
3510:
3483:
3456:
3431:
3407:
3372:
3348:
3321:
3297:
3272:
3253:
3194:
3159:
3140:
3051:
3026:
3002:
2945:
2910:
2886:
2829:
2767:
2740:
2716:
2691:
2639:
2587:
2524:
2453:
2420:
2398:
2337:
2312:
2281:
2256:
2143:
2119:"A Simple Method for Estimating the Strength of Natural Selection on Overlapping Genes"
2118:
2094:
2059:
1997:
1962:
1943:
1930:
1913:
1889:
1780:
1720:
1677:
1658:
1412:
1355:
1320:
1070:
1035:
942:
909:
874:
845:
785:
636:
580:
334:
109:
4088:
4071:
4000:
3957:
3623:
3598:
3127:
2227:
2192:
1588:
1507:
1482:
1036:"Origin, Evolution and Stability of Overlapping Genes in Viruses: A Systematic Review"
1004:
971:
4788:
4770:
4721:
4662:
4644:
4588:
4414:
4396:
4347:
4329:
4280:
4262:
4227:
4215:
4166:
4148:
4101:
4093:
4052:
4044:
4040:
4005:
3987:
3938:
3930:
3889:
3854:
3842:
3834:
3814:
3795:
3746:
3687:
3628:
3579:
3515:
3461:
3412:
3353:
3302:
3245:
3199:
3181:
3132:
3091:
3056:
3007:
2989:
2950:
2932:
2891:
2873:
2834:
2816:
2772:
2721:
2644:
2626:
2579:
2571:
2516:
2508:
2458:
2390:
2342:
2286:
2232:
2171:
2148:
2099:
2037:
2002:
1935:
1894:
1876:
1829:
1821:
1772:
1764:
1725:
1707:
1650:
1642:
1592:
1550:
1512:
1452:
1404:
1360:
1286:
1251:
1246:
1233:
1203:
1184:
1179:
1144:
1122:
1114:
1075:
1057:
1009:
947:
929:
879:
686:
258:
121:
4113:
3257:
3144:
1784:
1662:
846:"Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic"
4778:
4760:
4711:
4703:
4652:
4636:
4574:
4528:
4482:
4436:
4433:"Supplemental Information 2: NCBI genome database accession information (PDF file)"
4404:
4386:
4337:
4319:
4270:
4254:
4205:
4197:
4156:
4140:
4083:
4036:
3995:
3977:
3920:
3881:
3826:
3785:
3777:
3736:
3726:
3677:
3667:
3618:
3610:
3569:
3559:
3505:
3495:
3451:
3443:
3402:
3392:
3343:
3333:
3292:
3284:
3237:
3189:
3171:
3122:
3083:
3046:
3038:
2997:
2981:
2940:
2922:
2881:
2865:
2824:
2808:
2762:
2752:
2711:
2703:
2634:
2618:
2591:
2563:
2528:
2500:
2448:
2440:
2402:
2382:
2332:
2324:
2276:
2268:
2222:
2212:
2138:
2130:
2089:
2079:
2029:
1992:
1982:
1947:
1925:
1884:
1868:
1811:
1756:
1715:
1697:
1634:
1584:
1542:
1502:
1494:
1444:
1416:
1396:
1350:
1340:
1278:
1241:
1223:
1174:
1164:
1106:
1065:
1047:
999:
991:
937:
921:
869:
859:
802:
730:
The retention and evolution of overlapping genes within viruses may also be due to
694:
557:
528:
520:
431:
345:
196:
ends of the two genes overlap on opposite strands. This can be written as ← →.
182:
ends of the two genes overlap on opposite strands. This can be written as → ←.
93:
4579:
4570:
4024:
3110:
1270:
344:
By extension of an existing ORF upstream into a contiguous gene due to loss of an
96:. The alternative open reading frames (ORF) are thought to be created by critical
4391:
4324:
4242:
3731:
3087:
2969:
2084:
1702:
1345:
1282:
669:
562:
472:
399:
375:
130:
4624:
4441:
4432:
4258:
4201:
3925:
3908:
3273:"Origin and Length Distribution of Unidirectional Prokaryotic Overlapping Genes"
2985:
2664:"Scientists Just Found a Mysteriously Hidden 'Gene Within a Gene' in SARS-CoV-2"
2272:
1816:
1799:
698:, only four gene pairs are well validated as having long, overprinted overlaps.
4813:
4707:
4640:
2197:
Proceedings of the
National Academy of Sciences of the United States of America
2033:
1448:
1149:
Proceedings of the
National Academy of Sciences of the United States of America
925:
789:
777:
767:
690:
624:
386:
352:
294:
116:
97:
40:
4698:
3885:
3830:
3397:
3241:
2927:
2757:
1760:
4807:
4774:
4648:
4487:
4478:
4400:
4333:
4266:
4152:
4097:
4048:
3991:
3934:
3838:
3672:
3185:
3176:
2993:
2936:
2877:
2820:
2630:
2575:
2512:
2421:"Recognition of small interfering RNA by a viral suppressor of RNA silencing"
1880:
1825:
1768:
1744:
1711:
1646:
1237:
1118:
1061:
933:
747:
721:
681:
overlaps are more commonly read in phase 2. Long overlaps of greater than 60
628:
616:
536:
524:
504:
439:
394:
indicating the likely evolutionary trajectory of the gene-dense pX region in
359:
The use of the same nucleotide sequence to encode multiple genes may provide
298:
205:
89:
63:
59:
4765:
4533:
4524:
4243:"Gene Birth Contributes to Structural Disorder Encoded by Overlapping Genes"
3564:
3500:
3338:
2853:
2328:
2257:"Gene Birth Contributes to Structural Disorder Encoded by Overlapping Genes"
1532:
1228:
1169:
995:
771:
within another gene. Furthermore, some bioinformatics pipelines such as the
603:
have fewer than a quarter that contains them while almost three-quarters of
4792:
4725:
4666:
4418:
4351:
4284:
4219:
4170:
4009:
3982:
3942:
3893:
3846:
3799:
3750:
3691:
3632:
3583:
3519:
3465:
3447:
3416:
3357:
3306:
3249:
3203:
3136:
3095:
3060:
3011:
2954:
2895:
2869:
2838:
2776:
2725:
2707:
2668:
2648:
2480:
2462:
2346:
2290:
2217:
2152:
2103:
2041:
2006:
1987:
1939:
1898:
1833:
1776:
1729:
1596:
1516:
1456:
1364:
1079:
1013:
951:
883:
648:
584:
499:, a protein encoded by an overprinted gene. The protein specifically binds
443:
326:
36:
4105:
4025:"Packaging of the bacteriophage λ chromosome: Effect of chromosome length"
3781:
3288:
2606:
2583:
2520:
2236:
2168:
Evolution of Living
Organisms: Evidence for a New Theory of Transformation
1654:
1554:
1408:
1255:
1188:
1126:
1052:
16:
A gene whose sequence partially overlaps the reading frame of another gene
4741:"Definitive demonstration by synthesis of genome annotation completeness"
4128:
4072:"Effects of genome size on bacteriophage phi X174 DNA packaging in vitro"
4056:
3907:
Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D (December 2012).
3042:
2812:
2622:
2394:
2134:
1615:"Constrained evolution with respect to gene overlap of hepatitis B virus"
1613:
Mizokami M, Orito E, Ohba Ki, Ikeo K, Lau JY, Gojobori T (January 1997).
822:
793:
739:
644:
588:
541:
447:
435:
423:
273:
243:
4144:
2796:
2444:
1614:
1483:"Properties of overlapping genes are conserved across microbial genomes"
864:
438:. Which member of an overlapping gene pair is younger can be identified
124:
in databases. Overprinted genes are particularly common features of the
3614:
3025:
Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D (29 July 2009).
1848:
1638:
1498:
632:
604:
549:
487:
338:
251:
247:
238:
51:
3436:
Philosophical
Transactions of the Royal Society B: Biological Sciences
2543:
1872:
2567:
2504:
2386:
1963:"The origin of a novel gene through overprinting in Escherichia coli"
1400:
735:
682:
608:
600:
544:
within the genome replication gene A of ΦX174 was shown to express a
512:
511:
The existence of overlapping genes was first identified in the virus
391:
360:
67:
39:. Overlapping genes are present in and a fundamental feature of both
3111:"Size Selective Recognition of siRNA by an RNA Silencing Suppressor"
761:
1846:
664:
322:
71:
4739:
Jaschke PR, Dotson GA, Hung KS, Liu D, Endy D (12 November 2019).
724:
257:
However, these overlap regions are known to be less important for
2544:"Intragenic regulation of the synthesis of ΦX174 gene A proteins"
1676:
Allison JR, Lechner M, Hoeppner MP, Poole AM (12 February 2016).
969:
797:
772:
532:
523:
in 1977. Previous analysis of ΦX174, a small single-stranded DNA
475:, though with varying frequencies. They are especially common in
414:
proposed that one of the genes in the pair could have originated
125:
105:
2313:"Evolution of Viral Proteins Originated De Novo by Overprinting"
1092:
3906:
1377:
1319:
Pavesi A, Magiorkinis G, Karlin DG, Wilke CO (15 August 2013).
731:
612:
516:
364:
79:
47:
3813:
Wright BW, Ruan J, Molloy MP, Jaschke PR (20 November 2020).
1742:
850:
652:
576:
567:
500:
476:
351:
By generation of a novel ORF within an existing one due to a
337:(ORF) downstream into a contiguous gene due to the loss of a
318:
226:
193:
179:
165:
161:
55:
44:
3370:
3109:
Vargason JM, Szittya G, Burgyán J, Hall TM (December 2003).
3024:
1574:
1318:
3958:"Redundancy, antiredundancy, and the robustness of genomes"
3481:
2605:
Roznowski AP, Doore SM, Kemp SZ, Fane BA (6 January 2020).
1849:"Substitution rate and natural selection in parvovirus B19"
1675:
28:
3707:"De Novo Origin of Protein-Coding Genes in Murine Rodents"
3599:"Mammalian Overlapping Genes: The Comparative Perspective"
3108:
2911:"Gene overlapping and size constraints in the viral world"
2797:"Properties and abundance of overlapping genes in viruses"
2741:"Gene overlapping and size constraints in the viral world"
1960:
972:"Comparative study of overlapping genes in the genomes of
115:
from existing sequences, either older genes or previously
4183:
2060:"Evolutionary Dynamics of Overlapped Genes in Salmonella"
2058:
Luo Y, Battistuzzi F, Lin K, Gibas C (29 November 2013).
4297:
4622:
4364:
4022:
3812:
2858:
Proceedings of the Royal
Society B: Biological Sciences
2696:
Proceedings of the Royal
Society B: Biological Sciences
3537:
2851:
2689:
2604:
2057:
1204:"Origins of genes: "big bang" or continuous creation?"
1145:"Origins of genes: "big bang" or continuous creation?"
762:
Methods in identifying overlapping genes and ORFs
100:
within an expressible pre-existing gene, which can be
4365:
Pavesi A, Magiorkinis G, Karlin DG (15 August 2013).
3766:"Recent de novo origin of human protein-coding genes"
3705:
Murphy DN, McLysaght A, Carmel L (21 November 2012).
3704:
3319:
2364:
1612:
910:"Overlapping genes in natural and engineered genomes"
4738:
4023:
Feiss M, Fisher R, Crayton M, Egner C (March 1977).
2019:
1743:
Fujii Y, Kiyotani K, Yoshida T, Sakaguchi T (2001).
1434:
579:
genomes. Some studies attribute this observation to
575:
virus. Overlapping genes are particularly common in
3271:Fonseca MM, Harris DJ, Posada D (5 November 2013).
3270:
2478:
908:Wright BW, Molloy MP, Jaschke PR (5 October 2021).
907:
3533:
3531:
3529:
3484:"Overlapping genes in the human and mouse genomes"
3160:"Overlapping genes: a window on gene evolvability"
2479:Barrell BG, Air GM, Hutchison CA (November 1976).
2310:
450:. Younger members of the pair tend to have higher
3540:"Birth and death of gene overlaps in vertebrates"
3429:
1961:Delaye L, DeLuna A, Lazcano A, Becerra A (2008).
4805:
3763:
2852:Chirico N, Vianelli A, Belshaw R (7 July 2010).
2690:Chirico N, Vianelli A, Belshaw R (7 July 2010).
4745:Proceedings of the National Academy of Sciences
4129:"Effect of Genome Size on AAV Vector Packaging"
3962:Proceedings of the National Academy of Sciences
3955:
3526:
2418:
1570:
1568:
1566:
1564:
1528:
1526:
1480:
1476:
1474:
1472:
1470:
1468:
1466:
1208:Proceedings of the National Academy of Sciences
844:Nelson, Chase W, et al. (1 October 2020).
843:
4679:
3645:
3596:
3423:
2419:Ye K, Malinina L, Patel DJ (3 December 2003).
2360:
2358:
2356:
2306:
2304:
2302:
2300:
2250:
2248:
2246:
1914:"Stability and Evolution of Overlapping Genes"
1430:
1428:
1426:
1277:, Cambridge University Press, pp. 76–90,
4069:
3867:
3477:
3475:
3219:
3102:
3018:
2908:
2738:
2541:
2311:Sabath N, Wagner A, Karlin D (19 July 2012).
1905:
1797:
1371:
1314:
1312:
1310:
1308:
965:
963:
961:
839:
837:
738:by >1% results in almost complete loss of
4126:
3764:Knowles DG, McLysaght A (2 September 2009).
3757:
3364:
3313:
3264:
3215:
3213:
2794:
2732:
2683:
1561:
1523:
1463:
1138:
1136:
200:Overlapping genes can also be classified by
4240:
3956:Krakauer DC, Plotkin JB (29 January 2002).
3698:
3639:
3157:
2655:
2353:
2297:
2254:
2243:
2159:
2053:
2051:
1954:
1423:
635:distributions and high levels of intrinsic
519:was the first DNA genome ever sequenced by
3590:
3538:Makałowska I, Lin CF, Hernandez K (2007).
3472:
3430:McLysaght A, Guerzoni D (31 August 2015).
3151:
3067:
2481:"Overlapping genes in bacteriophage φX174"
2414:
2412:
1305:
1268:
1201:
1142:
970:Y. Fukuda, M. Tomita et T. Washio (1999).
958:
834:
811:catalytically dead Cas9 (dCas9) disruption
631:. Overprinted proteins often have unusual
627:to viral proliferation, but contribute to
4782:
4764:
4715:
4697:
4656:
4578:
4568:
4532:
4486:
4440:
4408:
4390:
4341:
4323:
4274:
4209:
4160:
4087:
3999:
3981:
3924:
3789:
3740:
3730:
3681:
3671:
3622:
3573:
3563:
3509:
3499:
3455:
3406:
3396:
3347:
3337:
3320:Pallejà A, Harrington ED, Bork P (2008).
3296:
3210:
3193:
3175:
3126:
3050:
3001:
2944:
2926:
2885:
2828:
2766:
2756:
2715:
2638:
2452:
2336:
2280:
2226:
2216:
2184:
2142:
2093:
2083:
2013:
1996:
1986:
1929:
1888:
1815:
1719:
1701:
1506:
1354:
1344:
1271:"In search of the origins of viral genes"
1245:
1227:
1178:
1168:
1133:
1069:
1051:
1003:
941:
873:
863:
548:with an identical coding sequence to the
458:
3868:Pradhan P, Li W, Kaur P (January 2009).
2661:
2116:
2110:
2048:
1911:
672:, permitting the overlapped genes to be
651:and a novel binding mode in recognizing
486:
462:
385:
140:
4522:
4127:Wu Z, Yang H, Colosi P (January 2010).
2795:Schlub TE, Holmes EC (1 January 2020).
2409:
381:
4806:
4070:Aoyama A, Hayashi M (September 1985).
2967:
2165:
1033:
329:. They may originate in three ways:
4476:
3158:Huvet M, Stumpf MP (1 January 2014).
3073:
2790:
2788:
2786:
2474:
2472:
1608:
1606:
1269:Gibbs A, Keese PK (19 October 1995),
1202:Keese PK, Gibbs A (15 October 1992).
1143:Keese PK, Gibbs A (15 October 1992).
3597:Veeramachaneni V (1 February 2004).
3482:C. Sanna, W. Li et L. Zhang (2008).
2190:
1798:Guyader S, Ducray DG (1 July 2002).
1029:
1027:
1025:
1023:
903:
901:
899:
897:
895:
893:
565:of viruses. Another example is the
434:, contributing to the prevalence of
367:size and due to the opportunity for
2909:Brandes N, Linial M (21 May 2016).
2739:Brandes N, Linial M (21 May 2016).
2117:Wei X, Zhang J (31 December 2014).
1547:10.1146/annurev.ge.17.120183.002435
1111:10.1146/annurev.ge.17.120183.002435
535:produced during infection required
446:distribution, or by less optimized
13:
4241:Willis S, Masel J (19 July 2018).
2783:
2469:
2255:Willis S, Masel J (19 July 2018).
1931:10.1111/j.0014-3820.2000.tb00075.x
1603:
1275:Molecular Basis of Virus Evolution
164:end of one gene overlaps with the
14:
4825:
1020:
890:
305:for one frame while mostly being
136:
4569:Bazzini A, Wu Q (6 March 2020).
2542:LINNEY E, HAYASHI M (May 1974).
503:produced as part of the plant's
422:. It was later substantiated by
204:, which describe their relative
4732:
4673:
4616:
4562:
4523:Ben-Tal N, ed. (23 June 2017).
4516:
4470:
4425:
4358:
4291:
4234:
4177:
4120:
4076:Journal of Biological Chemistry
4063:
4016:
3949:
3900:
3861:
3806:
3646:Behura SK, Severson DW (2013).
2961:
2902:
2845:
2662:Dockrill P (11 November 2020).
2598:
2535:
2317:Molecular Biology and Evolution
1840:
1791:
1736:
1669:
711:in a given eukaryotic lineage.
471:Overlapping genes occur in all
3222:Journal of Molecular Evolution
2854:"Why genes overlap in viruses"
2692:"Why genes overlap in viruses"
1619:Journal of Molecular Evolution
1481:Johnson Z, Chisholm S (2004).
1262:
1195:
1086:
658:
363:advantage due to reduction in
92:from another gene at the same
1:
4580:10.3410/f.737484924.793572056
4089:10.1016/s0021-9258(17)39144-5
3128:10.1016/S0092-8674(03)00984-X
1589:10.1016/S0168-9525(02)02649-5
828:
766:Standardized methods such as
701:
663:Estimates of gene overlap in
452:intrinsic structural disorder
4392:10.1371/journal.pcbi.1003162
4325:10.1371/journal.pone.0202513
4041:10.1016/0042-6822(77)90425-1
3874:Journal of Molecular Biology
3732:10.1371/journal.pone.0048650
3277:G3: Genes, Genomes, Genetics
3088:10.1016/j.biochi.2015.08.008
2123:Genome Biology and Evolution
2085:10.1371/journal.pone.0081016
1703:10.1371/journal.pone.0147619
1346:10.1371/journal.pcbi.1003162
1283:10.1017/cbo9780511661686.008
641:RNA silencing suppressor p19
493:RNA silencing suppressor p19
442:either by a more restricted
396:human T-lymphotropic virus 1
333:By extension of an existing
312:
7:
4259:10.1534/genetics.118.301249
4202:10.1016/j.ymthe.2021.03.023
3926:10.1016/j.virol.2012.09.020
2986:10.1016/j.virol.2020.03.007
2273:10.1534/genetics.118.301249
1817:10.1099/0022-1317-83-7-1799
1804:Journal of General Virology
816:
714:
609:single-stranded DNA genomes
527:that infected the bacteria
10:
4830:
4708:10.1038/s41467-020-20841-6
4641:10.1038/s41587-020-00806-2
4371:PLOS Computational Biology
2034:10.1016/j.gene.2016.02.002
1449:10.1016/j.gene.2003.09.021
1325:PLOS Computational Biology
926:10.1038/s41576-021-00417-w
482:
4699:10.1101/2020.03.04.977314
4477:Ahmed N (27 March 2009).
4442:10.7717/peerj.6447/supp-2
3886:10.1016/j.jmb.2008.11.027
3831:10.1021/acssynbio.0c00323
3398:10.1186/s12862-015-0558-z
3242:10.1007/s00239-006-0180-1
2928:10.1186/s13062-016-0128-3
2758:10.1186/s13062-016-0128-3
1912:Krakauer DC (June 2000).
1535:Annual Review of Genetics
1099:Annual Review of Genetics
647:, which has both a novel
268:while the other is under
4488:10.3410/f.1157743.618965
3673:10.1186/1471-2148-13-124
3652:BMC Evolutionary Biology
3544:BMC Evolutionary Biology
3377:BMC Evolutionary Biology
3177:10.1186/1471-2164-15-721
1967:BMC Evolutionary Biology
1034:Pavesi A (26 May 2021).
744:adeno-associated viruses
507:defense against viruses.
497:tomato bushy stunt virus
98:nucleotide substitutions
58:overlap must be between
4766:10.1073/pnas.1905990116
4534:10.7554/elife.27860.082
3565:10.1186/1471-2148-7-193
3501:10.1186/1471-2164-9-169
3339:10.1186/1471-2164-9-335
1761:10.1023/a:1008130318633
1229:10.1073/pnas.89.20.9489
1170:10.1073/pnas.89.20.9489
914:Nature Reviews Genetics
217:alternative start sites
4604:Cite journal requires
4550:Cite journal requires
4504:Cite journal requires
4458:Cite journal requires
3983:10.1073/pnas.032668599
3448:10.1098/rstb.2014.0332
2968:Pavesi A (July 2020).
2870:10.1098/rspb.2010.1052
2708:10.1098/rspb.2010.1052
2218:10.1073/pnas.81.8.2421
1988:10.1186/1471-2148-8-31
587:, particularly one of
542:alternative start site
508:
468:
459:Taxonomic distribution
407:
146:
113:emergence of new genes
4686:Nature Communications
3819:ACS Synthetic Biology
3782:10.1101/gr.095026.109
3289:10.1534/g3.113.005652
2329:10.1093/molbev/mss179
2191:Ohno S (April 1984).
1053:10.3390/genes12060809
996:10.1093/nar/27.8.1847
978:Mycoplasma pneumoniae
974:Mycoplasma genitalium
554:polypeptide synthesis
531:, suggested that the
490:
466:
406:through overprinting.
389:
309:for the other frame.
291:potato leafroll virus
223:Out-of-phase overlaps
144:
4629:Nature Biotechnology
3043:10.1128/JVI.00595-09
2623:10.1128/jvi.01593-19
805:techniques, such as
782:bottom-up proteomics
382:Origins of new genes
131:evolutionary history
76:untranslated regions
4757:2019PNAS..11624206J
4751:(48): 24206–24213.
4383:2013PLSCB...9E3162P
4316:2018PLoSO..1302513P
4145:10.1038/mt.2009.255
4082:(20): 11033–11038.
3974:2002PNAS...99.1405K
3723:2012PLoSO...748650M
3664:2013BMCEE..13..124B
3556:2007BMCEE...7..193M
3389:2015BMCEE..15..283F
3234:2007JMolE..64..457C
3037:(20): 10719–10736.
3031:Journal of Virology
2864:(1701): 3809–3817.
2702:(1701): 3809–3817.
2611:Journal of Virology
2560:1974Natur.249..345L
2497:1976Natur.264...34B
2445:10.1038/nature02213
2437:2003Natur.426..874Y
2379:1977Natur.265..687S
2209:1984PNAS...81.2421O
2076:2013PLoSO...881016L
1979:2008BMCEE...8...31D
1865:2016NatSR...635759S
1694:2016PLoSO..1147619A
1631:1997JMolE..44S..83M
1393:1981Natur.290..457A
1337:2013PLSCB...9E3162P
1220:1992PNAS...89.9489K
1161:1992PNAS...89.9489K
1095:"Overlapping Genes"
865:10.7554/eLife.59633
270:purifying selection
235:selection pressures
104:to express a novel
33:nucleotide sequence
3615:10.1101/gr.1590904
3442:(1678): 20140332.
2813:10.1093/ve/veaa009
2170:. Academic Press.
2166:Grassé PP (1977).
2135:10.1093/gbe/evu294
1853:Scientific Reports
1639:10.1007/pl00000061
1577:Trends in Genetics
1499:10.1101/gr.2433104
786:ribosome profiling
596:genes in viruses.
581:selective pressure
509:
469:
412:Pierre-Paul Grassé
408:
335:open reading frame
266:positive selection
147:
78:(UTRs) along with
31:whose expressible
4190:Molecular Therapy
4133:Molecular Therapy
3825:(11): 3079–3090.
3776:(10): 1752–1759.
2554:(5455): 345–348.
2431:(6968): 874–878.
2323:(12): 3767–3780.
2177:978-1-4832-7409-6
1873:10.1038/srep35759
1387:(5806): 457–465.
1292:978-0-521-45533-6
1214:(20): 9489–9493.
984:Nucleic Acids Res
768:genome annotation
674:transcriptionally
607:and viruses with
546:truncated protein
440:bioinformatically
244:hepatitis B virus
219:of the same gene.
4821:
4797:
4796:
4786:
4768:
4736:
4730:
4729:
4719:
4701:
4677:
4671:
4670:
4660:
4620:
4614:
4613:
4607:
4602:
4600:
4592:
4582:
4566:
4560:
4559:
4553:
4548:
4546:
4538:
4536:
4520:
4514:
4513:
4507:
4502:
4500:
4492:
4490:
4474:
4468:
4467:
4461:
4456:
4454:
4446:
4444:
4429:
4423:
4422:
4412:
4394:
4362:
4356:
4355:
4345:
4327:
4310:(10): e0202513.
4295:
4289:
4288:
4278:
4238:
4232:
4231:
4213:
4181:
4175:
4174:
4164:
4124:
4118:
4117:
4091:
4067:
4061:
4060:
4020:
4014:
4013:
4003:
3985:
3968:(3): 1405–1409.
3953:
3947:
3946:
3928:
3904:
3898:
3897:
3865:
3859:
3858:
3810:
3804:
3803:
3793:
3761:
3755:
3754:
3744:
3734:
3702:
3696:
3695:
3685:
3675:
3643:
3637:
3636:
3626:
3594:
3588:
3587:
3577:
3567:
3535:
3524:
3523:
3513:
3503:
3479:
3470:
3469:
3459:
3427:
3421:
3420:
3410:
3400:
3368:
3362:
3361:
3351:
3341:
3317:
3311:
3310:
3300:
3268:
3262:
3261:
3217:
3208:
3207:
3197:
3179:
3155:
3149:
3148:
3130:
3106:
3100:
3099:
3071:
3065:
3064:
3054:
3022:
3016:
3015:
3005:
2965:
2959:
2958:
2948:
2930:
2906:
2900:
2899:
2889:
2849:
2843:
2842:
2832:
2792:
2781:
2780:
2770:
2760:
2736:
2730:
2729:
2719:
2687:
2681:
2680:
2678:
2676:
2659:
2653:
2652:
2642:
2602:
2596:
2595:
2568:10.1038/249345a0
2539:
2533:
2532:
2505:10.1038/264034a0
2476:
2467:
2466:
2456:
2416:
2407:
2406:
2387:10.1038/265687a0
2373:(5596): 687–95.
2362:
2351:
2350:
2340:
2308:
2295:
2294:
2284:
2252:
2241:
2240:
2230:
2220:
2188:
2182:
2181:
2163:
2157:
2156:
2146:
2114:
2108:
2107:
2097:
2087:
2055:
2046:
2045:
2017:
2011:
2010:
2000:
1990:
1958:
1952:
1951:
1933:
1909:
1903:
1902:
1892:
1844:
1838:
1837:
1819:
1810:(7): 1799–1807.
1795:
1789:
1788:
1740:
1734:
1733:
1723:
1705:
1673:
1667:
1666:
1610:
1601:
1600:
1572:
1559:
1558:
1530:
1521:
1520:
1510:
1478:
1461:
1460:
1432:
1421:
1420:
1401:10.1038/290457a0
1375:
1369:
1368:
1358:
1348:
1316:
1303:
1302:
1301:
1299:
1266:
1260:
1259:
1249:
1231:
1199:
1193:
1192:
1182:
1172:
1140:
1131:
1130:
1090:
1084:
1083:
1073:
1055:
1031:
1018:
1017:
1007:
990:(8): 1847–1853.
967:
956:
955:
945:
905:
888:
887:
877:
867:
841:
803:reverse genetics
695:Escherichia coli
537:coding sequences
529:Escherichia coli
521:Frederick Sanger
432:gene duplication
428:subfunctionalize
346:initiation codon
213:In-phase overlap
60:coding sequences
21:overlapping gene
4829:
4828:
4824:
4823:
4822:
4820:
4819:
4818:
4804:
4803:
4800:
4737:
4733:
4678:
4674:
4621:
4617:
4605:
4603:
4594:
4593:
4567:
4563:
4551:
4549:
4540:
4539:
4521:
4517:
4505:
4503:
4494:
4493:
4475:
4471:
4459:
4457:
4448:
4447:
4431:
4430:
4426:
4377:(8): e1003162.
4363:
4359:
4296:
4292:
4239:
4235:
4182:
4178:
4125:
4121:
4068:
4064:
4021:
4017:
3954:
3950:
3905:
3901:
3866:
3862:
3811:
3807:
3770:Genome Research
3762:
3758:
3703:
3699:
3644:
3640:
3603:Genome Research
3595:
3591:
3536:
3527:
3480:
3473:
3428:
3424:
3369:
3365:
3318:
3314:
3269:
3265:
3218:
3211:
3156:
3152:
3107:
3103:
3072:
3068:
3023:
3019:
2966:
2962:
2907:
2903:
2850:
2846:
2801:Virus Evolution
2793:
2784:
2737:
2733:
2688:
2684:
2674:
2672:
2660:
2656:
2603:
2599:
2540:
2536:
2491:(5581): 34–41.
2477:
2470:
2417:
2410:
2363:
2354:
2309:
2298:
2253:
2244:
2189:
2185:
2178:
2164:
2160:
2115:
2111:
2056:
2049:
2018:
2014:
1959:
1955:
1910:
1906:
1845:
1841:
1796:
1792:
1741:
1737:
1688:(2): e0147619.
1674:
1670:
1625:(S1): S83–S90.
1611:
1604:
1573:
1562:
1531:
1524:
1493:(11): 2268–72.
1479:
1464:
1433:
1424:
1376:
1372:
1331:(8): e1003162.
1317:
1306:
1297:
1295:
1293:
1267:
1263:
1200:
1196:
1155:(20): 9489–93.
1141:
1134:
1091:
1087:
1032:
1021:
968:
959:
906:
891:
842:
835:
831:
819:
764:
717:
704:
678:translationally
670:gene regulation
661:
485:
473:domains of life
461:
400:deltaretrovirus
384:
369:transcriptional
315:
276:, the proteins
139:
17:
12:
11:
5:
4827:
4817:
4816:
4799:
4798:
4731:
4672:
4635:(6): 697–704.
4615:
4606:|journal=
4561:
4552:|journal=
4515:
4506:|journal=
4469:
4460:|journal=
4424:
4357:
4290:
4253:(1): 303–313.
4233:
4196:(1): 223–237.
4176:
4119:
4062:
4035:(1): 281–293.
4015:
3948:
3919:(2): 278–284.
3899:
3880:(3): 831–842.
3860:
3805:
3756:
3717:(11): e48650.
3697:
3638:
3609:(2): 280–286.
3589:
3525:
3471:
3422:
3363:
3312:
3263:
3228:(4): 457–462.
3209:
3150:
3121:(7): 799–811.
3101:
3066:
3017:
2960:
2915:Biology Direct
2901:
2844:
2807:(1): veaa009.
2782:
2745:Biology Direct
2731:
2682:
2654:
2597:
2534:
2468:
2408:
2352:
2296:
2267:(1): 303–313.
2242:
2183:
2176:
2158:
2129:(1): 381–390.
2109:
2070:(11): e81016.
2047:
2028:(2): 143–147.
2012:
1953:
1924:(3): 731–739.
1904:
1839:
1790:
1735:
1668:
1602:
1583:(5): 228–232.
1560:
1522:
1462:
1422:
1370:
1304:
1291:
1261:
1194:
1132:
1105:(1): 499–525.
1085:
1019:
957:
920:(3): 154–168.
889:
832:
830:
827:
826:
825:
818:
815:
798:RNA sequencing
790:DNA sequencing
763:
760:
716:
713:
703:
700:
691:model organism
660:
657:
617:mutation rates
484:
481:
460:
457:
383:
380:
357:
356:
353:point mutation
349:
342:
314:
311:
303:non-synonymous
295:parvovirus B19
250:domain of the
231:
230:
220:
206:reading frames
198:
197:
183:
169:
154:Unidirectional
138:
137:Classification
135:
15:
9:
6:
4:
3:
2:
4826:
4815:
4812:
4811:
4809:
4802:
4794:
4790:
4785:
4780:
4776:
4772:
4767:
4762:
4758:
4754:
4750:
4746:
4742:
4735:
4727:
4723:
4718:
4713:
4709:
4705:
4700:
4695:
4691:
4687:
4683:
4676:
4668:
4664:
4659:
4654:
4650:
4646:
4642:
4638:
4634:
4630:
4626:
4619:
4611:
4598:
4590:
4586:
4581:
4576:
4572:
4565:
4557:
4544:
4535:
4530:
4526:
4519:
4511:
4498:
4489:
4484:
4480:
4473:
4465:
4452:
4443:
4438:
4434:
4428:
4420:
4416:
4411:
4406:
4402:
4398:
4393:
4388:
4384:
4380:
4376:
4372:
4368:
4361:
4353:
4349:
4344:
4339:
4335:
4331:
4326:
4321:
4317:
4313:
4309:
4305:
4301:
4294:
4286:
4282:
4277:
4272:
4268:
4264:
4260:
4256:
4252:
4248:
4244:
4237:
4229:
4225:
4221:
4217:
4212:
4207:
4203:
4199:
4195:
4191:
4187:
4180:
4172:
4168:
4163:
4158:
4154:
4150:
4146:
4142:
4138:
4134:
4130:
4123:
4115:
4111:
4107:
4103:
4099:
4095:
4090:
4085:
4081:
4077:
4073:
4066:
4058:
4054:
4050:
4046:
4042:
4038:
4034:
4030:
4026:
4019:
4011:
4007:
4002:
3997:
3993:
3989:
3984:
3979:
3975:
3971:
3967:
3963:
3959:
3952:
3944:
3940:
3936:
3932:
3927:
3922:
3918:
3914:
3910:
3903:
3895:
3891:
3887:
3883:
3879:
3875:
3871:
3864:
3856:
3852:
3848:
3844:
3840:
3836:
3832:
3828:
3824:
3820:
3816:
3809:
3801:
3797:
3792:
3787:
3783:
3779:
3775:
3771:
3767:
3760:
3752:
3748:
3743:
3738:
3733:
3728:
3724:
3720:
3716:
3712:
3708:
3701:
3693:
3689:
3684:
3679:
3674:
3669:
3665:
3661:
3657:
3653:
3649:
3642:
3634:
3630:
3625:
3620:
3616:
3612:
3608:
3604:
3600:
3593:
3585:
3581:
3576:
3571:
3566:
3561:
3557:
3553:
3549:
3545:
3541:
3534:
3532:
3530:
3521:
3517:
3512:
3507:
3502:
3497:
3493:
3489:
3485:
3478:
3476:
3467:
3463:
3458:
3453:
3449:
3445:
3441:
3437:
3433:
3426:
3418:
3414:
3409:
3404:
3399:
3394:
3390:
3386:
3382:
3378:
3374:
3367:
3359:
3355:
3350:
3345:
3340:
3335:
3331:
3327:
3323:
3316:
3308:
3304:
3299:
3294:
3290:
3286:
3282:
3278:
3274:
3267:
3259:
3255:
3251:
3247:
3243:
3239:
3235:
3231:
3227:
3223:
3216:
3214:
3205:
3201:
3196:
3191:
3187:
3183:
3178:
3173:
3169:
3165:
3161:
3154:
3146:
3142:
3138:
3134:
3129:
3124:
3120:
3116:
3112:
3105:
3097:
3093:
3089:
3085:
3081:
3077:
3070:
3062:
3058:
3053:
3048:
3044:
3040:
3036:
3032:
3028:
3021:
3013:
3009:
3004:
2999:
2995:
2991:
2987:
2983:
2979:
2975:
2971:
2964:
2956:
2952:
2947:
2942:
2938:
2934:
2929:
2924:
2920:
2916:
2912:
2905:
2897:
2893:
2888:
2883:
2879:
2875:
2871:
2867:
2863:
2859:
2855:
2848:
2840:
2836:
2831:
2826:
2822:
2818:
2814:
2810:
2806:
2802:
2798:
2791:
2789:
2787:
2778:
2774:
2769:
2764:
2759:
2754:
2750:
2746:
2742:
2735:
2727:
2723:
2718:
2713:
2709:
2705:
2701:
2697:
2693:
2686:
2671:
2670:
2665:
2658:
2650:
2646:
2641:
2636:
2632:
2628:
2624:
2620:
2616:
2612:
2608:
2601:
2593:
2589:
2585:
2581:
2577:
2573:
2569:
2565:
2561:
2557:
2553:
2549:
2545:
2538:
2530:
2526:
2522:
2518:
2514:
2510:
2506:
2502:
2498:
2494:
2490:
2486:
2482:
2475:
2473:
2464:
2460:
2455:
2450:
2446:
2442:
2438:
2434:
2430:
2426:
2422:
2415:
2413:
2404:
2400:
2396:
2392:
2388:
2384:
2380:
2376:
2372:
2368:
2361:
2359:
2357:
2348:
2344:
2339:
2334:
2330:
2326:
2322:
2318:
2314:
2307:
2305:
2303:
2301:
2292:
2288:
2283:
2278:
2274:
2270:
2266:
2262:
2258:
2251:
2249:
2247:
2238:
2234:
2229:
2224:
2219:
2214:
2210:
2206:
2203:(8): 2421–5.
2202:
2198:
2194:
2187:
2179:
2173:
2169:
2162:
2154:
2150:
2145:
2140:
2136:
2132:
2128:
2124:
2120:
2113:
2105:
2101:
2096:
2091:
2086:
2081:
2077:
2073:
2069:
2065:
2061:
2054:
2052:
2043:
2039:
2035:
2031:
2027:
2023:
2016:
2008:
2004:
1999:
1994:
1989:
1984:
1980:
1976:
1972:
1968:
1964:
1957:
1949:
1945:
1941:
1937:
1932:
1927:
1923:
1919:
1915:
1908:
1900:
1896:
1891:
1886:
1882:
1878:
1874:
1870:
1866:
1862:
1858:
1854:
1850:
1843:
1835:
1831:
1827:
1823:
1818:
1813:
1809:
1805:
1801:
1794:
1786:
1782:
1778:
1774:
1770:
1766:
1762:
1758:
1754:
1750:
1746:
1739:
1731:
1727:
1722:
1717:
1713:
1709:
1704:
1699:
1695:
1691:
1687:
1683:
1679:
1672:
1664:
1660:
1656:
1652:
1648:
1644:
1640:
1636:
1632:
1628:
1624:
1620:
1616:
1609:
1607:
1598:
1594:
1590:
1586:
1582:
1578:
1571:
1569:
1567:
1565:
1556:
1552:
1548:
1544:
1540:
1536:
1529:
1527:
1518:
1514:
1509:
1504:
1500:
1496:
1492:
1488:
1484:
1477:
1475:
1473:
1471:
1469:
1467:
1458:
1454:
1450:
1446:
1442:
1438:
1431:
1429:
1427:
1418:
1414:
1410:
1406:
1402:
1398:
1394:
1390:
1386:
1382:
1374:
1366:
1362:
1357:
1352:
1347:
1342:
1338:
1334:
1330:
1326:
1322:
1315:
1313:
1311:
1309:
1294:
1288:
1284:
1280:
1276:
1272:
1265:
1257:
1253:
1248:
1243:
1239:
1235:
1230:
1225:
1221:
1217:
1213:
1209:
1205:
1198:
1190:
1186:
1181:
1176:
1171:
1166:
1162:
1158:
1154:
1150:
1146:
1139:
1137:
1128:
1124:
1120:
1116:
1112:
1108:
1104:
1100:
1096:
1089:
1081:
1077:
1072:
1067:
1063:
1059:
1054:
1049:
1045:
1041:
1037:
1030:
1028:
1026:
1024:
1015:
1011:
1006:
1001:
997:
993:
989:
985:
981:
979:
975:
966:
964:
962:
953:
949:
944:
939:
935:
931:
927:
923:
919:
915:
911:
904:
902:
900:
898:
896:
894:
885:
881:
876:
871:
866:
861:
857:
853:
852:
847:
840:
838:
833:
824:
821:
820:
814:
812:
808:
804:
799:
795:
791:
787:
783:
779:
778:Proteogenomic
774:
773:RAST pipeline
769:
759:
756:
752:
749:
748:gene delivery
745:
741:
737:
733:
728:
726:
723:
722:bacteriophage
712:
710:
699:
697:
696:
692:
688:
687:misannotation
684:
679:
675:
671:
666:
656:
654:
650:
646:
645:Tombusviruses
642:
638:
634:
630:
629:pathogenicity
626:
621:
618:
614:
610:
606:
602:
597:
595:
590:
586:
582:
578:
574:
570:
569:
564:
563:pathogenicity
559:
555:
551:
547:
543:
538:
534:
530:
526:
525:bacteriophage
522:
518:
514:
506:
505:RNA silencing
502:
498:
494:
489:
480:
478:
474:
465:
456:
453:
449:
445:
441:
437:
433:
429:
425:
421:
417:
413:
405:
401:
397:
393:
388:
379:
377:
376:co-regulation
374:
373:translational
370:
366:
362:
354:
350:
347:
343:
340:
336:
332:
331:
330:
328:
324:
320:
310:
308:
304:
300:
296:
292:
288:
283:
279:
275:
274:tombusviruses
271:
267:
262:
260:
256:
253:
249:
245:
240:
236:
228:
224:
221:
218:
214:
211:
210:
209:
207:
203:
195:
192:overlap: the
191:
187:
184:
181:
178:overlap: the
177:
173:
170:
167:
163:
160:overlap: the
159:
155:
152:
151:
150:
143:
134:
132:
127:
123:
118:
114:
112:
107:
103:
99:
95:
91:
90:reading frame
87:
83:
81:
77:
73:
69:
65:
61:
57:
53:
49:
46:
42:
38:
37:gene products
34:
30:
26:
22:
4801:
4748:
4744:
4734:
4689:
4685:
4675:
4632:
4628:
4618:
4597:cite journal
4564:
4543:cite journal
4518:
4497:cite journal
4472:
4451:cite journal
4427:
4374:
4370:
4360:
4307:
4303:
4293:
4250:
4246:
4236:
4193:
4189:
4179:
4139:(1): 80–86.
4136:
4132:
4122:
4079:
4075:
4065:
4032:
4028:
4018:
3965:
3961:
3951:
3916:
3912:
3902:
3877:
3873:
3863:
3822:
3818:
3808:
3773:
3769:
3759:
3714:
3710:
3700:
3655:
3651:
3641:
3606:
3602:
3592:
3547:
3543:
3494:(169): 169.
3491:
3488:BMC Genomics
3487:
3439:
3435:
3425:
3380:
3376:
3366:
3329:
3326:BMC Genomics
3325:
3315:
3283:(1): 19–27.
3280:
3276:
3266:
3225:
3221:
3167:
3164:BMC Genomics
3163:
3153:
3118:
3114:
3104:
3079:
3075:
3069:
3034:
3030:
3020:
2977:
2973:
2963:
2918:
2914:
2904:
2861:
2857:
2847:
2804:
2800:
2748:
2744:
2734:
2699:
2695:
2685:
2673:. Retrieved
2669:ScienceAlert
2667:
2657:
2614:
2610:
2600:
2551:
2547:
2537:
2488:
2484:
2428:
2424:
2370:
2366:
2320:
2316:
2264:
2260:
2200:
2196:
2186:
2167:
2161:
2126:
2122:
2112:
2067:
2063:
2025:
2021:
2015:
1970:
1966:
1956:
1921:
1917:
1907:
1859:(1): 35759.
1856:
1852:
1842:
1807:
1803:
1793:
1755:(1): 47–52.
1752:
1748:
1738:
1685:
1681:
1671:
1622:
1618:
1580:
1576:
1538:
1534:
1490:
1486:
1440:
1436:
1384:
1380:
1373:
1328:
1324:
1296:, retrieved
1274:
1264:
1211:
1207:
1197:
1152:
1148:
1102:
1098:
1088:
1043:
1039:
987:
983:
977:
973:
917:
913:
855:
849:
794:perturbation
765:
729:
718:
708:
705:
693:
662:
649:protein fold
622:
605:retroviridae
598:
593:
585:viral capsid
571:gene in the
566:
510:
470:
444:phylogenetic
436:orphan genes
420:overprinting
419:
415:
409:
403:
361:evolutionary
358:
327:mitochondria
316:
293:, and human
287:Sendai virus
263:
255:
232:
222:
212:
201:
199:
189:
185:
175:
171:
157:
153:
148:
110:
86:Overprinting
85:
84:
24:
20:
18:
3082:: 231–243.
2675:11 November
1749:Virus Genes
1541:: 499–525.
1443:: 181–187.
823:Nested gene
807:CRISPR-Cas9
740:infectivity
659:Prokaryotes
601:RNA viruses
589:icosahedral
448:codon usage
424:Susumu Ohno
398:(HTLV1), a
259:replication
52:prokaryotes
4692:(1): 508.
3658:(1): 124.
3550:(1): 193.
3383:(1): 283.
3332:(1): 335.
3170:(1): 721.
1487:Genome Res
1298:3 December
1046:(6): 809.
829:References
702:Eukaryotes
683:base pairs
633:amino acid
573:SARS-CoV 2
558:gene locus
550:C-terminus
430:following
339:stop codon
307:synonymous
252:polymerase
239:amino acid
172:Convergent
117:non-coding
68:eukaryotes
4775:0027-8424
4649:1087-0156
4589:215850701
4401:1553-7358
4334:1932-6203
4267:1943-2631
4228:232761334
4153:1525-0016
4098:0021-9258
4049:0042-6822
3992:0027-8424
3935:0042-6822
3855:222300240
3839:2161-5063
3186:1471-2164
3076:Biochimie
2994:0042-6822
2980:: 51–66.
2937:1745-6150
2921:(1): 26.
2878:0962-8452
2821:2057-1577
2751:(1): 26.
2631:0022-538X
2576:0028-0836
2513:1476-4687
1973:(1): 31.
1918:Evolution
1881:2045-2322
1826:0022-1317
1769:0920-8569
1712:1932-6203
1647:0022-2844
1238:0027-8424
1119:0066-4197
1062:2073-4425
934:1471-0064
665:bacterial
643:found in
625:essential
479:genomes.
410:In 1977,
392:cladogram
313:Evolution
186:Divergent
4808:Category
4793:31719208
4726:33479206
4667:33510483
4419:23966842
4352:30339683
4304:PLOS ONE
4285:30026186
4247:Genetics
4220:33794364
4171:19904234
4114:32443408
4029:Virology
4010:11818563
3943:23079106
3913:Virology
3894:19063901
3847:33044064
3800:19726446
3751:23185269
3711:PLOS ONE
3692:23777277
3633:14762064
3584:17939861
3520:18410680
3466:26323763
3417:26677845
3358:18627618
3307:24192837
3258:21612308
3250:17479344
3204:25159814
3145:12993441
3137:14697199
3096:26296474
3061:19640978
3012:32452417
2974:Virology
2955:27209091
2896:20610432
2839:32071766
2777:27209091
2726:20610432
2649:31666371
2463:14661029
2347:22821011
2291:30026186
2261:Genetics
2153:25552532
2104:24312259
2064:PLOS ONE
2042:26853049
2007:18226237
1940:10937248
1899:27775080
1834:12075102
1785:12869504
1777:11210938
1730:26871901
1682:PLOS ONE
1663:22644652
1597:12047938
1517:15520290
1457:14659892
1365:23966842
1080:34073395
1014:10101192
952:34611352
884:33001029
817:See also
715:Function
637:disorder
533:proteins
515:, whose
323:bacteria
122:homologs
72:mutation
62:but not
41:cellular
4784:6883844
4753:Bibcode
4717:7820019
4694:bioRxiv
4658:8195866
4410:3744397
4379:Bibcode
4343:6195259
4312:Bibcode
4276:6116962
4211:8753290
4162:2839202
4106:3161888
3970:Bibcode
3791:2765279
3742:3504067
3719:Bibcode
3683:3689595
3660:Bibcode
3575:2151771
3552:Bibcode
3511:2335118
3457:4571571
3408:4683798
3385:Bibcode
3349:2478687
3298:3887535
3230:Bibcode
3195:4161906
3052:2753099
3003:7157939
2946:4875738
2887:2992710
2830:7017920
2768:4875738
2717:2992710
2640:6955274
2592:4175651
2584:4601823
2556:Bibcode
2529:4264796
2521:1004533
2493:Bibcode
2454:4694583
2433:Bibcode
2403:4206886
2375:Bibcode
2338:3494269
2282:6116962
2237:6585807
2205:Bibcode
2144:4316641
2095:3843671
2072:Bibcode
1998:2268670
1975:Bibcode
1948:8818055
1890:5075947
1861:Bibcode
1721:4752228
1690:Bibcode
1655:9071016
1627:Bibcode
1555:6198955
1417:4355527
1409:7219534
1389:Bibcode
1356:3744397
1333:Bibcode
1256:1329098
1216:Bibcode
1189:1329098
1157:Bibcode
1127:6198955
1071:8227390
943:8490965
875:7655111
755:vectors
751:vectors
709:de novo
594:de novo
483:Viruses
416:de novo
404:de novo
319:viruses
190:tail-on
126:genomic
111:de novo
106:protein
102:induced
80:introns
56:viruses
48:genomes
27:) is a
4791:
4781:
4773:
4724:
4714:
4696:
4665:
4655:
4647:
4587:
4417:
4407:
4399:
4350:
4340:
4332:
4283:
4273:
4265:
4226:
4218:
4208:
4169:
4159:
4151:
4112:
4104:
4096:
4057:841861
4055:
4047:
4008:
4001:122203
3998:
3990:
3941:
3933:
3892:
3853:
3845:
3837:
3798:
3788:
3749:
3739:
3690:
3680:
3631:
3624:327103
3621:
3582:
3572:
3518:
3508:
3464:
3454:
3415:
3405:
3356:
3346:
3305:
3295:
3256:
3248:
3202:
3192:
3184:
3143:
3135:
3094:
3059:
3049:
3010:
3000:
2992:
2953:
2943:
2935:
2894:
2884:
2876:
2837:
2827:
2819:
2775:
2765:
2724:
2714:
2647:
2637:
2629:
2590:
2582:
2574:
2548:Nature
2527:
2519:
2511:
2485:Nature
2461:
2451:
2425:Nature
2401:
2395:870828
2393:
2367:Nature
2345:
2335:
2289:
2279:
2235:
2228:345072
2225:
2174:
2151:
2141:
2102:
2092:
2040:
2005:
1995:
1946:
1938:
1897:
1887:
1879:
1832:
1824:
1783:
1775:
1767:
1728:
1718:
1710:
1661:
1653:
1645:
1595:
1553:
1515:
1508:525685
1505:
1455:
1415:
1407:
1381:Nature
1363:
1353:
1289:
1254:
1244:
1236:
1187:
1177:
1125:
1117:
1078:
1068:
1060:
1012:
1005:148392
1002:
950:
940:
932:
882:
872:
792:, and
732:capsid
653:siRNAs
613:capsid
517:genome
501:siRNAs
365:genome
325:, and
248:spacer
202:phases
176:end-on
158:tandem
4814:Genes
4585:S2CID
4224:S2CID
4110:S2CID
3851:S2CID
3254:S2CID
3141:S2CID
2617:(2).
2588:S2CID
2525:S2CID
2399:S2CID
1944:S2CID
1781:S2CID
1659:S2CID
1413:S2CID
1247:50157
1180:50157
1040:Genes
851:eLife
736:ΦX174
725:ΦX174
577:viral
568:ORF3d
513:ΦX174
495:from
477:viral
272:. In
227:codon
94:locus
45:viral
4789:PMID
4771:ISSN
4722:PMID
4663:PMID
4645:ISSN
4610:help
4556:help
4510:help
4464:help
4415:PMID
4397:ISSN
4348:PMID
4330:ISSN
4281:PMID
4263:ISSN
4216:PMID
4167:PMID
4149:ISSN
4102:PMID
4094:ISSN
4053:PMID
4045:ISSN
4006:PMID
3988:ISSN
3939:PMID
3931:ISSN
3890:PMID
3843:PMID
3835:ISSN
3796:PMID
3747:PMID
3688:PMID
3629:PMID
3580:PMID
3516:PMID
3462:PMID
3413:PMID
3354:PMID
3303:PMID
3246:PMID
3200:PMID
3182:ISSN
3133:PMID
3115:Cell
3092:PMID
3057:PMID
3008:PMID
2990:ISSN
2951:PMID
2933:ISSN
2892:PMID
2874:ISSN
2835:PMID
2817:ISSN
2773:PMID
2722:PMID
2677:2020
2645:PMID
2627:ISSN
2580:PMID
2572:ISSN
2517:PMID
2509:ISSN
2459:PMID
2391:PMID
2343:PMID
2287:PMID
2233:PMID
2172:ISBN
2149:PMID
2100:PMID
2038:PMID
2022:Gene
2003:PMID
1936:PMID
1895:PMID
1877:ISSN
1830:PMID
1822:ISSN
1773:PMID
1765:ISSN
1726:PMID
1708:ISSN
1651:PMID
1643:ISSN
1593:PMID
1551:PMID
1513:PMID
1453:PMID
1437:Gene
1405:PMID
1361:PMID
1300:2021
1287:ISBN
1252:PMID
1234:ISSN
1185:PMID
1123:PMID
1115:ISSN
1076:PMID
1058:ISSN
1010:PMID
976:and
948:PMID
930:ISSN
880:PMID
809:and
676:and
491:The
371:and
299:rate
280:and
64:mRNA
54:and
43:and
29:gene
23:(or
4779:PMC
4761:doi
4749:116
4712:PMC
4704:doi
4653:PMC
4637:doi
4575:doi
4529:doi
4483:doi
4437:doi
4405:PMC
4387:doi
4338:PMC
4320:doi
4271:PMC
4255:doi
4251:210
4206:PMC
4198:doi
4157:PMC
4141:doi
4084:doi
4080:260
4037:doi
3996:PMC
3978:doi
3921:doi
3917:434
3882:doi
3878:385
3827:doi
3786:PMC
3778:doi
3737:PMC
3727:doi
3678:PMC
3668:doi
3619:PMC
3611:doi
3570:PMC
3560:doi
3506:PMC
3496:doi
3452:PMC
3444:doi
3440:370
3403:PMC
3393:doi
3344:PMC
3334:doi
3293:PMC
3285:doi
3238:doi
3190:PMC
3172:doi
3123:doi
3119:115
3084:doi
3080:119
3047:PMC
3039:doi
2998:PMC
2982:doi
2978:546
2941:PMC
2923:doi
2882:PMC
2866:doi
2862:277
2825:PMC
2809:doi
2763:PMC
2753:doi
2712:PMC
2704:doi
2700:277
2635:PMC
2619:doi
2564:doi
2552:249
2501:doi
2489:264
2449:PMC
2441:doi
2429:426
2383:doi
2371:265
2333:PMC
2325:doi
2277:PMC
2269:doi
2265:210
2223:PMC
2213:doi
2139:PMC
2131:doi
2090:PMC
2080:doi
2030:doi
2026:582
1993:PMC
1983:doi
1926:doi
1885:PMC
1869:doi
1812:doi
1757:doi
1716:PMC
1698:doi
1635:doi
1585:doi
1543:doi
1503:PMC
1495:doi
1445:doi
1441:323
1397:doi
1385:290
1351:PMC
1341:doi
1279:doi
1242:PMC
1224:doi
1175:PMC
1165:doi
1107:doi
1066:PMC
1048:doi
1000:PMC
992:doi
938:PMC
922:doi
870:PMC
860:doi
746:as
540:An
282:p22
278:p19
188:or
174:or
156:or
25:OLG
19:An
4810::
4787:.
4777:.
4769:.
4759:.
4747:.
4743:.
4720:.
4710:.
4702:.
4690:12
4688:.
4684:.
4661:.
4651:.
4643:.
4633:39
4631:.
4627:.
4601::
4599:}}
4595:{{
4583:.
4573:.
4547::
4545:}}
4541:{{
4527:.
4501::
4499:}}
4495:{{
4481:.
4455::
4453:}}
4449:{{
4435:.
4413:.
4403:.
4395:.
4385:.
4373:.
4369:.
4346:.
4336:.
4328:.
4318:.
4308:13
4306:.
4302:.
4279:.
4269:.
4261:.
4249:.
4245:.
4222:.
4214:.
4204:.
4194:30
4192:.
4188:.
4165:.
4155:.
4147:.
4137:18
4135:.
4131:.
4108:.
4100:.
4092:.
4078:.
4074:.
4051:.
4043:.
4033:77
4031:.
4027:.
4004:.
3994:.
3986:.
3976:.
3966:99
3964:.
3960:.
3937:.
3929:.
3915:.
3911:.
3888:.
3876:.
3872:.
3849:.
3841:.
3833:.
3821:.
3817:.
3794:.
3784:.
3774:19
3772:.
3768:.
3745:.
3735:.
3725:.
3713:.
3709:.
3686:.
3676:.
3666:.
3656:13
3654:.
3650:.
3627:.
3617:.
3607:14
3605:.
3601:.
3578:.
3568:.
3558:.
3546:.
3542:.
3528:^
3514:.
3504:.
3490:.
3486:.
3474:^
3460:.
3450:.
3438:.
3434:.
3411:.
3401:.
3391:.
3381:15
3379:.
3375:.
3352:.
3342:.
3328:.
3324:.
3301:.
3291:.
3279:.
3275:.
3252:.
3244:.
3236:.
3226:64
3224:.
3212:^
3198:.
3188:.
3180:.
3168:15
3166:.
3162:.
3139:.
3131:.
3117:.
3113:.
3090:.
3078:.
3055:.
3045:.
3035:83
3033:.
3029:.
3006:.
2996:.
2988:.
2976:.
2972:.
2949:.
2939:.
2931:.
2919:11
2917:.
2913:.
2890:.
2880:.
2872:.
2860:.
2856:.
2833:.
2823:.
2815:.
2803:.
2799:.
2785:^
2771:.
2761:.
2749:11
2747:.
2743:.
2720:.
2710:.
2698:.
2694:.
2666:.
2643:.
2633:.
2625:.
2615:94
2613:.
2609:.
2586:.
2578:.
2570:.
2562:.
2550:.
2546:.
2523:.
2515:.
2507:.
2499:.
2487:.
2483:.
2471:^
2457:.
2447:.
2439:.
2427:.
2423:.
2411:^
2397:.
2389:.
2381:.
2369:.
2355:^
2341:.
2331:.
2321:29
2319:.
2315:.
2299:^
2285:.
2275:.
2263:.
2259:.
2245:^
2231:.
2221:.
2211:.
2201:81
2199:.
2195:.
2147:.
2137:.
2125:.
2121:.
2098:.
2088:.
2078:.
2066:.
2062:.
2050:^
2036:.
2024:.
2001:.
1991:.
1981:.
1969:.
1965:.
1942:.
1934:.
1922:54
1920:.
1916:.
1893:.
1883:.
1875:.
1867:.
1855:.
1851:.
1828:.
1820:.
1808:83
1806:.
1802:.
1779:.
1771:.
1763:.
1753:22
1751:.
1747:.
1724:.
1714:.
1706:.
1696:.
1686:11
1684:.
1680:.
1657:.
1649:.
1641:.
1633:.
1623:44
1621:.
1617:.
1605:^
1591:.
1581:18
1579:.
1563:^
1549:.
1539:17
1537:.
1525:^
1511:.
1501:.
1491:14
1489:.
1485:.
1465:^
1451:.
1439:.
1425:^
1411:.
1403:.
1395:.
1383:.
1359:.
1349:.
1339:.
1327:.
1323:.
1307:^
1285:,
1273:,
1250:.
1240:.
1232:.
1222:.
1212:89
1210:.
1206:.
1183:.
1173:.
1163:.
1153:89
1151:.
1147:.
1135:^
1121:.
1113:.
1103:17
1101:.
1097:.
1074:.
1064:.
1056:.
1044:12
1042:.
1038:.
1022:^
1008:.
998:.
988:27
986:.
982:.
960:^
946:.
936:.
928:.
918:23
916:.
912:.
892:^
878:.
868:.
858:.
854:.
848:.
836:^
796:.
788:,
784:,
655:.
390:A
321:,
289:,
208::
194:5'
180:3'
166:5'
162:3'
133:.
82:.
4795:.
4763::
4755::
4728:.
4706::
4669:.
4639::
4612:)
4608:(
4591:.
4577::
4558:)
4554:(
4537:.
4531::
4512:)
4508:(
4491:.
4485::
4466:)
4462:(
4445:.
4439::
4421:.
4389::
4381::
4375:9
4354:.
4322::
4314::
4287:.
4257::
4230:.
4200::
4173:.
4143::
4116:.
4086::
4059:.
4039::
4012:.
3980::
3972::
3945:.
3923::
3896:.
3884::
3857:.
3829::
3823:9
3802:.
3780::
3753:.
3729::
3721::
3715:7
3694:.
3670::
3662::
3635:.
3613::
3586:.
3562::
3554::
3548:7
3522:.
3498::
3492:9
3468:.
3446::
3419:.
3395::
3387::
3360:.
3336::
3330:9
3309:.
3287::
3281:4
3260:.
3240::
3232::
3206:.
3174::
3147:.
3125::
3098:.
3086::
3063:.
3041::
3014:.
2984::
2957:.
2925::
2898:.
2868::
2841:.
2811::
2805:6
2779:.
2755::
2728:.
2706::
2679:.
2651:.
2621::
2594:.
2566::
2558::
2531:.
2503::
2495::
2465:.
2443::
2435::
2405:.
2385::
2377::
2349:.
2327::
2293:.
2271::
2239:.
2215::
2207::
2180:.
2155:.
2133::
2127:7
2106:.
2082::
2074::
2068:8
2044:.
2032::
2009:.
1985::
1977::
1971:8
1950:.
1928::
1901:.
1871::
1863::
1857:6
1836:.
1814::
1787:.
1759::
1732:.
1700::
1692::
1665:.
1637::
1629::
1599:.
1587::
1557:.
1545::
1519:.
1497::
1459:.
1447::
1419:.
1399::
1391::
1367:.
1343::
1335::
1329:9
1281::
1258:.
1226::
1218::
1191:.
1167::
1159::
1129:.
1109::
1082:.
1050::
1016:.
994::
980:"
954:.
924::
886:.
862::
856:9
355:.
348:;
341:;
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.