236:
Because of the asymmetry in pyrimidine and purine use in coding sequences, the strand with the greater coding content will tend to have the greater number of purine bases (Szybalski's rule). Because the number of purine bases will, to a very good approximation, equal the number of their complementary pyrimidines within the same strand and, because the coding sequences occupy 80–90% of the strand, there appears to be (1) a selective pressure on the third base to minimize the number of purine bases in the strand with the greater coding content; and (2) that this pressure is proportional to the mismatch in the length of the coding sequences between the two strands.
177:
240:
279:; length ≤ 10) is present in equal numbers to its reverse complementary nucleotide. Because of the computational requirements this has not been verified in all genomes for all oligonucleotides. It has been verified for triplet oligonucleotides for a large data set. Albrecht-Buehler has suggested that this rule is the consequence of genomes evolving by a process of
287:. This process does not appear to have acted on the mitochondrial genomes. Chargaff's second parity rule appears to be extended from the nucleotide-level to populations of codon triplets, in the case of whole single-stranded Human genome DNA. A kind of "codon-level second Chargaff's parity rule" is proposed as follows:
686:
The following table is a representative sample of Erwin
Chargaff's 1952 data, listing the base composition of DNA from various organisms and support both of Chargaff's rules. An organism such as φX174 with significant variation from A/T and G/C equal to one, is indicative of single stranded DNA.
235:
Multivariate statistical analysis of codon use within genomes with unequal quantities of coding sequences on the two strands has shown that codon use in the third position depends on the strand on which the gene is located. This seems likely to be the result of
Szybalski's and Chargaff's rules.
677:
In 2020, it is suggested that the physical properties of the dsDNA (double stranded DNA) and the tendency to maximum entropy of all the physical systems are the cause of
Chargaff's second parity rule. The symmetries and patterns present in the dsDNA sequences can emerge from the physical
130:
The first empirical generalization of
Chargaff's second parity rule, called the Symmetry Principle, was proposed by Vinayakumar V. Prabhu in 1993. This principle states that for any given oligonucleotide, its frequency is approximately equal to the frequency of its complementary reverse
260:). The longer the strands are separated the greater the quantity of deamination. For reasons that are not yet clear the strands tend to exist longer in single form in mitochondria than in chromosomal DNA. This process tends to yield one strand that is enriched in
247:
The origin of the deviation from
Chargaff's rule in the organelles has been suggested to be a consequence of the mechanism of replication. During replication the DNA strands separate. In single stranded DNA,
232:—TGA and TAG respectively.) The mismatch between the number of codons and amino acids allows several codons to code for a single amino acid—such codons normally differ only at the third codon base position.
184:
The rule itself has consequences. In most bacterial genomes (which are generally 80-90% coding) genes are arranged in such a fashion that approximately 50% of the coding sequence lies on either strand.
103:
The second rule holds that both Α% ≈ Τ% and G% ≈ C% are valid for each of the two DNA strands. This describes only a global feature of the base composition in a single DNA strand.
20:
643:
583:
523:
463:
403:
343:
1943:
Szybalski W, Kubinski H, Sheldrick P (1966). "Pyrimidine clusters on the transcribing strands of DNA and their possible role in the initiation of RNA synthesis".
1552:
Szybalski W, Kubinski H, Sheldrick O (1966). "Pyrimidine clusters on the transcribing strand of DNA and their possible role in the initiation of RNA synthesis".
208:
The combined effect of
Chargaff's second rule and Szybalski's rule can be seen in bacterial genomes where the coding sequences are not equally distributed. The
1694:
Nikolaou C, Almirantis Y (2006). "Deviations from
Chargaff's second parity rule in organellar DNA. Insights into the evolution of organellar genomes".
268:(T) with its complement enriched in cytosine (C) and adenosine (A), and this process may have given rise to the deviations found in the mitochondria.
1885:
1791:
Perez, J.-C. (September 2010). "Codon populations in single-stranded whole human genome DNA are fractal and fine-tuned by the Golden Ratio 1.618".
678:
peculiarities of the dsDNA molecule and the maximum entropy principle alone, rather than from biological or environmental evolutionary pressure.
1647:"Thermophilic Bacteria Strictly Obey Szybalski's Transcription Direction Rule and Politely Purine-Load RNAs with Both Adenine and Guanine"
2050:
McLean MJ, Wolfe KH, Devine KM (1998). "Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes".
271:
Chargaff's second rule appears to be the consequence of a more complex parity rule: within a single strand of DNA any oligonucleotide (
205:". While Szybalski's rule generally holds, exceptions are known to exist. The biological basis for Szybalski's rule is not yet known.
1734:"Asymptotically increasing compliance of genomes with Chargaff's second parity rules through inversions and inverted transpositions"
134:
In 2006, it was shown that this rule applies to four of the five types of double stranded genomes; specifically it applies to the
131:
oligonucleotide. A theoretical generalization was mathematically derived by Michel E. B. Yamagishi and
Roberto H. Herai in 2011.
1493:
1442:
1900:
1602:
Bell SJ, Forsdyke DR (1999). "Deviations from
Chargaff's second parity rule correlate with direction of transcription".
2167:
111:
The second parity rule was discovered in 1968. It states that, in single-stranded DNA, the number of adenine units is
674:
36530115 TTT and 36381293 AAA (ratio % = 1.00409). 2087242 TCG and 2085226 CGA (ratio % = 1.00096), etc...
91:
has percentage base pair equality: A% = T% and G% = C%. The rigorous validation of the rule constitutes the basis of
2009:"Proteome composition and codon usage in spirochaetes: species-specific and DNA strand-specific mutational biases"
2310:
66:) should exist. This pattern is found in both strands of the DNA. They were discovered by Austrian-born chemist
2305:
2300:
2285:
2280:
2170:
2016-05-16 at the
Portuguese Web Archive — contains hundreds of examples of base skews and had problems.
2315:
2173:
92:
2295:
201:(C and T). This rule has since been confirmed in other organisms and should probably be now termed "
173:
genome. The basis for this rule is still under investigation, although genome size may play a role.
2072:
1833:
2290:
2067:
1879:
628:
568:
508:
448:
388:
328:
280:
2118:
2059:
1745:
1611:
1283:
2190:"CBS Genome Atlas Database: A dynamic storage for bioinformatic results and sequence data"
1990:
1973:
1836:"DNA sequence symmetries from randomness: the origin of the Chargaff's second parity rule"
1168:
Elson D, Chargaff E (1952). "On the deoxyribonucleic acid content of sea urchin gametes".
8:
202:
2122:
2063:
1749:
1615:
1287:
186:
2093:
1862:
1835:
1816:
1768:
1733:
1499:
1471:
1448:
1420:
1245:
1193:
671:
Examples — computing whole human genome using the first codons reading frame provides:
2033:
2008:
1671:
1646:
1392:
1367:
1306:
1267:
1232:
1215:
2252:
2211:
2146:
2141:
2106:
2085:
2038:
1995:
1960:
1867:
1808:
1773:
1711:
1676:
1627:
1569:
1534:
1489:
1438:
1397:
1348:
1311:
1237:
1185:
2247:
2230:
2206:
2189:
2107:"Replicational and transcriptional selection on codon usage in Borrelia burgdorferi"
2097:
1820:
1503:
1452:
1343:
1330:
1249:
1197:
2242:
2201:
2136:
2126:
2077:
2028:
2020:
1985:
1952:
1857:
1847:
1800:
1763:
1753:
1703:
1666:
1658:
1619:
1561:
1526:
1481:
1430:
1387:
1379:
1338:
1301:
1291:
1227:
1177:
1064:
1956:
1834:
Piero Farisell, Cristian Taccioli, Luca Pagani & Amos Maritan (April 2020).
1565:
1707:
1530:
1276:
Proceedings of the National Academy of Sciences of the United States of America
807:
221:
180:
Histogram showing how 20309 chromosomes adhere to Chargaff's second parity rule
154:
67:
31:
2164:
1915:
1804:
1485:
1434:
2269:
2131:
1383:
1148:
1031:
190:
2024:
1758:
1216:"Composition of the deoxypentose nucleic acids of four genera of sea-urchin"
2275:
2256:
2215:
2042:
1871:
1812:
1777:
1715:
1680:
1631:
1623:
1538:
1352:
1241:
1189:
257:
209:
158:
23:
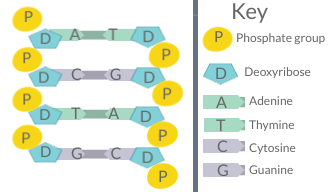
A diagram of DNA base pairing, demonstrating the basis for Chargaff's rules
2150:
2089:
1999:
1964:
1852:
1573:
1401:
1315:
1296:
169:, nor does it apply to single stranded DNA (viral) genomes or any type of
1071:
915:
731:
225:
198:
176:
138:
1662:
2081:
1181:
956:
951:
920:
284:
239:
229:
217:
59:
1974:"Asymmetric substitution patterns in the two DNA strands of bacteria"
253:
135:
2231:"The Z curve database: a graphic representation of genome sequences"
2007:
Lafay B, Lloyd AT, McLean MJ, Devine KM, Sharp PM, Wolfe KH (1999).
1331:"The Z curve database: a oraphic representation of genome sequences"
993:
249:
166:
142:
43:
1476:
1425:
1103:
220:
normally present in proteins. (There are two uncommon amino acids—
1517:
Mitchell D, Bridge R (2006). "A test of Chargaff's second rule".
1109:
801:
769:
763:
265:
261:
194:
162:
150:
51:
47:
39:
1589:
Characterization of G0/G1 switch genes in cultured T lymphocytes
845:
276:
213:
55:
16:
Two rules about the percentage of A, C, G, and T in DNA strands
19:
2176:— a 3-dimensional visualization and analysis tool of genomes.
1025:
987:
877:
725:
291:
Intra-strand relation among percentages of codon populations
272:
216:
of which 3 function as termination codons: there are only 20
883:
1551:
228:—found in a limited number of proteins and encoded by the
1901:"DNA structure: Revisiting the Watson-Crick double helix"
839:
170:
146:
84:
35:
2222:
1793:
Interdisciplinary Sciences: Computational Life Sciences
1727:
1725:
1468:
Chargaff's "Grammar of Biology": New Fractal-like Rules
1322:
1213:
631:
571:
511:
451:
391:
331:
1722:
1265:
1945:
Cold Spring Harbor Symposia on Quantitative Biology
1368:"Symmetry observation in long nucleotide sequences"
1272:
DNA into complementary strands. 3. Direct analysis"
2187:
1731:
1693:
637:
577:
517:
457:
397:
337:
243:Chargaff's 2nd parity rule for prokaryotic 6-mers
2267:
1591:. Kingston, Ontario, Canada: Queen's University.
38:of any species and any organism, the amount of
1516:
1465:
1209:
1207:
681:
1580:
1414:
1167:
2228:
1884:: CS1 maint: multiple names: authors list (
1790:
1687:
1601:
1586:
1545:
1328:
1261:
1259:
83:The first rule holds that a double-stranded
1644:
1419:. SpringerBriefs in Mathematics. Springer.
1266:Rudner, R; Karkas, JD; Chargaff, E (1968).
1204:
2181:
1892:
2246:
2205:
2140:
2130:
2071:
2032:
1989:
1861:
1851:
1767:
1757:
1670:
1475:
1424:
1391:
1365:
1342:
1305:
1295:
1256:
1231:
119:%T), and the number of cytosine units is
54:. Further, a 1:1 stoichiometric ratio of
1898:
1595:
1214:Chargaff E, Lipshitz R, Green C (1952).
1161:
238:
175:
18:
1638:
2268:
1991:10.1093/oxfordjournals.molbev.a025626
98:
78:
252:spontaneously slowly deaminates to
13:
2188:Hallin PF, David Ussery D (2004).
1936:
153:chromosomes. It does not apply to
14:
2327:
2229:Zhang CT, Zhang R, Ou HY (2003).
2158:
1470:. SpringerBriefs in Mathematics.
1329:Zhang CT, Zhang R, Ou HY (2003).
145:chromosomes, the double stranded
50:should be equal to the amount of
42:should be equal to the amount of
1554:Cold Spring Harb Symp Quant Biol
2174:The Z curve database of genomes
1914:(11): 1556–1563. Archived from
1827:
1784:
1466:Yamagishi ME, Herai RH (2011).
1417:Mathematical Grammar of Biology
189:, in the 1960s, showed that in
95:in the DNA double helix model.
1510:
1459:
1408:
1359:
73:
1:
2248:10.1093/bioinformatics/btg041
2207:10.1093/bioinformatics/bth423
1344:10.1093/bioinformatics/btg041
1233:10.1016/S0021-9258(19)50884-5
1154:
123:equal to that of guanine (%C
115:equal to that of thymine (%A
1645:Lao PJ, Forsdyke DR (2000).
1337:. 19 [issue=5 (5): 590–599.
7:
1957:10.1101/SQB.1966.031.01.019
1840:Briefings in Bioinformatics
1732:Albrecht-Buehler G (2006).
1566:10.1101/SQB.1966.031.01.019
1142:
682:Percentages of bases in DNA
106:
10:
2332:
1708:10.1016/j.gene.2006.06.010
1531:10.1016/j.bbrc.2005.11.160
1519:Biochem Biophys Res Commun
2165:CBS Genome Atlas Database
1805:10.1007/s12539-010-0022-0
1486:10.1007/978-3-319-62689-5
1435:10.1007/978-3-319-62689-5
719:
716:
713:
710:
707:
704:
701:
698:
695:
692:
304:
301:
298:
295:
2132:10.1073/pnas.95.18.10698
658:are mirror codons, e.g.
619:(1st base position is G)
613:(3rd base position is C)
598:are mirror codons, e.g.
559:(1st base position is A)
553:(3rd base position is T)
538:are mirror codons, e.g.
499:(2nd base position is G)
493:(2nd base position is C)
478:are mirror codons, e.g.
439:(2nd base position is A)
433:(2nd base position is T)
418:are mirror codons, e.g.
379:(3rd base position is G)
373:(1st base position is C)
358:are mirror codons, e.g.
319:(3rd base position is A)
313:(1st base position is T)
1759:10.1073/pnas.0605553103
638:{\displaystyle \simeq }
578:{\displaystyle \simeq }
518:{\displaystyle \simeq }
458:{\displaystyle \simeq }
398:{\displaystyle \simeq }
338:{\displaystyle \simeq }
149:viral genomes, and the
93:Watson–Crick base pairs
2311:Biological engineering
2111:Proc Natl Acad Sci USA
1738:Proc Natl Acad Sci USA
1624:10.1006/jtbi.1998.0858
1415:Yamagishi MEB (2017).
1384:10.1093/nar/21.12.2797
1372:Nucleic Acids Research
639:
579:
519:
459:
399:
339:
244:
181:
165:) smaller than ~20-30
24:
2306:Laboratory techniques
2105:McInerney JO (1998).
2025:10.1093/nar/27.7.1642
1846:(bbaa04): 2172–2181.
1587:Cristillo AD (1998).
1297:10.1073/pnas.60.3.921
640:
580:
520:
460:
400:
340:
242:
179:
22:
629:
569:
509:
449:
389:
329:
34:) state that in the
2301:Biology experiments
2286:History of genetics
2281:Genetics techniques
2123:1998PNAS...9510698M
2117:(18): 10698–10703.
2064:1998JMolE..47..691M
1853:10.1093/bib/bbaa041
1750:2006PNAS..10317828A
1744:(47): 17828–17833.
1663:10.1101/gr.10.2.228
1616:1999JThBi.197...63B
1288:1968PNAS...60..921R
292:
70:in the late 1940s.
2082:10.1007/PL00006428
1366:Prabhu VV (1993).
1182:10.1007/BF02170221
635:
575:
515:
455:
395:
335:
290:
245:
182:
155:organellar genomes
99:Second parity rule
46:and the amount of
25:
2316:Molecular biology
2200:(18): 3682–3686.
2013:Nucleic Acids Res
1972:Lobry JR (1996).
1899:Bansal M (2003).
1495:978-3-319-62688-8
1444:978-3-319-62688-8
1378:(12): 2797–2800.
1140:
1139:
669:
668:
302:Relation proposed
197:(A and G) exceed
193:coding sequences
79:First parity rule
2323:
2296:Medical research
2261:
2260:
2250:
2226:
2220:
2219:
2209:
2185:
2154:
2144:
2134:
2101:
2075:
2046:
2036:
2019:(7): 1642–1649.
2003:
1993:
1968:
1930:
1929:
1927:
1926:
1920:
1905:
1896:
1890:
1889:
1883:
1875:
1865:
1855:
1831:
1825:
1824:
1788:
1782:
1781:
1771:
1761:
1729:
1720:
1719:
1691:
1685:
1684:
1674:
1642:
1636:
1635:
1599:
1593:
1592:
1584:
1578:
1577:
1549:
1543:
1542:
1514:
1508:
1507:
1479:
1463:
1457:
1456:
1428:
1412:
1406:
1405:
1395:
1363:
1357:
1356:
1346:
1326:
1320:
1319:
1309:
1299:
1263:
1254:
1253:
1235:
1211:
1202:
1201:
1165:
690:
689:
665:
661:
657:
653:
648:
644:
642:
641:
636:
625:
618:
612:
605:
601:
597:
593:
588:
584:
582:
581:
576:
565:
558:
552:
545:
541:
537:
533:
528:
524:
522:
521:
516:
505:
498:
492:
485:
481:
477:
473:
468:
464:
462:
461:
456:
445:
438:
432:
425:
421:
417:
413:
408:
404:
402:
401:
396:
385:
378:
372:
365:
361:
357:
353:
348:
344:
342:
341:
336:
325:
318:
312:
293:
289:
203:Szybalski's rule
187:Wacław Szybalski
126:
118:
65:
28:Chargaff's rules
2331:
2330:
2326:
2325:
2324:
2322:
2321:
2320:
2266:
2265:
2264:
2227:
2223:
2186:
2182:
2161:
2104:
2049:
2006:
1978:Mol. Biol. Evol
1971:
1942:
1939:
1937:Further reading
1934:
1933:
1924:
1922:
1918:
1908:Current Science
1903:
1897:
1893:
1877:
1876:
1832:
1828:
1789:
1785:
1730:
1723:
1692:
1688:
1651:Genome Research
1643:
1639:
1600:
1596:
1585:
1581:
1550:
1546:
1515:
1511:
1496:
1464:
1460:
1445:
1413:
1409:
1364:
1360:
1327:
1323:
1268:"Separation of
1264:
1257:
1212:
1205:
1166:
1162:
1157:
1145:
684:
675:
663:
659:
655:
651:
646:
630:
627:
626:
623:
616:
610:
603:
599:
595:
591:
586:
570:
567:
566:
563:
556:
550:
543:
539:
535:
531:
526:
510:
507:
506:
503:
496:
490:
483:
479:
475:
471:
466:
450:
447:
446:
443:
436:
430:
423:
419:
415:
411:
406:
390:
387:
386:
383:
376:
370:
363:
359:
355:
351:
346:
330:
327:
326:
323:
316:
310:
124:
116:
109:
101:
81:
76:
63:
17:
12:
11:
5:
2329:
2319:
2318:
2313:
2308:
2303:
2298:
2293:
2288:
2283:
2278:
2263:
2262:
2241:(5): 593–599.
2235:Bioinformatics
2221:
2194:Bioinformatics
2179:
2178:
2177:
2171:
2160:
2159:External links
2157:
2156:
2155:
2102:
2073:10.1.1.28.9035
2058:(6): 691–696.
2047:
2004:
1984:(5): 660–665.
1969:
1938:
1935:
1932:
1931:
1891:
1826:
1799:(3): 228–240.
1783:
1721:
1686:
1657:(2): 228–236.
1637:
1594:
1579:
1544:
1509:
1494:
1458:
1443:
1407:
1358:
1335:Bioinformatics
1321:
1255:
1226:(1): 155–160.
1203:
1176:(4): 143–145.
1159:
1158:
1156:
1153:
1152:
1151:
1144:
1141:
1138:
1137:
1134:
1131:
1128:
1125:
1122:
1119:
1116:
1113:
1106:
1100:
1099:
1096:
1093:
1090:
1087:
1084:
1081:
1078:
1075:
1068:
1060:
1059:
1056:
1053:
1050:
1047:
1044:
1041:
1038:
1035:
1028:
1022:
1021:
1018:
1015:
1012:
1009:
1006:
1003:
1000:
997:
990:
984:
983:
980:
977:
974:
971:
968:
965:
962:
959:
954:
948:
947:
944:
941:
938:
935:
932:
929:
926:
923:
918:
912:
911:
908:
905:
902:
899:
896:
893:
890:
887:
880:
874:
873:
870:
867:
864:
861:
858:
855:
852:
849:
842:
836:
835:
832:
829:
826:
823:
820:
817:
814:
811:
804:
798:
797:
794:
791:
788:
785:
782:
779:
776:
773:
766:
760:
759:
756:
753:
750:
747:
744:
741:
738:
735:
728:
722:
721:
718:
715:
712:
709:
706:
703:
700:
697:
694:
683:
680:
673:
667:
666:
649:
634:
620:
614:
607:
606:
589:
574:
560:
554:
547:
546:
529:
514:
500:
494:
487:
486:
469:
454:
440:
434:
427:
426:
409:
394:
380:
374:
367:
366:
349:
334:
320:
314:
307:
306:
303:
300:
297:
222:selenocysteine
108:
105:
100:
97:
80:
77:
75:
72:
68:Erwin Chargaff
32:Erwin Chargaff
15:
9:
6:
4:
3:
2:
2328:
2317:
2314:
2312:
2309:
2307:
2304:
2302:
2299:
2297:
2294:
2292:
2291:Biotechnology
2289:
2287:
2284:
2282:
2279:
2277:
2274:
2273:
2271:
2258:
2254:
2249:
2244:
2240:
2236:
2232:
2225:
2217:
2213:
2208:
2203:
2199:
2195:
2191:
2184:
2180:
2175:
2172:
2169:
2166:
2163:
2162:
2152:
2148:
2143:
2138:
2133:
2128:
2124:
2120:
2116:
2112:
2108:
2103:
2099:
2095:
2091:
2087:
2083:
2079:
2074:
2069:
2065:
2061:
2057:
2053:
2048:
2044:
2040:
2035:
2030:
2026:
2022:
2018:
2014:
2010:
2005:
2001:
1997:
1992:
1987:
1983:
1979:
1975:
1970:
1966:
1962:
1958:
1954:
1950:
1946:
1941:
1940:
1921:on 2014-07-26
1917:
1913:
1909:
1902:
1895:
1887:
1881:
1873:
1869:
1864:
1859:
1854:
1849:
1845:
1841:
1837:
1830:
1822:
1818:
1814:
1810:
1806:
1802:
1798:
1794:
1787:
1779:
1775:
1770:
1765:
1760:
1755:
1751:
1747:
1743:
1739:
1735:
1728:
1726:
1717:
1713:
1709:
1705:
1701:
1697:
1690:
1682:
1678:
1673:
1668:
1664:
1660:
1656:
1652:
1648:
1641:
1633:
1629:
1625:
1621:
1617:
1613:
1609:
1605:
1598:
1590:
1583:
1575:
1571:
1567:
1563:
1559:
1555:
1548:
1540:
1536:
1532:
1528:
1524:
1520:
1513:
1505:
1501:
1497:
1491:
1487:
1483:
1478:
1473:
1469:
1462:
1454:
1450:
1446:
1440:
1436:
1432:
1427:
1422:
1418:
1411:
1403:
1399:
1394:
1389:
1385:
1381:
1377:
1373:
1369:
1362:
1354:
1350:
1345:
1340:
1336:
1332:
1325:
1317:
1313:
1308:
1303:
1298:
1293:
1289:
1285:
1281:
1277:
1273:
1271:
1262:
1260:
1251:
1247:
1243:
1239:
1234:
1229:
1225:
1221:
1217:
1210:
1208:
1199:
1195:
1191:
1187:
1183:
1179:
1175:
1171:
1164:
1160:
1150:
1149:Genetic codes
1147:
1146:
1135:
1132:
1129:
1126:
1123:
1120:
1117:
1114:
1112:
1111:
1107:
1105:
1102:
1101:
1097:
1094:
1091:
1088:
1085:
1082:
1079:
1076:
1074:
1073:
1069:
1067:
1066:
1062:
1061:
1057:
1054:
1051:
1048:
1045:
1042:
1039:
1036:
1034:
1033:
1032:Saccharomyces
1029:
1027:
1024:
1023:
1019:
1016:
1013:
1010:
1007:
1004:
1001:
998:
996:
995:
991:
989:
986:
985:
981:
978:
975:
972:
969:
966:
963:
960:
958:
955:
953:
950:
949:
945:
942:
939:
936:
933:
930:
927:
924:
922:
919:
917:
914:
913:
909:
906:
903:
900:
897:
894:
891:
888:
886:
885:
881:
879:
876:
875:
871:
868:
865:
862:
859:
856:
853:
850:
848:
847:
843:
841:
838:
837:
833:
830:
827:
824:
821:
818:
815:
812:
810:
809:
805:
803:
800:
799:
795:
792:
789:
786:
783:
780:
777:
774:
772:
771:
767:
765:
762:
761:
757:
754:
751:
748:
745:
742:
739:
736:
734:
733:
729:
727:
724:
723:
691:
688:
679:
672:
650:
632:
621:
615:
609:
608:
590:
572:
561:
555:
549:
548:
530:
512:
501:
495:
489:
488:
470:
452:
441:
435:
429:
428:
410:
392:
381:
375:
369:
368:
350:
332:
321:
315:
309:
308:
294:
288:
286:
285:transposition
282:
278:
274:
269:
267:
263:
259:
255:
251:
241:
237:
233:
231:
227:
223:
219:
215:
211:
206:
204:
200:
196:
192:
191:bacteriophage
188:
178:
174:
172:
168:
164:
160:
156:
152:
148:
144:
140:
137:
132:
128:
122:
121:approximately
114:
113:approximately
104:
96:
94:
90:
86:
71:
69:
62:bases (i.e.,
61:
57:
53:
49:
45:
41:
37:
33:
29:
21:
2238:
2234:
2224:
2197:
2193:
2183:
2114:
2110:
2055:
2051:
2016:
2012:
1981:
1977:
1948:
1944:
1923:. Retrieved
1916:the original
1911:
1907:
1894:
1880:cite journal
1843:
1839:
1829:
1796:
1792:
1786:
1741:
1737:
1699:
1695:
1689:
1654:
1650:
1640:
1610:(1): 63–76.
1607:
1604:J Theor Biol
1603:
1597:
1588:
1582:
1557:
1553:
1547:
1525:(1): 90–94.
1522:
1518:
1512:
1467:
1461:
1416:
1410:
1375:
1371:
1361:
1334:
1324:
1282:(3): 921–2.
1279:
1275:
1269:
1223:
1219:
1173:
1169:
1163:
1108:
1070:
1063:
1030:
992:
882:
844:
806:
768:
730:
685:
676:
670:
299:Second codon
270:
258:transversion
246:
234:
210:genetic code
207:
183:
159:mitochondria
133:
129:
120:
112:
110:
102:
88:
82:
27:
26:
1951:: 123–127.
1560:: 123–127.
1270:B. Subtilis
1220:J Biol Chem
1170:Experientia
1072:Escherichia
916:Grasshopper
296:First codon
230:stop codons
226:pyrrolysine
218:amino acids
199:pyrimidines
139:chromosomes
74:Definitions
2270:Categories
2052:J Mol Evol
1925:2013-07-26
1155:References
957:Echinoidea
952:Sea urchin
921:Orthoptera
256:(a C to A
136:eukaryotic
87:molecule,
60:pyrimidine
30:(given by
2068:CiteSeerX
1702:: 34–41.
1477:1112.1528
1426:1112.1528
633:≃
573:≃
513:≃
453:≃
393:≃
333:≃
281:inversion
254:adenosine
143:bacterial
2257:12651717
2216:15256401
2168:Archived
2098:12917481
2043:10075995
1872:32266404
1821:54565279
1813:20658335
1778:17093051
1716:16893615
1681:10673280
1632:10036208
1539:16364245
1504:16742066
1453:16742066
1353:12651717
1250:11358561
1242:14938364
1198:36803326
1190:14945441
1143:See also
994:Triticum
693:Organism
645: %
585: %
525: %
465: %
405: %
345: %
305:Details
264:(G) and
250:cytosine
163:plastids
151:archaeal
107:Research
89:globally
44:cytosine
2151:9724767
2119:Bibcode
2090:9847411
2060:Bibcode
2000:8676740
1965:4966069
1863:7986665
1769:1635160
1746:Bibcode
1612:Bibcode
1574:4966069
1402:8332488
1316:4970114
1284:Bibcode
1110:PhiX174
1065:E. coli
802:Chicken
770:Octopus
764:Octopus
266:thymine
262:guanine
212:has 64
195:purines
64:A+G=T+C
52:thymine
48:adenine
40:guanine
2255:
2214:
2149:
2139:
2096:
2088:
2070:
2041:
2034:148367
2031:
1998:
1963:
1870:
1860:
1819:
1811:
1776:
1766:
1714:
1679:
1672:310832
1669:
1630:
1572:
1537:
1502:
1492:
1451:
1441:
1400:
1393:309655
1390:
1351:
1314:
1307:225140
1304:
1248:
1240:
1196:
1188:
846:Rattus
808:Gallus
277:n-gram
214:codons
141:, the
56:purine
2142:27958
2094:S2CID
1919:(PDF)
1904:(PDF)
1817:S2CID
1500:S2CID
1472:arXiv
1449:S2CID
1421:arXiv
1246:S2CID
1194:S2CID
1136:55.2
1104:φX174
1098:48.3
1058:64.4
1026:Yeast
1020:54.4
988:Wheat
982:64.9
946:58.6
910:59.3
878:Human
872:57.0
834:56.4
796:64.8
758:54.0
726:Maize
714:G / C
711:A / T
696:Taxon
273:k-mer
127:%G).
2253:PMID
2212:PMID
2147:PMID
2086:PMID
2039:PMID
1996:PMID
1961:PMID
1886:link
1868:PMID
1809:PMID
1774:PMID
1712:PMID
1696:Gene
1677:PMID
1628:PMID
1570:PMID
1535:PMID
1490:ISBN
1439:ISBN
1398:PMID
1349:PMID
1312:PMID
1238:PMID
1186:PMID
1133:44.8
1130:1.08
1127:0.77
1124:31.2
1121:21.5
1118:23.3
1115:24.0
1095:51.7
1092:1.01
1089:1.05
1086:23.6
1083:25.7
1080:26.0
1077:24.7
1055:35.8
1052:1.09
1049:0.95
1046:32.9
1043:17.1
1040:18.7
1037:31.3
1017:45.5
1014:1.00
1011:1.01
1008:27.1
1005:22.8
1002:22.7
999:27.3
979:35.0
976:1.02
973:1.02
970:32.1
967:17.3
964:17.7
961:32.8
943:41.2
940:0.99
937:1.00
934:29.3
931:20.7
928:20.5
925:29.3
907:40.7
904:1.04
901:0.98
898:30.0
895:20.0
892:20.7
889:29.3
884:Homo
869:42.9
866:1.00
863:1.01
860:28.4
857:20.5
854:21.4
851:28.6
831:43.7
828:1.02
825:0.99
822:28.4
819:21.6
816:22.0
813:28.0
793:35.2
790:1.00
787:1.05
784:31.6
781:17.6
778:17.6
775:33.2
755:46.1
752:0.98
749:0.99
746:27.2
743:23.2
740:22.8
737:26.8
720:%AT
662:and
654:and
602:and
594:and
542:and
534:and
482:and
474:and
422:and
414:and
362:and
354:and
283:and
224:and
161:and
58:and
2276:DNA
2243:doi
2202:doi
2137:PMC
2127:doi
2078:doi
2029:PMC
2021:doi
1986:doi
1953:doi
1858:PMC
1848:doi
1801:doi
1764:PMC
1754:doi
1742:103
1704:doi
1700:381
1667:PMC
1659:doi
1620:doi
1608:197
1562:doi
1527:doi
1523:340
1482:doi
1431:doi
1388:PMC
1380:doi
1339:doi
1302:PMC
1292:doi
1228:doi
1224:195
1178:doi
840:Rat
732:Zea
717:%GC
664:GCC
660:GGC
656:Gyz
652:wxC
647:Gyz
624:wxC
617:Gyz
611:wxC
604:AAG
600:CTT
596:Ayz
592:wxT
587:Ayz
564:wxT
557:Ayz
551:wxT
544:AGA
540:TCT
536:yGz
532:wCx
527:yGz
504:wCx
497:yGz
491:wCx
484:CAG
480:CTG
476:yAz
472:wTx
467:yAz
444:wTx
437:yAz
431:wTx
424:TAG
420:CTA
416:yzG
412:Cwx
407:yzG
384:Cwx
377:yzG
371:Cwx
364:CGA
360:TCG
356:yzA
352:Twx
347:yzA
324:Twx
317:yzA
311:Twx
275:or
171:RNA
167:kbp
147:DNA
85:DNA
36:DNA
2272::
2251:.
2239:19
2237:.
2233:.
2210:.
2198:20
2196:.
2192:.
2145:.
2135:.
2125:.
2115:95
2113:.
2109:.
2092:.
2084:.
2076:.
2066:.
2056:47
2054:.
2037:.
2027:.
2017:27
2015:.
2011:.
1994:.
1982:13
1980:.
1976:.
1959:.
1949:31
1947:.
1912:85
1910:.
1906:.
1882:}}
1878:{{
1866:.
1856:.
1844:22
1842:.
1838:.
1815:.
1807:.
1795:.
1772:.
1762:.
1752:.
1740:.
1736:.
1724:^
1710:.
1698:.
1675:.
1665:.
1655:10
1653:.
1649:.
1626:.
1618:.
1606:.
1568:.
1558:31
1556:.
1533:.
1521:.
1498:.
1488:.
1480:.
1447:.
1437:.
1429:.
1396:.
1386:.
1376:21
1374:.
1370:.
1347:.
1333:.
1310:.
1300:.
1290:.
1280:60
1278:.
1274:.
1258:^
1244:.
1236:.
1222:.
1218:.
1206:^
1192:.
1184:.
1172:.
708:%T
705:%C
702:%G
699:%A
622:%
562:%
502:%
442:%
382:%
322:%
2259:.
2245::
2218:.
2204::
2153:.
2129::
2121::
2100:.
2080::
2062::
2045:.
2023::
2002:.
1988::
1967:.
1955::
1928:.
1888:)
1874:.
1850::
1823:.
1803::
1797:2
1780:.
1756::
1748::
1718:.
1706::
1683:.
1661::
1634:.
1622::
1614::
1576:.
1564::
1541:.
1529::
1506:.
1484::
1474::
1455:.
1433::
1423::
1404:.
1382::
1355:.
1341::
1318:.
1294::
1286::
1252:.
1230::
1200:.
1180::
1174:8
157:(
125:≈
117:≈
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.