31:
531:. Trisomy is caused by the presence of one additional chromosome in the zygote as compared to the normal number, and monosomy is characterized by the presence of one fewer chromosome in the zygote as compared to the normal number. If this uneven division occurs in meiosis I, then none of the daughter cells will have proper chromosomal distribution and non-typical effects can ensue, including Down's syndrome. Unequal division can also occur during the second meiotic division. Nondisjunction which occurs at this stage can result in normal daughter cells and deformed cells.
290:
282:
540:
486:
212:. Both of these properties (i.e., the length of chromosomal arms, and the placement of the chromosomal centromere) are the main factors for creating structural homology between chromosomes. Therefore, when two chromosomes containing the relatively same structure exist (e.g., maternal chromosome 15 and paternal chromosome 15), they are able to pair together via the process of synapsis to form homologous chromosomes.
429:
daughter cells that have arisen from the single diploid parent cell by meiosis I) resulting from meiosis I undergo another cell division in meiosis II but without another round of chromosomal replication. The sister chromatids in the two daughter cells are pulled apart during anaphase II by nuclear spindle fibers, resulting in four haploid daughter cells.
438:
with each other. Instead, the replicants, or sister chromatids, will line up along the metaphase plate and then separate in the same way as meiosis II – by being pulled apart at their centromeres by nuclear mitotic spindles. If any crossing over does occur between sister chromatids during mitosis, it does not produce any new recombinant genotypes.
567:
this damage by aligning themselves with chromosomes of the same genetic sequence. Once the base pairs have been matched and oriented correctly between the two strands, the homologous chromosomes perform a process that is very similar to recombination, or crossing over as seen in meiosis. Part of the
437:
Homologous chromosomes do not function the same in mitosis as they do in meiosis. Prior to every single mitotic division a cell undergoes, the chromosomes in the parent cell replicate themselves. The homologous chromosomes within the cell will ordinarily not pair up and undergo genetic recombination
589:
There is ongoing research concerning the ability of homologous chromosomes to repair double-strand DNA breaks. Researchers are investigating the possibility of exploiting this capability for regenerative medicine. This medicine could be very prevalent in relation to cancer, as DNA damage is thought
585:
Current and future research on the subject of homologous chromosome is heavily focused on the roles of various proteins during recombination or during DNA repair. In a recently published article by Pezza et al. the protein known as HOP2 is responsible for both homologous chromosome synapsis as well
415:
to release the cohesin that held the homologous chromosome arms together. This allows the chiasmata to release and the homologs to move to opposite poles of the cell. The homologous chromosomes are now randomly segregated into two daughter cells that will undergo meiosis II to produce four haploid
489:
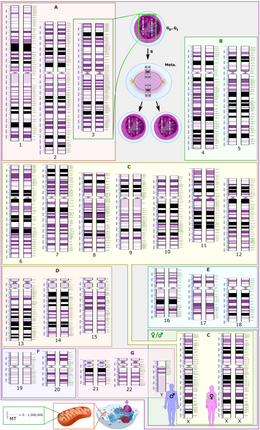
1. Meiosis I 2. Meiosis II 3. Fertilization 4. Zygote
Nondisjunction is when chromosomes fail to separate normally resulting in a gain or loss of chromosomes. In the left image the blue arrow indicates nondisjunction taking place during meiosis II. In the right image the green arrow is indicating
260:
So, humans have two sets of 23 chromosomes in each cell that contains a nucleus. One set of 23 chromosomes (n) is from the mother (22 autosomes, 1 sex chromosome (X only)) and one set of 23 chromosomes (n) is from the father (22 autosomes, 1 sex chromosome (X or Y)). Ultimately, this means that
452:
Homologous pairing in most contexts will refer to germline cells, however also takes place in somatic cells. For example, in humans, somatic cells have very tightly regulated homologous pairing (separated into chromosomal territories, and pairing at specific loci under control of developmental
428:
After the tetrads of homologous chromosomes are separated in meiosis I, the sister chromatids from each pair are separated. The two haploid daughter cells (the number of chromosomes has been reduced to half: earlier two sets of chromosomes were present, but now each set exists in two different
334:
of meiosis I, each chromosome is aligned with its homologous partner and pairs completely. In prophase I, the DNA has already undergone replication so each chromosome consists of two identical chromatids connected by a common centromere. During the zygotene stage of prophase I, the homologous
313:
in meiosis I. During meiosis, genetic recombination (by random segregation) and crossing over produces daughter cells that each contain different combinations of maternally and paternally coded genes. This recombination of genes allows for the introduction of new allele pairings and genetic
253:
may or may not be homologous, depending on the sex of the individual. For instance, females contain XX, thus have a homologous pair of sex chromosomes. This means that females have 23 pairs of homologous chromosomes in total (i.e., 22 pairs of non-sex chromosomes (autosomes), 1 pair of sex
182:
were studying meiosis in corn cells and examining gene loci on corn chromosomes. Creighton and McClintock discovered that the new allele combinations present in the offspring and the event of crossing over were directly related. This proved interchromosomal genetic recombination.
173:
experiments, he revealed that, for a single parent, the alleles of genes near to one another along the length of the chromosome move together. Using this logic he concluded that the two genes he was studying were located on homologous chromosomes. Later on during the 1930s,
590:
to be contributor to carcinogenesis. Manipulating the repair function of homologous chromosomes might allow for bettering a cell's damage response system. While research has not yet confirmed the effectiveness of such treatment, it may become a useful therapy for cancer.
42:
human cell contains 22 pairs of homologous chromosomes and 2 sex chromosomes. The cell has two sets of each chromosome; one of the pair is derived from the mother and the other from the father. The maternal and paternal chromosomes in a homologous pair have the same
191:
Homologous chromosomes are pairs of chromosomes in a diploid organism that have similar genes, although not necessarily identical. There are two main properties of homologous chromosomes: 1) the length of chromosomal arms and 2) the placement of the centromere.
375:, particularly double-strand breaks. At the diplotene stage of prophase I the synaptonemal complex disassembles before which will allow the homologous chromosomes to separate, while the sister chromatids stay associated by their centromeres.
309:. The process of meiosis I is generally longer than meiosis II because it takes more time for the chromatin to replicate and for the homologous chromosomes to be properly oriented and segregated by the processes of pairing and
522:
Proper homologous chromosome separation in meiosis I is crucial for sister chromatid separation in meiosis II. A failure to separate properly is known as nondisjunction. There are two main types of nondisjunction that occur:
366:
are the site of the exchange. Chiasmata physically link the homologous chromosomes once crossing over occurs and throughout the process of chromosomal segregation during meiosis. Both the non-crossover and crossover types of
586:
as double-strand break repair via homologous recombination. The deletion of HOP2 in mice has large repercussions in meiosis. Other current studies focus on specific proteins involved in homologous recombination as well.
195:
The actual length of the arm, in accordance with the gene locations, is critically important for proper alignment. Centromere placement on the chromosome can be characterized by four main arrangements, either
399:. The random orientation is another way for cells to introduce genetic variation. Meiotic spindles emanating from opposite spindle poles attach to each of the homologs (each pair of sister chromatids) at the
576:
and complexes are then recruited to the site of damage, allowing for repair and proper replication to occur. Through this functioning, double-strand breaks can be repaired and DNA can function normally.
257:
In humans, the 22 pairs of homologous autosomal chromosomes contain the same genes but code for different traits in their allelic forms, as one was inherited from the mother and one from the father.
273:
Homologous chromosomes are important in the processes of meiosis and mitosis. They allow for the recombination and random segregation of genetic material from the mother and father into new cells.
297:
Meiosis is a round of two cell divisions that results in four haploid daughter cells that each contain half the number of chromosomes as the parent cell. It reduces the chromosome number in a
137:(2n) organisms, the genome is composed of one set of each homologous chromosome pair, as compared to tetraploid organisms which may have two sets of each homologous chromosome pair. The
141:
on the homologous chromosomes may be different, resulting in different phenotypes of the same genes. This mixing of maternal and paternal traits is enhanced by crossing over during
782:
Klug, William; Michael
Cummings; Charlotte Spencer; Michael Pallodino (2009). "Chromosome Mutations: Variation in chromosome number and arrangement". In Beth Wilbur (ed.).
510:. Though the mechanisms for pairing and adhering homologous chromosomes vary among organisms, proper functioning of those mechanisms is imperative in order for the final
469:
in which an allele on one chromosome affects the expression of the homologous allele on the homologous chromosome. One notable function of this is the
878:
362:
In the process of crossing-over, genes are exchanged by the breaking and union of homologous portions of the chromosomes' lengths. Structures called
254:
chromosomes). Conversely, males contain XY, which means that they have a non-homologous pair of sex chromosomes as their 23rd pair of chromosomes.
942:
1381:
89:, where they provide points along each chromosome that enable a pair of chromosomes to align correctly with each other before separating during
165:
and they noted that some combinations of alleles appeared more frequently than others. That data and information was further explored by
351:, a type of recombination, occurs during the pachytene stage of prophase I. In addition, another type of recombination referred to as
659:
145:, wherein lengths of chromosomal arms and the DNA they contain within a homologous chromosome pair are exchanged with one another.
1000:
1074:
1144:
285:
During the process of meiosis, homologous chromosomes can recombine and produce new combinations of genes in the daughter cells.
411:
In anaphase I of meiosis I the homologous chromosomes are pulled apart from each other. The homologs are cleaved by the enzyme
129:. One homologous chromosome is inherited from the organism's mother; the other is inherited from the organism's father. After
918:
861:
827:
653:
352:
133:
occurs within the daughter cells, they have the correct number of genes which are a mix of the two parents' genes. In
1366:
1173:"Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination"
791:
739:
710:
547:
While the main function of homologous chromosomes is their use in nuclear division, they are also used in repairing
215:
Since homologous chromosomes are not identical and do not originate from the same organism, they are different from
372:
1100:
Gerton JL, Hawley RS (June 2005). "Homologous chromosome interactions in meiosis: diversity amidst conservation".
543:
Diagram of the general process for double-stranded break repair as well as synthesis-dependent strand annealing.
965:"The Theory and Application of a New Method of Detecting Chromosomal Rearrangements in Drosophila melanogaster"
934:
494:
There are severe repercussions when chromosomes do not segregate properly. Faulty segregation can lead to
1263:"Homologous Recombination DNA Repair Genes Play a Critical Role in Reprogramming to a Pluripotent State"
882:
559:
and are most often the result of interaction of DNA with naturally occurring damaging molecules such as
1310:
Khanna KK, Jackson SP (2001). "DNA double-strand breaks: Signaling, repair and the cancer connection".
447:
355:(SDSA) frequently occurs. SDSA recombination involves information exchange between paired homologous
343:
crosslinking occurs between the homologous chromosomes and helps them resist being pulled apart until
318:
among organisms helps make a population more stable by providing a wider range of genetic traits for
1377:
599:
17:
560:
466:
1222:"The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis"
569:
573:
392:
339:– a protein scaffold – is assembled and joins the homologous chromosomes along their lengths.
1261:
González F, Georgieva D, Vanoli F, Shi ZD, Stadtfeld M, Ludwig T, Jasin M, Huangfu D (2013).
609:
604:
368:
348:
94:
1407:
1034:
548:
336:
121:. Homologous chromosomes are made up of chromosome pairs of approximately the same length,
30:
643:
8:
503:
388:
335:
chromosomes pair up with each other. This pairing occurs by a synapsis process where the
1038:
1335:
1287:
1262:
1125:
1066:
992:
964:
363:
179:
166:
1238:
1221:
1197:
1172:
1022:
781:
1362:
1327:
1292:
1243:
1202:
1152:
1117:
1070:
1058:
1050:
996:
984:
914:
857:
823:
787:
735:
706:
649:
470:
396:
319:
315:
216:
175:
1129:
1339:
1319:
1282:
1274:
1233:
1192:
1184:
1109:
1042:
976:
197:
158:
126:
86:
48:
1278:
619:
556:
220:
154:
701:
Griffiths JF, Gelbart WM, Lewontin RC, Wessler SR, Suzuki DT, Miller JH (2005).
359:, but not physical exchange. SDSA recombination does not cause crossing-over.
201:
1401:
1054:
988:
786:(9 ed.). San Francisco, CA: Pearson Benjamin Cumming. pp. 213–214.
728:
223:
has occurred, and thus are identical, side-by-side duplicates of each other.
78:
1046:
344:
1331:
1296:
1247:
1121:
1062:
499:
243:
239:
1206:
881:. Clinton Community College – State University of New York. Archived from
485:
234:
have a total of 46 chromosomes, but there are only 22 pairs of homologous
1188:
1023:"Transvection regulates the sex-biased expression of a fly X-linked gene"
1021:
Galouzis, Charalampos
Chrysovalantis; Prud'homme, Benjamin (2021-01-22).
495:
474:
400:
384:
209:
205:
927:
564:
539:
461:
455:
417:
331:
306:
289:
170:
122:
101:
to its offspring parent developmental cell at the given time and area.
97:, which characterizes inheritance patterns of genetic material from an
74:
822:(2 ed.). Philadelphia: Saunders/Elsevier. pp. 815, 821–822.
125:
position, and staining pattern, for genes with the same corresponding
356:
302:
298:
118:
58:
35:
1113:
465:
the homologous pairing supports a gene regulatory phenomenon called
980:
648:. 2. Philadelphia: Saunders/Elsevier. 2008. pp. 815, 821–822.
624:
614:
528:
412:
310:
235:
162:
98:
1323:
817:
1219:
524:
511:
387:
of meiosis I, the pairs of homologous chromosomes, also known as
340:
262:
142:
130:
114:
90:
281:
700:
507:
231:
138:
134:
52:
39:
238:
chromosomes. The additional 23rd pair is the sex chromosomes,
913:. Sunderland, MA: Sinauer Associates, Inc. pp. 606–610.
1220:
Petukhova GV, Romanienko PJ, Camerini-Otero RD (Dec 2003).
82:
44:
1260:
1136:
301:
by half by first separating the homologous chromosomes in
1142:
705:. W.H. Freeman and Co. pp. 34–40, 473–476, 626–629.
552:
110:
818:
Pollard TD, Earnshaw WC, Lippincott-Schwartz J (2008).
1170:
1020:
459:) exhibit homologous pairing much more frequently. In
1171:
Sargent RG, Brenneman MA, Wilson JH (January 1997).
856:. New York: W.H. Freeman and Co. pp. 355, 891.
1151:. University of Illinois at Chicago. Archived from
727:
77:that pair up with each other inside a cell during
870:
293:Sorting of homologous chromosomes during meiosis.
109:Chromosomes are linear arrangements of condensed
1399:
766:
1375:
568:intact DNA sequence overlaps with that of the
1309:
725:
490:nondisjunction taking place during meiosis I.
1099:
696:
555:. These double-stranded breaks may occur in
453:signalling). Other species however (notably
73:, is a set of one maternal and one paternal
694:
692:
690:
688:
686:
684:
682:
680:
678:
676:
1356:
1166:
1164:
1162:
908:
904:
902:
900:
898:
896:
894:
892:
1361:. Sunderland, Mass.: Sinauer Associates.
1286:
1237:
1196:
1095:
1093:
1091:
851:
813:
811:
809:
807:
805:
803:
847:
845:
843:
841:
839:
673:
538:
484:
288:
280:
29:
1159:
935:"The Cell Cycle & Mitosis Tutorial"
889:
14:
1400:
1088:
876:
800:
762:
760:
395:, line up in a random order along the
117:proteins, which form a complex called
27:Chromosomes that pair in fertilization
1384:from the original on 16 November 2013
962:
836:
775:
734:. San Francisco: Benjamin Cummings.
580:
371:function as processes for repairing
353:synthesis-dependent strand annealing
941:. University of Arizona. Oct 2004.
757:
441:
24:
1349:
771:. Boston: Pearson. pp. 21–22.
305:and then the sister chromatids in
25:
1419:
1143:Tissot, Robert; Kaufman, Elliot.
517:
219:. Sister chromatids result after
703:Introduction to Genetic Analysis
1376:OpenStaxCollege (25 Apr 2013).
1303:
1254:
1213:
1077:from the original on 2021-12-27
1014:
1003:from the original on 2020-10-17
956:
945:from the original on 2018-09-22
662:from the original on 2015-11-29
748:
726:Campbell NA, Reece JB (2002).
719:
636:
378:
13:
1:
1239:10.1016/s1534-5807(03)00369-1
630:
563:. Homologous chromosomes can
534:
432:
423:
406:
325:
276:
1279:10.1016/j.celrep.2013.02.005
268:
226:
186:
7:
593:
480:
104:
10:
1424:
963:Lewis, E. B. (July 1954).
754:Himabindu Sreenivasulu 23
448:Homologous somatic pairing
445:
148:
56:
1145:"Chromosomal Inheritance"
767:Klug, William S. (2012).
81:. Homologs have the same
51:, but possibly different
600:Homologous recombination
514:to be sorted correctly.
93:. This is the basis for
1047:10.1126/science.abc2745
969:The American Naturalist
561:reactive oxygen species
645:Homologous chromosomes
544:
491:
294:
286:
249:Note that the pair of
161:were studying genetic
67:homologous chromosomes
62:
911:Developmental Biology
854:Molecular cell biolog
610:Developmental biology
605:Mendelian inheritance
542:
488:
292:
284:
111:deoxyribonucleic acid
95:Mendelian inheritance
57:Further information:
33:
1359:Developmental biolog
1189:10.1128/MCB.17.1.267
784:Concepts of Genetics
769:Concepts of Genetics
574:Replication proteins
570:damaged chromosome's
549:double-strand breaks
337:synaptonemal complex
153:Early in the 1900s,
1380:. Rice University.
1357:Gilbert SF (2003).
1039:2021Sci...371..396G
939:The Biology Project
909:Gilbert SF (2014).
852:Lodish HF (2013).
545:
492:
471:sexually dimorphic
295:
287:
180:Barbara McClintock
63:
1033:(6527): 396–400.
920:978-0-87893-978-7
879:"The Biology Web"
863:978-1-4292-3413-9
829:978-1-4160-2255-8
655:978-1-4160-2255-8
581:Relevant research
320:natural selection
316:Genetic variation
265:(2n) organisms.
217:sister chromatids
176:Harriet Creighton
16:(Redirected from
1415:
1393:
1391:
1389:
1372:
1344:
1343:
1307:
1301:
1300:
1290:
1258:
1252:
1251:
1241:
1217:
1211:
1210:
1200:
1168:
1157:
1156:
1140:
1134:
1133:
1097:
1086:
1085:
1083:
1082:
1018:
1012:
1011:
1009:
1008:
975:(841): 225–239.
960:
954:
953:
951:
950:
931:
925:
924:
906:
887:
886:
874:
868:
867:
849:
834:
833:
815:
798:
797:
779:
773:
772:
764:
755:
752:
746:
745:
733:
723:
717:
716:
698:
671:
670:
668:
667:
640:
512:genetic material
442:In somatic cells
159:Reginald Punnett
21:
1423:
1422:
1418:
1417:
1416:
1414:
1413:
1412:
1398:
1397:
1396:
1387:
1385:
1369:
1352:
1350:Further reading
1347:
1312:Nature Genetics
1308:
1304:
1259:
1255:
1218:
1214:
1177:Mol. Cell. Biol
1169:
1160:
1141:
1137:
1114:10.1038/nrg1614
1102:Nat. Rev. Genet
1098:
1089:
1080:
1078:
1019:
1015:
1006:
1004:
961:
957:
948:
946:
933:
932:
928:
921:
907:
890:
875:
871:
864:
850:
837:
830:
816:
801:
794:
780:
776:
765:
758:
753:
749:
742:
724:
720:
713:
699:
674:
665:
663:
656:
642:
641:
637:
633:
620:Non-disjunction
596:
583:
557:replicating DNA
537:
520:
483:
450:
444:
435:
426:
409:
397:metaphase plate
381:
328:
279:
271:
251:sex chromosomes
229:
221:DNA replication
189:
155:William Bateson
151:
107:
61:
28:
23:
22:
15:
12:
11:
5:
1421:
1411:
1410:
1395:
1394:
1373:
1367:
1353:
1351:
1348:
1346:
1345:
1318:(3): 247–254.
1302:
1273:(3): 651–660.
1253:
1212:
1158:
1155:on 1999-10-10.
1149:Human Genetics
1135:
1087:
1013:
981:10.1086/281833
955:
926:
919:
888:
885:on 2001-11-16.
869:
862:
835:
828:
799:
792:
774:
756:
747:
740:
718:
711:
672:
654:
634:
632:
629:
628:
627:
622:
617:
612:
607:
602:
595:
592:
582:
579:
536:
533:
519:
518:Nondisjunction
516:
482:
479:
473:regulation of
446:Main article:
443:
440:
434:
431:
425:
422:
408:
405:
380:
377:
327:
324:
278:
275:
270:
267:
228:
225:
202:submetacentric
188:
185:
150:
147:
106:
103:
26:
9:
6:
4:
3:
2:
1420:
1409:
1406:
1405:
1403:
1383:
1379:
1374:
1370:
1368:0-87893-258-5
1364:
1360:
1355:
1354:
1341:
1337:
1333:
1329:
1325:
1324:10.1038/85798
1321:
1317:
1313:
1306:
1298:
1294:
1289:
1284:
1280:
1276:
1272:
1268:
1264:
1257:
1249:
1245:
1240:
1235:
1232:(6): 927–36.
1231:
1227:
1223:
1216:
1208:
1204:
1199:
1194:
1190:
1186:
1183:(1): 267–77.
1182:
1178:
1174:
1167:
1165:
1163:
1154:
1150:
1146:
1139:
1131:
1127:
1123:
1119:
1115:
1111:
1108:(6): 477–87.
1107:
1103:
1096:
1094:
1092:
1076:
1072:
1068:
1064:
1060:
1056:
1052:
1048:
1044:
1040:
1036:
1032:
1028:
1024:
1017:
1002:
998:
994:
990:
986:
982:
978:
974:
970:
966:
959:
944:
940:
936:
930:
922:
916:
912:
905:
903:
901:
899:
897:
895:
893:
884:
880:
873:
865:
859:
855:
848:
846:
844:
842:
840:
831:
825:
821:
814:
812:
810:
808:
806:
804:
795:
793:9780321540980
789:
785:
778:
770:
763:
761:
751:
743:
741:0-8053-6624-5
737:
732:
731:
722:
714:
712:0-7167-4939-4
708:
704:
697:
695:
693:
691:
689:
687:
685:
683:
681:
679:
677:
661:
657:
651:
647:
646:
639:
635:
626:
623:
621:
618:
616:
613:
611:
608:
606:
603:
601:
598:
597:
591:
587:
578:
575:
571:
566:
562:
558:
554:
550:
541:
532:
530:
526:
515:
513:
509:
505:
504:birth defects
501:
497:
487:
478:
476:
472:
468:
464:
463:
458:
457:
449:
439:
430:
421:
419:
414:
404:
402:
398:
394:
390:
386:
376:
374:
370:
369:recombination
365:
360:
358:
354:
350:
349:crossing-over
346:
342:
338:
333:
323:
321:
317:
312:
308:
304:
300:
291:
283:
274:
266:
264:
258:
255:
252:
247:
245:
241:
237:
233:
224:
222:
218:
213:
211:
207:
203:
199:
193:
184:
181:
177:
172:
168:
167:Thomas Morgan
164:
160:
156:
146:
144:
140:
136:
132:
128:
124:
120:
116:
112:
102:
100:
96:
92:
88:
84:
80:
79:fertilization
76:
72:
68:
60:
54:
50:
46:
41:
37:
32:
19:
1386:. Retrieved
1358:
1315:
1311:
1305:
1270:
1267:Cell Reports
1266:
1256:
1229:
1225:
1215:
1180:
1176:
1153:the original
1148:
1138:
1105:
1101:
1079:. Retrieved
1030:
1026:
1016:
1005:. Retrieved
972:
968:
958:
947:. Retrieved
938:
929:
910:
883:the original
877:Gregory MJ.
872:
853:
820:Cell Biology
819:
783:
777:
768:
750:
729:
721:
702:
664:. Retrieved
644:
638:
588:
584:
546:
521:
500:embryo death
493:
467:transvection
460:
454:
451:
436:
427:
410:
382:
361:
329:
296:
272:
259:
256:
250:
248:
230:
214:
194:
190:
152:
108:
85:in the same
70:
66:
64:
47:at the same
1408:Chromosomes
401:kinetochore
385:metaphase I
379:Metaphase I
322:to act on.
314:variation.
261:humans are
210:telocentric
206:acrocentric
198:metacentric
163:inheritance
38:displays a
1388:1 November
1081:2022-06-30
1007:2021-03-23
949:2013-11-01
666:2013-11-01
631:References
572:sequence.
535:Other uses
498:problems,
462:Drosophila
456:Drosophila
433:In mitosis
424:Meiosis II
418:germ cells
407:Anaphase I
373:DNA damage
357:chromatids
347:. Genetic
332:prophase I
326:Prophase I
307:meiosis II
277:In meiosis
171:test cross
123:centromere
113:(DNA) and
75:chromosome
65:A pair of
1378:"Meiosis"
1071:231666458
1055:0036-8075
997:222327165
989:0003-0147
496:fertility
416:daughter
389:bivalents
364:chiasmata
303:meiosis I
299:germ cell
269:Functions
236:autosomal
227:In humans
187:Structure
119:chromatin
59:Karyotype
36:karyotype
1402:Category
1382:Archived
1332:11242102
1297:23478019
1248:14667414
1226:Dev Cell
1130:31929047
1122:15931171
1075:Archived
1063:33479152
1001:Archived
943:Archived
660:Archived
625:Heredity
615:Synapsis
594:See also
529:monosomy
481:Problems
475:X-linked
413:separase
345:anaphase
311:synapsis
169:. Using
105:Overview
99:organism
71:homologs
34:As this
18:Homologs
1340:3012823
1288:4315363
1207:8972207
1035:Bibcode
1027:Science
730:Biology
525:trisomy
477:genes.
393:tetrads
341:Cohesin
263:diploid
149:History
143:meiosis
139:alleles
135:diploid
131:mitosis
115:histone
91:meiosis
53:alleles
40:diploid
1365:
1338:
1330:
1295:
1285:
1246:
1205:
1198:231751
1195:
1128:
1120:
1069:
1061:
1053:
995:
987:
917:
860:
826:
790:
738:
709:
652:
565:repair
508:cancer
506:, and
232:Humans
1336:S2CID
1126:S2CID
1067:S2CID
993:S2CID
208:, or
83:genes
69:, or
49:locus
45:genes
1390:2013
1363:ISBN
1328:PMID
1293:PMID
1244:PMID
1203:PMID
1118:PMID
1059:PMID
1051:ISSN
985:ISSN
915:ISBN
858:ISBN
824:ISBN
788:ISBN
736:ISBN
707:ISBN
650:ISBN
527:and
242:and
178:and
157:and
127:loci
87:loci
1320:doi
1283:PMC
1275:doi
1234:doi
1193:PMC
1185:doi
1110:doi
1043:doi
1031:371
977:doi
553:DNA
551:of
391:or
383:In
330:In
1404::
1334:.
1326:.
1316:27
1314:.
1291:.
1281:.
1269:.
1265:.
1242:.
1228:.
1224:.
1201:.
1191:.
1181:17
1179:.
1175:.
1161:^
1147:.
1124:.
1116:.
1104:.
1090:^
1073:.
1065:.
1057:.
1049:.
1041:.
1029:.
1025:.
999:.
991:.
983:.
973:88
971:.
967:.
937:.
891:^
838:^
802:^
759:^
675:^
658:.
502:,
420:.
403:.
246:.
204:,
200:,
55:.
1392:.
1371:.
1342:.
1322::
1299:.
1277::
1271:3
1250:.
1236::
1230:5
1209:.
1187::
1132:.
1112::
1106:6
1084:.
1045::
1037::
1010:.
979::
952:.
923:.
866:.
832:.
796:.
744:.
715:.
669:.
244:Y
240:X
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.