370:
171:
725:. This strongly suggests that rheumatoid arthritis is caused by unidentified arthritogenic antigens. The antigen could be any exogenous antigen, such as viral proteins, or an endogenous protein. Recently, a number of possible endogenous antigens have been identified, for example, human cartilage glycoprotein 39, heavy chain binding protein and citrullinated protein. Activated CD4+ T lymphocytes stimulate monocytes, macrophages and synovial fibroblasts to elaborate the cytokines
38:
807:
2342:
260:, in which those double-positive T cells that bind to foreign antigen in the presence of self MHC. They will differentiate into either CD4 or CD8 depending on which MHC is associated with the antigen presented (MHC1 for CD8, MHC2 for CD4). In this case, the cells would have been presented antigen in the context of MHC1. Positive selection means selecting those TCRs capable of recognizing self MHC molecules.
316:
379:
that have been in contact with the antigen at least once but have returned subsequently to a quiescent or inactive state, ready to respond again to the antigen against which they were stimulated. Finally, when the specific immune response is triggered, these naive and memory T cells are activated, giving rise to effector T cells that have the capacity to kill pathogens or tumor cells.
721:, increased vascularity and infiltration of inflammatory cells; mainly CD4+ T lymphocytes, which are the main organisers of cell-mediated immune responses. In different studies, rheumatoid arthritis is strongly linked to major histocompatibility complex (MHC) class II antigens. The only cells in the body that express MHC class II antigens are constitutive
227:), which recognize non-protein antigens. The latter are characterised by their ability to recognise antigens that are not presented. In addition, they can recognise microbial toxic shock proteins and self-cell stress proteins. T γδ cells possess a wide functional plasticity after recognising infected or transformed cells, as they are able to produce
390:). In the thymus-independent pathway, because the APC is infected, it is highly activated and expresses a large number of co-receptors for coactivation. If APCs are not infected, CD4 cells need to be involved: either to activate the APC by co-stimulation (more common) or to directly activate the Tc cell by secreting
651:(DNA-activated protein kinase). The final result is apoptosis of the cell that expressed Fas. CD8 T cells can also show Activation Induced Cell Death or AICD which is mediated by CD3 receptor complex. Recently, a platelet released protein TLT-1 has been shown to induce AICD like cell death in CD8 T cells
638:
effector cells. Engagement of Fas with FasL allows for recruitment of the death-induced signaling complex (DISC). The Fas-associated death domain (FADD) translocates with the DISC, allowing recruitment of procaspases 8 and 10. These caspases then activate the effector caspases 3, 6, and 7, leading to
178:
The immune system must recognize millions of potential antigens. There are fewer than 30,000 genes in the human body, so it is impossible to have one gene for every antigen. Instead, the DNA in millions of white blood cells in the bone marrow is shuffled to create cells with unique receptors, each of
658:
is suggested to play a key role in CD8 T cell function, acting as a regulatory gene in the adaptive immune response. Studies investigating the effect of loss-of-function
Eomesodermin found that a decrease in expression of this transcription factor resulted in decreased amount of perforin produced by
378:
T cells go through different stages, depending on the number of times they have been in contact with the antigen. In the first place, naïve T-lymphocytes are those cells that have not yet encountered an antigen in the thymus. Then, T-lymphocytes become memory T cells. This type of T cells are those
293:
Only those T cells that bind to the MHC-self-antigen complexes weakly are positively selected. Those cells that survive positive and negative selection differentiate into single-positive T cells (either CD4 or CD8), depending on whether their TCR recognizes an MHC class I-presented antigen (CD8) or
741:
activation through immune complex formation. Moreover, several animal studies suggest that cytotoxic T cells may have a predominantly proinflammatory effect in the disease. It is also studied that the production of cytokines by the CD8+ cells may accelerate the progresses of the arthritis disease.
214:
DNA to form a developmental form of the TCR protein, known as pre-TCR. If that rearrangement is successful, the cells then rearrange their alpha-chain TCR DNA to create a functional alpha-beta TCR complex. This highly-variable genetic rearrangement product in the TCR genes helps create millions of
749:
infection. HIV over time has developed many strategies to evade the host cell immune system. For example, HIV has adopted very high mutation rates to allow them to escape recognition by CD8 T cells. They are also able to down-regulate expression of surface MHC Class I proteins of cells that they
541:
and CD8 T cells. During this process, the CD4 helper T cells "license" the dendritic cells to give a potent activating signal to the naive CD8 T cells. This licensing of antigen-presenting cells by the CD4 T helper cells proceeds by signaling between CD154/CD40L on the T helper cell and the CD40
373:
In this immunofluorescence image, a group of killer T cells (outer three) is engaging a cancer cell (centered one). A patch of signaling molecules (pink) that gathers at the site of cell-cell contact indicates that the CTL has identified a target. Lytic granules (red) that contain cytotoxic
397:
If activation occurs, the lymphocyte polarizes its granules towards the site of the synapse and releases them, producing a "lethal hit". At this point, it separates from the target cell, and can move on to another, and another. The target cell dies in about 6 hours, usually by apoptosis.
737:(TNFa), and to secrete metalloproteinases. The first three of which are key in driving inflammation in rheumatoid arthritis. These activated lymphocytes also stimulate B cells to produce immunoglobulins, including rheumatoid factor. Their pathogenic role is unknown, but may be due to
545:
While in most cases activation is dependent on TCR recognition of antigen, alternative pathways for activation have been described. For example, cytotoxic T cells have been shown to become activated when targeted by other CD8 T cells leading to tolerization of the latter.
882:
Helper T cells/CD4+ •express CD4 glycoproteins on their cell surface, which activate in the presence of peptide antigens on the surface of invading pathogens; •respond immediately to protect the immune system; •secrete different cytokine proteins according to the immune
119:
molecules, and brought to the surface of the cell by the class I MHC molecule, where they can be recognized by the T cell. If the TCR is specific for that antigen, it binds to the complex of the class I MHC molecule and the antigen, and the T cell destroys the cell.
686:(HBV) infection, cytotoxic T cells kill infected cells and produce antiviral cytokines capable of purging HBV from viable hepatocytes. They also play an important pathogenic role, contributing to nearly all of the liver injury associated with HBV infection.
179:
which can bind to a different antigen. Some receptors bind to tissues in the human body itself, so to prevent the body from attacking itself, those self-reactive white blood cells are destroyed during further development in the
235:(IP-10, lymphotactin), trigger cytolysis of target cells (perforins, granzymes...), and interact with other cells, such as epithelial cells, monocytes, dendritic cells, neutrophils and B cells. In some infections, such as
601:(programmed cell death). This is called a "lethal hit” and allows to observe a wave-like death of the target cells. Due to high lipid order and negatively charged phosphatidylserine present in their plasma membrane, T
761:. However, CD8+ cells have been shown to play an effector role, responsible for the ultimate destruction of islet beta cells. However, in studies with NOD mice carrying a null mutation at the
253:
and are therefore termed "double-positive" (DP) T cells (CD4CD8). The double-positive T cells are exposed to a wide variety of self-antigens in the thymus and undergo two selection criteria:
750:
infect, in order to further evade destruction by CD8 T cells. If CD8 T cells cannot find, recognize and bind to infected cells, the virus will not be destroyed and will continue to grow.
1581:
Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, et al. (November 2003). "Control of effector CD8+ T cell function by the transcription factor
Eomesodermin".
150:
cells once they become activated and are generally classified as having a pre-defined cytotoxic role within the immune system. However, CD8 T cells also have the ability to make some
431:
The activation of cytotoxic T cells is dependent on several simultaneous interactions between molecules expressed on the surface of the T cell and molecules on the surface of the
779:(cytotoxic T-lymphocytes attack the new organ after detecting it as foreign, due to HLA variation between donor and recipient); in excessive cytokine production in severe
2465:
239:, there is a clonal expansion of peripheral γδ T cells that have specific TCRs, indicating the adaptive nature of the immune response mediated by these cells.
561:
factor for T cells. This increases the number of cells specific for the target antigen that can then travel throughout the body in search of antigen-positive
702:
levels facilitated the recruitment of intrahepatic CXCR5+CD8+T cells and, these types of cells produced high levels of HBV-specific interferon (IFN)-γ and
690:
have been shown to facilitate the accumulation of virus-specific cytotoxic T cells into the infected liver. In some studies with mice, the injection with
626:(Apo1)(CD95) molecules expressed on the target cell. However, this Fas-Fas ligand interaction is thought to be more important to the disposal of unwanted
2749:
830:
1285:
Bennett SR, Carbone FR, Karamalis F, Flavell RA, Miller JF, Heath WR (June 1998). "Help for cytotoxic-T-cell responses is mediated by CD40 signalling".
765:(B2M) locus and thus lacking major histocompatibility complex class I molecules and CD8+ T cells, it was found that they did not develop diabetes.
1187:
2010:
Wang B, Gonzalez A, Benoist C, Mathis D (August 1996). "The role of CD8+ T cells in the initiation of insulin-dependent diabetes mellitus".
772:(CIPN). Mice without CD8 T cells show prolonged CIPN compared to normal mice and injection of educated CD8 T cells resolve or prevent CIPN.
769:
757:. Studies in a diabetic mouse model showed that CD4+ cells are responsible for the massive infiltration of mononuclear leukocytes into
382:
The threshold for activation of these cells is very high, and the process can occur via two pathways: thymus-independent (by infected
190:
TCRs have two parts, usually an alpha and a beta chain. (Some TCRs have a gamma and a delta chain. They are inherent to act against
2153:
Wiebe C, Nickerson PW (February 2020). "Human leukocyte antigen molecular mismatch to risk stratify kidney transplant recipients".
529:
A simple activation of naive CD8 T cells requires the interaction with professional antigen-presenting cells, mainly with matured
96:
cells, cells that are infected by intracellular pathogens (such as viruses or bacteria), or cells that are damaged in other ways.
2742:
374:
components then travel along the microtubule cytoskeleton (green) to the contact site and are secreted, thus killing the target.
2294:
Neumann H, Medana IM, Bauer J, Lassmann H (June 2002). "Cytotoxic T lymphocytes in autoimmune and degenerative CNS diseases".
2379:
1511:
1163:
1076:
867:
1896:
Carvalheiro H, da Silva JA, Souto-Carneiro MM (January 2013). "Potential roles for CD8(+) T cells in rheumatoid arthritis".
930:
Kabelitz D, Wesch D (2003). "Features and functions of gamma delta T lymphocytes: focus on chemokines and their receptors".
295:
2598:
1105:"Concurrent interaction of DCs with CD4(+) and CD8(+) T cells improves secondary CTL expansion: It takes three to tango"
2735:
2346:
2352:
223:
express alpha-beta TCRs (αβ T cells), but some T cells in epithelial tissues (like the gut) express gamma-delta TCRs (
1986:
356:
787:
are generated, damaging the subject); inflammatory and degenerative diseases of the central nervous system, such as
537:
and to allow repetitive stimulation of cytotoxic T cells, dendritic cells have to interact with both, activated CD4
3081:
2634:
1681:
Iannacone M, Sitia G, Guidotti LG (2006). "Pathogenetic and antiviral immune responses against hepatitis B virus".
271:
146:
cell and the target cell bound closely together during antigen-specific activation. CD8 T cells are recognized as T
775:
Cytotoxic T-lymphocytes have been implicated in the development of various diseases and disorders, for example in
3076:
3019:
2557:
1759:"CXCL13-mediated recruitment of intrahepatic CXCR5CD8 T cells favors viral control in chronic HBV infection"
2679:
2649:
2629:
2055:"Cisplatin educates CD8+ T cells to prevent and resolve chemotherapy-induced peripheral neuropathy in mice"
1201:
Hivroz C, Chemin K, Tourret M, Bohineust A (2012). "Crosstalk between T lymphocytes and dendritic cells".
1103:
Hoyer S, Prommersberger S, Pfeiffer IA, Schuler-Thurner B, Schuler G, Dörrie J, Schaft N (December 2014).
1016:"Characterization of Adaptive-like γδ T Cells in Ugandan Infants during Primary Cytomegalovirus Infection"
2667:
2662:
2502:
2198:"Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology"
2104:"CD8+ T Cells and Endogenous IL-10 Are Required for Resolution of Chemotherapy-Induced Neuropathic Pain"
2905:
644:
558:
131:, which binds to the constant portion of the class I MHC molecule. Therefore, these T cells are called
2247:"COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome?"
2245:
Sarzi-Puttini P, Giorgi V, Sirotti S, Marotto D, Ardizzone S, Rizzardini G, et al. (March 2020).
2372:
820:
784:
2727:
1708:
Iannacone M, Sitia G, Isogawa M, Marchese P, Castro MG, Lowenstein PR, et al. (November 2005).
1387:"Preparing the lethal hit: interplay between exo- and endocytic pathways in cytotoxic T lymphocytes"
1338:"CTLs respond with activation and granule secretion when serving as targets for T-cell recognition"
1014:
Tuengel J, Ranchal S, Maslova A, Aulakh G, Papadopoulou M, Drissler S, et al. (October 2021).
754:
334:
195:
1385:
Chang HF, Bzeih H, Chitirala P, Ravichandran K, Sleiman M, Krause E, et al. (February 2017).
3085:
2843:
2706:
722:
432:
383:
215:
different T cells with different TCRs, helping the body's immune system respond to virtually any
139:
1434:
Rudd-Schmidt JA, Hodel AW, Noori T, Lopez JA, Cho HJ, Verschoor S, et al. (November 2019).
3107:
2825:
2761:
2549:
967:"Ligand recognition by the γδ TCR and discrimination between homeostasis and stress conditions"
2102:
Krukowski K, Eijkelkamp N, Laumet G, Hack CE, Li Y, Dougherty PM, et al. (October 2016).
1934:
Gulzar N, Copeland KF (January 2004). "CD8+ T-cells: function and response to HIV infection".
123:
In order for the TCR to bind to the class I MHC molecule, the former must be accompanied by a
3054:
2910:
2888:
2588:
2565:
2541:
1336:
Milstein O, Hagin D, Lask A, Reich-Zeliger S, Shezen E, Ophir E, et al. (January 2011).
734:
589:. Through the action of perforin, granzymes enter the cytoplasm of the target cell and their
338:
236:
31:
17:
1634:"Type B coxsackieviruses and their interactions with the innate and adaptive immune systems"
515:
for T cell activation. This second signal can be assisted (or replaced) by stimulating the T
3183:
3071:
3004:
2915:
2839:
2612:
2429:
2365:
1590:
1447:
1294:
1249:
776:
762:
726:
8:
3188:
3131:
3066:
3049:
2883:
2801:
2621:
2460:
326:
207:
1594:
1451:
1298:
1253:
369:
2951:
2593:
2580:
2319:
2276:
2263:
2246:
2222:
2197:
2178:
2130:
2103:
2079:
2054:
2035:
1847:
1834:
1807:
1788:
1734:
1709:
1658:
1633:
1614:
1555:
1528:
1468:
1435:
1411:
1386:
1362:
1337:
1318:
1181:
1134:
1042:
1015:
991:
966:
873:
859:
788:
424:. These result in peptide fragments, some of which are presented by MHC Class I to the
421:
224:
2307:
1978:
943:
795:, attacking healthy cells and recruiting more immune cells, aggravating the disease).
2999:
2765:
2644:
2525:
2444:
2424:
2311:
2280:
2268:
2227:
2182:
2170:
2135:
2084:
2027:
1992:
1982:
1969:
Tsai S, Shameli A, Santamaria P (2008). "Chapter 4 CD8+ T Cells in Type 1 Diabetes".
1951:
1913:
1878:
1839:
1792:
1780:
1739:
1663:
1606:
1560:
1507:
1473:
1416:
1367:
1310:
1267:
1218:
1214:
1169:
1159:
1126:
1082:
1072:
1047:
996:
947:
912:
877:
863:
758:
738:
714:
683:
298:-presented antigen (CD4). It is the CD8 T-cells that will mature and go on to become
170:
2323:
2070:
2039:
1851:
1618:
3178:
3119:
3059:
3031:
3026:
2994:
2981:
2971:
2517:
2303:
2258:
2217:
2209:
2162:
2125:
2120:
2115:
2074:
2066:
2019:
1974:
1943:
1905:
1829:
1819:
1770:
1729:
1721:
1690:
1653:
1645:
1598:
1550:
1540:
1499:
1463:
1455:
1406:
1398:
1357:
1349:
1322:
1302:
1257:
1210:
1138:
1116:
1037:
1027:
986:
978:
939:
904:
855:
191:
159:
89:
41:
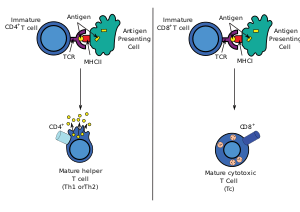
Antigen presentation stimulates T cells to become either "cytotoxic" CD8+ cells or
115:, viruses, bacteria or intracellular signals. Antigens inside a cell are bound to
3014:
2509:
2166:
1909:
1503:
1353:
590:
542:
receptor on the antigen-presenting cell during immunological synapse formation.
530:
468:
425:
211:
108:
100:
3041:
2986:
2927:
2847:
2820:
1824:
1775:
1758:
1529:"Platelet-derived TLT-1 promotes tumor progression by suppressing CD8+ T cells"
1459:
812:
703:
538:
534:
511:
402:
2213:
1402:
1086:
982:
908:
608:
A second way to induce apoptosis is via cell-surface interaction between the T
605:
cells are resistant to the effects of their perforin and granzyme cytotoxins.
3172:
3009:
2898:
2716:
2698:
2536:
2498:
2419:
1947:
1694:
1173:
825:
730:
713:. The main involvement of rheumatoid arthritis is its joint involvement. The
627:
554:
520:
414:
391:
387:
42:
1602:
2976:
2961:
2956:
2893:
2791:
2531:
2439:
2434:
2315:
2272:
2231:
2174:
2139:
2088:
2023:
1996:
1955:
1917:
1882:
1866:
1843:
1784:
1743:
1667:
1610:
1564:
1477:
1420:
1371:
1222:
1130:
1066:
1051:
1000:
951:
916:
655:
622:
562:
410:
406:
286:
124:
112:
2031:
1808:"Antibody-dependent and -independent mechanisms of inflammatory arthritis"
1314:
1271:
1153:
1121:
1104:
1102:
2711:
2414:
2409:
1545:
718:
597:
cascade, which is a series of cysteine proteases that eventually lead to
474:
250:
199:
116:
783:
infection (due to an exaggerated lymphocyte response, a large amount of
3099:
2852:
2758:
2388:
1649:
895:
Venturi S, Venturi M (September 2009). "Iodine, thymus, and immunity".
780:
617:
586:
37:
2053:
Laumet G, Edralin JD, Dantzer R, Heijnen CJ, Kavelaars A (June 2019).
1032:
3157:
2862:
2757:
2684:
2639:
2487:
1757:
Li Y, Tang L, Guo L, Chen C, Gu S, Zhou Y, et al. (March 2020).
1436:"Lipid order and charge protect killer T cells from accidental death"
710:
687:
598:
582:
282:
232:
679:
infections, cytotoxic T cells are mostly effective against viruses.
3147:
2932:
2920:
2878:
2832:
2796:
2453:
2404:
1725:
806:
676:
668:
648:
578:
417:
228:
151:
1306:
1262:
1237:
1194:
1071:(Cuarta edición ed.). Ciudad de México: Médica Panamericana.
706:, which can help to improve the control of chronic HBV infection.
484:
coreceptor and the class I MHC molecule to stabilize this signal.
155:
3152:
2966:
2815:
2786:
2672:
2244:
1895:
852:
Advanced
Hematology in Integrated Cardiovascular Chinese Medicine
640:
594:
277:
220:
216:
104:
3124:
3112:
2857:
2808:
2475:
2396:
2341:
1710:"Platelets mediate cytotoxic T lymphocyte-induced liver damage"
1494:
Bakshi RK, Cox MA, Zajac AJ (2014). "Cytotoxic T Lymphocytes".
1384:
792:
699:
203:
184:
180:
93:
85:
2357:
1335:
1284:
630:
during their development or to the lytic activity of certain T
553:
cell undergoes clonal expansion with the help of the cytokine
302:
following their activation with a class I-restricted antigen.
2101:
1707:
709:
Cytotoxic T cells have been implicated in the progression of
695:
691:
672:
285:
because they could otherwise become autoreactive, leading to
1200:
2052:
1580:
1433:
1013:
503:
499:
492:
2293:
1973:. Advances in Immunology. Vol. 100. pp. 79–124.
1098:
1096:
2009:
746:
481:
247:
243:
128:
1864:
791:(T cells become sensitised to certain proteins, such as
1631:
1093:
573:
When exposed to infected/dysfunctional somatic cells, T
242:
T cells with functionally stable TCRs express both the
1865:
Cope AP, Schulze-Koops H, Aringer M (September 2007).
1680:
1632:
Kemball CC, Alirezaei M, Whitton JL (September 2010).
843:
616:
is activated it starts to express the surface protein
27:
T cell that kills infected, damaged or cancerous cells
2195:
1968:
1867:"The central role of T cells in rheumatoid arthritis"
107:. An antigen is a molecule capable of stimulating an
802:
174:
Development of single positive T cells in the thymus
1151:
831:
List of distinct cell types in the adult human body
266:, in which those double-positive T cells that bind
3170:
1489:
1487:
662:
634:cells than it is to the cytolytic activity of T
187:is necessary for its development and activity.
2152:
1971:Immunopathogenesis of Type 1 Diabetes Mellitus
1933:
1493:
1064:
894:
745:CD8 T cells have been found to play a role in
2743:
2373:
1805:
1756:
1484:
929:
162:, with antitumour and antimicrobial effects.
1625:
1329:
1235:
753:Furthermore, CD8 T cells may be involved in
337:. There might be a discussion about this on
142:between CD8 and the MHC molecule keeps the T
1229:
964:
888:
694:CD8+T cells show a significant decrease of
2750:
2736:
2380:
2366:
1186:: CS1 maint: location missing publisher (
770:chemotherapy-induced peripheral neuropathy
480:There is a second interaction between the
194:and form part of the epithelial barrier).
2262:
2221:
2196:Channappanavar R, Perlman S (July 2017).
2129:
2119:
2078:
1833:
1823:
1774:
1733:
1657:
1554:
1544:
1467:
1410:
1361:
1261:
1120:
1041:
1031:
990:
420:, the cells degrade foreign proteins via
357:Learn how and when to remove this message
2155:Current Opinion in Organ Transplantation
1152:Abbas AK, Lichtman AH, Pillai S (2018).
849:
768:CD8 T cells may be necessary to resolve
620:(FasL)(Apo1L)(CD95L), which can bind to
368:
169:
36:
2620:
413:. When these cells are infected with a
14:
3171:
2251:Clinical and Experimental Rheumatology
1871:Clinical and Experimental Rheumatology
2731:
2361:
1929:
1927:
1576:
1574:
1526:
639:cleavage of death substrates such as
568:
103:(TCRs) that can recognize a specific
1806:Chang MH, Nigrovic PA (March 2019).
1391:Cellular and Molecular Life Sciences
1158:(Ninth ed.). Philadelphia, PA.
965:Deseke M, Prinz I (September 2020).
309:
219:of an invader. The vast majority of
2599:Mucosal associated invariant T cell
971:Cellular & Molecular Immunology
850:Al-Shura AN (2020). "Lymphocytes".
671:, which are effective against both
24:
1962:
1924:
1571:
1527:Tyagi, Tarun; et al. (2023).
1496:Encyclopedia of Medical Immunology
1278:
860:10.1016/b978-0-12-817572-9.00007-0
519:cell with cytokines released from
435:(APC). For instance, consider the
25:
3200:
2353:T-cell Group – Cardiff University
2334:
1155:Cellular and molecular immunology
944:10.1615/CritRevImmunol.v23.i56.10
2635:Lymphokine-activated killer cell
2340:
2264:10.55563/clinexprheumatol/xcdary
1533:Journal of Experimental Medicine
1215:10.1615/CritRevImmunol.v32.i2.30
805:
401:Class I MHC is expressed by all
314:
2387:
2287:
2238:
2189:
2146:
2095:
2071:10.1097/j.pain.0000000000001512
2046:
2003:
1889:
1858:
1799:
1750:
1701:
1674:
1520:
1427:
1378:
612:and the infected cell. When a T
99:Most cytotoxic T cells express
3077:Immunoglobulin class switching
2121:10.1523/JNEUROSCI.3708-15.2016
2012:European Journal of Immunology
1203:Critical Reviews in Immunology
1145:
1109:European Journal of Immunology
1058:
1007:
958:
932:Critical Reviews in Immunology
923:
557:(IL-2), which is a growth and
165:
13:
1:
2308:10.1016/S0166-2236(02)02154-9
1979:10.1016/S0065-2776(08)00804-3
1238:"Immunology. Licence to kill"
836:
577:cells release the cytotoxins
305:
2680:Type 3 innate lymphoid cells
2668:Type 2 innate lymphoid cells
2663:Type 1 innate lymphoid cells
2650:Uterine natural killer cells
2630:Cytokine-induced killer cell
2167:10.1097/MOT.0000000000000714
1910:10.1016/j.autrev.2012.07.011
1504:10.1007/978-0-387-84828-0_36
1354:10.1182/blood-2010-05-283770
1236:Lanzavecchia A (June 1998).
854:. Elsevier. pp. 41–46.
735:tumour necrosis factor alpha
663:Role in disease pathogenesis
643:, lamin B1, lamin B2, PARP (
7:
2202:Seminars in Immunopathology
2108:The Journal of Neuroscience
798:
509:CD80 and CD86 are known as
506:(also called B7-1 and B7-2)
10:
3205:
2906:Polyclonal B cell response
1825:10.1172/jci.insight.125278
1776:10.1016/j.jhep.2019.09.031
1460:10.1038/s41467-019-13385-x
785:pro-inflammatory cytokines
645:poly ADP ribose polymerase
533:. To generate longlasting
386:) or thymus-dependent (by
231:(IFN-γ, TNF-α, IL-17) and
29:
3140:
3098:
3040:
2941:
2871:
2779:
2772:
2697:
2658:
2611:
2579:
2483:
2474:
2395:
2214:10.1007/s00281-017-0629-x
1403:10.1007/s00018-016-2350-7
1065:Rojas-Espinosa O (2017).
983:10.1038/s41423-020-0503-y
909:10.1016/j.nut.2009.06.002
821:CTL-mediated cytotoxicity
654:The transcription factor
111:and is often produced by
1948:10.2174/1570162043485077
1695:10.2217/17460794.1.2.189
1068:Inmunología (de memoria)
723:antigen-presenting cells
196:Hematopoietic stem cells
30:Not to be confused with
2707:Hematopoietic stem cell
2466:Lymphoplasmacytoid cell
2296:Trends in Neurosciences
1603:10.1126/science.1090148
698:. Also, an increase of
433:antigen-presenting cell
426:T cell antigen receptor
3020:Tolerance in pregnancy
2762:adaptive immune system
2024:10.1002/eji.1830260815
593:function triggers the
495:molecule on the T cell
428:(TCR) on CD8 T cells.
405:cells, except for non-
375:
175:
62:cytotoxic T lymphocyte
46:
3055:Somatic hypermutation
2889:Polyclonal antibodies
2884:Monoclonal antibodies
2613:Innate lymphoid cells
2589:Natural killer T cell
1877:(5 Suppl 46): S4-11.
1763:Journal of Hepatology
1440:Nature Communications
1122:10.1002/eji.201444477
549:Once activated, the T
372:
237:human cytomegalovirus
206:, where they undergo
173:
40:
32:Natural killer T cell
3072:Junctional diversity
2840:Antigen presentation
2349:at Wikimedia Commons
1936:Current HIV Research
1898:Autoimmunity Reviews
1546:10.1084/jem.20212218
1498:. pp. 332–342.
777:transplant rejection
763:beta-2 microglobulin
717:is characterised by
327:confusing or unclear
210:of their beta-chain
3067:V(D)J recombination
3050:Affinity maturation
2802:Antigenic variation
2581:Innate-like T cells
2461:Transitional B cell
2114:(43): 11074–11083.
1638:Future Microbiology
1595:2003Sci...302.1041P
1589:(5647): 1041–1043.
1452:2019NatCo..10.5396R
1299:1998Natur.393..478B
1254:1998Natur.393..413L
335:clarify the section
225:gamma delta T cells
208:V(D)J recombination
43:"helper" CD4+ cells
1650:10.2217/fmb.10.101
789:multiple sclerosis
569:Effector functions
422:antigen processing
376:
264:negative selection
258:positive selection
176:
47:
3166:
3165:
3094:
3093:
2844:professional APCs
2725:
2724:
2693:
2692:
2607:
2606:
2347:Cytotoxic T cells
2345:Media related to
1720:(11): 1167–1169.
1513:978-0-387-84827-3
1293:(6684): 478–480.
1248:(6684): 413–414.
1165:978-0-323-52323-3
1115:(12): 3543–3559.
1078:978-968-7988-28-3
1033:10.3390/v13101987
869:978-0-12-817572-9
759:pancreatic islets
715:synovial membrane
684:hepatitis B virus
527:
526:
445:cell activation.
367:
366:
359:
300:cytotoxic T cells
202:migrate into the
16:(Redirected from
3196:
3060:Clonal selection
3032:Immune privilege
3027:Immunodeficiency
2982:Cross-reactivity
2972:Hypersensitivity
2777:
2776:
2752:
2745:
2738:
2729:
2728:
2645:Adaptive NK cell
2618:
2617:
2481:
2480:
2382:
2375:
2368:
2359:
2358:
2344:
2328:
2327:
2291:
2285:
2284:
2266:
2242:
2236:
2235:
2225:
2193:
2187:
2186:
2150:
2144:
2143:
2133:
2123:
2099:
2093:
2092:
2082:
2065:(6): 1459–1468.
2050:
2044:
2043:
2018:(8): 1762–1769.
2007:
2001:
2000:
1966:
1960:
1959:
1931:
1922:
1921:
1893:
1887:
1886:
1862:
1856:
1855:
1837:
1827:
1803:
1797:
1796:
1778:
1754:
1748:
1747:
1737:
1705:
1699:
1698:
1678:
1672:
1671:
1661:
1644:(9): 1329–1347.
1629:
1623:
1622:
1578:
1569:
1568:
1558:
1548:
1524:
1518:
1517:
1491:
1482:
1481:
1471:
1431:
1425:
1424:
1414:
1382:
1376:
1375:
1365:
1348:(3): 1042–1052.
1333:
1327:
1326:
1282:
1276:
1275:
1265:
1233:
1227:
1226:
1198:
1192:
1191:
1185:
1177:
1149:
1143:
1142:
1124:
1100:
1091:
1090:
1062:
1056:
1055:
1045:
1035:
1011:
1005:
1004:
994:
962:
956:
955:
938:(5–6): 339–370.
927:
921:
920:
892:
886:
885:
847:
815:
810:
809:
448:
447:
438:two signal model
362:
355:
351:
348:
342:
318:
317:
310:
101:T-cell receptors
90:white blood cell
74:cytolytic T cell
51:cytotoxic T cell
21:
3204:
3203:
3199:
3198:
3197:
3195:
3194:
3193:
3169:
3168:
3167:
3162:
3136:
3090:
3036:
3015:Clonal deletion
2943:
2937:
2867:
2768:
2756:
2726:
2721:
2689:
2654:
2603:
2575:
2569:
2561:
2553:
2545:
2521:
2513:
2506:
2470:
2448:
2391:
2386:
2337:
2332:
2331:
2292:
2288:
2243:
2239:
2194:
2190:
2151:
2147:
2100:
2096:
2051:
2047:
2008:
2004:
1989:
1967:
1963:
1932:
1925:
1894:
1890:
1863:
1859:
1804:
1800:
1755:
1751:
1714:Nature Medicine
1706:
1702:
1683:Future Virology
1679:
1675:
1630:
1626:
1579:
1572:
1525:
1521:
1514:
1492:
1485:
1432:
1428:
1383:
1379:
1334:
1330:
1283:
1279:
1234:
1230:
1199:
1195:
1179:
1178:
1166:
1150:
1146:
1101:
1094:
1079:
1063:
1059:
1012:
1008:
963:
959:
928:
924:
893:
889:
870:
848:
844:
839:
811:
804:
801:
755:Type 1 diabetes
665:
637:
633:
615:
611:
604:
591:serine protease
576:
571:
559:differentiation
552:
531:dendritic cells
518:
444:
363:
352:
346:
343:
332:
319:
315:
308:
168:
149:
145:
109:immune response
58:
53:(also known as
35:
28:
23:
22:
15:
12:
11:
5:
3202:
3192:
3191:
3186:
3181:
3164:
3163:
3161:
3160:
3155:
3150:
3144:
3142:
3138:
3137:
3135:
3134:
3129:
3128:
3127:
3117:
3116:
3115:
3104:
3102:
3096:
3095:
3092:
3091:
3089:
3088:
3079:
3074:
3069:
3064:
3063:
3062:
3057:
3046:
3044:
3042:Immunogenetics
3038:
3037:
3035:
3034:
3029:
3024:
3023:
3022:
3017:
3012:
3007:
3002:
2990:
2989:
2987:Co-stimulation
2984:
2979:
2974:
2969:
2964:
2959:
2954:
2947:
2945:
2939:
2938:
2936:
2935:
2930:
2928:Immune complex
2924:
2923:
2918:
2913:
2908:
2903:
2902:
2901:
2896:
2891:
2886:
2875:
2873:
2869:
2868:
2866:
2865:
2860:
2855:
2850:
2848:Dendritic cell
2836:
2835:
2830:
2829:
2828:
2826:Conformational
2823:
2812:
2811:
2806:
2805:
2804:
2799:
2794:
2783:
2781:
2774:
2770:
2769:
2755:
2754:
2747:
2740:
2732:
2723:
2722:
2720:
2719:
2714:
2709:
2703:
2701:
2695:
2694:
2691:
2690:
2688:
2687:
2682:
2677:
2676:
2675:
2665:
2659:
2656:
2655:
2653:
2652:
2647:
2642:
2637:
2632:
2626:
2624:
2615:
2609:
2608:
2605:
2604:
2602:
2601:
2596:
2591:
2585:
2583:
2577:
2576:
2574:
2573:
2572:
2571:
2567:
2563:
2559:
2555:
2551:
2547:
2543:
2534:
2529:
2519:
2511:
2504:
2496:
2490:
2484:
2478:
2472:
2471:
2469:
2468:
2463:
2458:
2457:
2456:
2446:
2442:
2437:
2432:
2427:
2422:
2417:
2412:
2407:
2401:
2399:
2393:
2392:
2385:
2384:
2377:
2370:
2362:
2356:
2355:
2350:
2336:
2335:External links
2333:
2330:
2329:
2302:(6): 313–319.
2286:
2257:(2): 337–342.
2237:
2208:(5): 529–539.
2188:
2145:
2094:
2045:
2002:
1987:
1961:
1923:
1904:(3): 401–409.
1888:
1857:
1818:(5): e125278.
1798:
1769:(3): 420–430.
1749:
1726:10.1038/nm1317
1700:
1673:
1624:
1570:
1519:
1512:
1483:
1426:
1397:(3): 399–408.
1377:
1328:
1277:
1228:
1209:(2): 139–155.
1193:
1164:
1144:
1092:
1077:
1057:
1006:
977:(9): 914–924.
957:
922:
903:(9): 977–979.
887:
868:
841:
840:
838:
835:
834:
833:
828:
823:
817:
816:
813:Biology portal
800:
797:
664:
661:
635:
631:
613:
609:
602:
574:
570:
567:
550:
539:helper T cells
535:memory T cells
525:
524:
521:T helper cells
516:
507:
496:
490:
486:
485:
478:
473:peptide-bound
471:
466:
462:
461:
458:
455:
452:
442:
409:ones, such as
365:
364:
322:
320:
313:
307:
304:
291:
290:
261:
167:
164:
147:
143:
56:
26:
9:
6:
4:
3:
2:
3201:
3190:
3187:
3185:
3182:
3180:
3177:
3176:
3174:
3159:
3156:
3154:
3151:
3149:
3146:
3145:
3143:
3139:
3133:
3130:
3126:
3123:
3122:
3121:
3118:
3114:
3111:
3110:
3109:
3106:
3105:
3103:
3101:
3097:
3087:
3083:
3080:
3078:
3075:
3073:
3070:
3068:
3065:
3061:
3058:
3056:
3053:
3052:
3051:
3048:
3047:
3045:
3043:
3039:
3033:
3030:
3028:
3025:
3021:
3018:
3016:
3013:
3011:
3010:Clonal anergy
3008:
3006:
3003:
3001:
2998:
2997:
2996:
2992:
2991:
2988:
2985:
2983:
2980:
2978:
2975:
2973:
2970:
2968:
2965:
2963:
2960:
2958:
2955:
2953:
2949:
2948:
2946:
2940:
2934:
2931:
2929:
2926:
2925:
2922:
2919:
2917:
2914:
2912:
2909:
2907:
2904:
2900:
2899:Microantibody
2897:
2895:
2892:
2890:
2887:
2885:
2882:
2881:
2880:
2877:
2876:
2874:
2870:
2864:
2861:
2859:
2856:
2854:
2851:
2849:
2845:
2841:
2838:
2837:
2834:
2831:
2827:
2824:
2822:
2819:
2818:
2817:
2814:
2813:
2810:
2807:
2803:
2800:
2798:
2795:
2793:
2790:
2789:
2788:
2785:
2784:
2782:
2778:
2775:
2771:
2767:
2763:
2760:
2753:
2748:
2746:
2741:
2739:
2734:
2733:
2730:
2718:
2717:Prolymphocyte
2715:
2713:
2710:
2708:
2705:
2704:
2702:
2700:
2699:Lymphopoiesis
2696:
2686:
2683:
2681:
2678:
2674:
2671:
2670:
2669:
2666:
2664:
2661:
2660:
2657:
2651:
2648:
2646:
2643:
2641:
2638:
2636:
2633:
2631:
2628:
2627:
2625:
2623:
2619:
2616:
2614:
2610:
2600:
2597:
2595:
2592:
2590:
2587:
2586:
2584:
2582:
2578:
2570:
2564:
2562:
2556:
2554:
2548:
2546:
2540:
2539:
2538:
2537:Memory T cell
2535:
2533:
2530:
2527:
2523:
2515:
2507:
2500:
2497:
2495:
2494:Cytotoxic CD8
2491:
2489:
2486:
2485:
2482:
2479:
2477:
2473:
2467:
2464:
2462:
2459:
2455:
2452:
2451:
2450:
2443:
2441:
2438:
2436:
2433:
2431:
2430:Marginal zone
2428:
2426:
2423:
2421:
2418:
2416:
2413:
2411:
2408:
2406:
2403:
2402:
2400:
2398:
2394:
2390:
2383:
2378:
2376:
2371:
2369:
2364:
2363:
2360:
2354:
2351:
2348:
2343:
2339:
2338:
2325:
2321:
2317:
2313:
2309:
2305:
2301:
2297:
2290:
2282:
2278:
2274:
2270:
2265:
2260:
2256:
2252:
2248:
2241:
2233:
2229:
2224:
2219:
2215:
2211:
2207:
2203:
2199:
2192:
2184:
2180:
2176:
2172:
2168:
2164:
2160:
2156:
2149:
2141:
2137:
2132:
2127:
2122:
2117:
2113:
2109:
2105:
2098:
2090:
2086:
2081:
2076:
2072:
2068:
2064:
2060:
2056:
2049:
2041:
2037:
2033:
2029:
2025:
2021:
2017:
2013:
2006:
1998:
1994:
1990:
1988:9780123743268
1984:
1980:
1976:
1972:
1965:
1957:
1953:
1949:
1945:
1941:
1937:
1930:
1928:
1919:
1915:
1911:
1907:
1903:
1899:
1892:
1884:
1880:
1876:
1872:
1868:
1861:
1853:
1849:
1845:
1841:
1836:
1831:
1826:
1821:
1817:
1813:
1809:
1802:
1794:
1790:
1786:
1782:
1777:
1772:
1768:
1764:
1760:
1753:
1745:
1741:
1736:
1731:
1727:
1723:
1719:
1715:
1711:
1704:
1696:
1692:
1689:(2): 189–96.
1688:
1684:
1677:
1669:
1665:
1660:
1655:
1651:
1647:
1643:
1639:
1635:
1628:
1620:
1616:
1612:
1608:
1604:
1600:
1596:
1592:
1588:
1584:
1577:
1575:
1566:
1562:
1557:
1552:
1547:
1542:
1538:
1534:
1530:
1523:
1515:
1509:
1505:
1501:
1497:
1490:
1488:
1479:
1475:
1470:
1465:
1461:
1457:
1453:
1449:
1445:
1441:
1437:
1430:
1422:
1418:
1413:
1408:
1404:
1400:
1396:
1392:
1388:
1381:
1373:
1369:
1364:
1359:
1355:
1351:
1347:
1343:
1339:
1332:
1324:
1320:
1316:
1312:
1308:
1307:10.1038/30996
1304:
1300:
1296:
1292:
1288:
1281:
1273:
1269:
1264:
1263:10.1038/30845
1259:
1255:
1251:
1247:
1243:
1239:
1232:
1224:
1220:
1216:
1212:
1208:
1204:
1197:
1189:
1183:
1175:
1171:
1167:
1161:
1157:
1156:
1148:
1140:
1136:
1132:
1128:
1123:
1118:
1114:
1110:
1106:
1099:
1097:
1088:
1084:
1080:
1074:
1070:
1069:
1061:
1053:
1049:
1044:
1039:
1034:
1029:
1025:
1021:
1017:
1010:
1002:
998:
993:
988:
984:
980:
976:
972:
968:
961:
953:
949:
945:
941:
937:
933:
926:
918:
914:
910:
906:
902:
898:
891:
884:
879:
875:
871:
865:
861:
857:
853:
846:
842:
832:
829:
827:
824:
822:
819:
818:
814:
808:
803:
796:
794:
790:
786:
782:
778:
773:
771:
766:
764:
760:
756:
751:
748:
743:
740:
736:
732:
731:interleukin-6
728:
727:interleukin-1
724:
720:
716:
712:
707:
705:
701:
697:
693:
689:
685:
680:
678:
674:
670:
660:
659:CD8 T cells.
657:
652:
650:
646:
642:
629:
628:T lymphocytes
625:
624:
619:
606:
600:
596:
592:
588:
584:
580:
566:
564:
563:somatic cells
560:
556:
555:interleukin 2
547:
543:
540:
536:
532:
522:
514:
513:
512:costimulators
508:
505:
501:
497:
494:
491:
489:Second Signal
488:
487:
483:
479:
476:
472:
470:
467:
464:
463:
459:
456:
453:
450:
449:
446:
440:
439:
434:
429:
427:
423:
419:
416:
415:intracellular
412:
408:
404:
399:
395:
393:
389:
385:
380:
371:
361:
358:
350:
340:
339:the talk page
336:
330:
328:
323:This section
321:
312:
311:
303:
301:
297:
288:
284:
280:
279:
273:
269:
265:
262:
259:
256:
255:
254:
252:
249:
245:
240:
238:
234:
230:
226:
222:
218:
213:
209:
205:
201:
197:
193:
188:
186:
182:
172:
163:
161:
157:
153:
141:
136:
134:
130:
126:
121:
118:
114:
110:
106:
102:
97:
95:
92:) that kills
91:
87:
83:
82:killer T cell
79:
75:
71:
70:T-killer cell
67:
63:
59:
52:
44:
39:
33:
19:
2977:Inflammation
2962:Alloimmunity
2957:Autoimmunity
2942:Immunity vs.
2894:Autoantibody
2792:Superantigen
2493:
2299:
2295:
2289:
2254:
2250:
2240:
2205:
2201:
2191:
2158:
2154:
2148:
2111:
2107:
2097:
2062:
2058:
2048:
2015:
2011:
2005:
1970:
1964:
1942:(1): 23–37.
1939:
1935:
1901:
1897:
1891:
1874:
1870:
1860:
1815:
1811:
1801:
1766:
1762:
1752:
1717:
1713:
1703:
1686:
1682:
1676:
1641:
1637:
1627:
1586:
1582:
1536:
1532:
1522:
1495:
1443:
1439:
1429:
1394:
1390:
1380:
1345:
1341:
1331:
1290:
1286:
1280:
1245:
1241:
1231:
1206:
1202:
1196:
1154:
1147:
1112:
1108:
1067:
1060:
1026:(10): 1987.
1023:
1019:
1009:
974:
970:
960:
935:
931:
925:
900:
896:
890:
881:
851:
845:
774:
767:
752:
744:
708:
681:
666:
656:Eomesodermin
653:
621:
607:
572:
548:
544:
528:
510:
465:First Signal
460:Description
437:
436:
430:
411:erythrocytes
400:
396:
388:CD4+ T cells
381:
377:
353:
344:
333:Please help
324:
299:
296:MHC class II
292:
287:autoimmunity
275:
268:too strongly
267:
263:
257:
251:co-receptors
241:
189:
177:
137:
132:
125:glycoprotein
122:
113:cancer cells
98:
86:T lymphocyte
81:
77:
73:
69:
65:
61:
54:
50:
48:
3184:Human cells
3100:Lymphocytes
2759:Lymphocytic
2712:Lymphoblast
2410:Plasmablast
2389:Lymphocytes
2161:(1): 8–14.
1812:JCI Insight
1446:(1): 5396.
826:CD4 T cells
719:hyperplasia
475:MHC class I
347:August 2023
274:-presented
200:bone marrow
183:, in which
166:Development
133:CD8 T cells
117:class I MHC
88:(a type of
3189:Immunology
3173:Categories
3141:Substances
3005:Peripheral
2993:Inaction:
2872:Antibodies
2853:Macrophage
2766:complement
2526:Regulatory
2499:Helper CD4
2425:Follicular
1087:1022564980
837:References
781:SARS-CoV-2
739:complement
669:antibodies
618:FAS ligand
587:granulysin
329:to readers
306:Activation
233:chemokines
154:, such as
78:CD8 T-cell
3158:Cytolysin
3148:Cytokines
2995:Tolerance
2944:tolerance
2863:Immunogen
2685:LTi cells
2640:Null cell
2488:Thymocyte
2281:214609305
2183:208537995
1793:204702318
1182:cite book
1174:973917896
897:Nutrition
883:response.
878:241913878
711:arthritis
688:Platelets
677:bacterial
599:apoptosis
583:granzymes
407:nucleated
283:apoptosis
229:cytokines
152:cytokines
3108:Cellular
2952:Immunity
2950:Action:
2933:Paratope
2921:Idiotype
2911:Allotype
2879:Antibody
2833:Mimotope
2797:Allergen
2780:Antigens
2773:Lymphoid
2673:Nuocytes
2622:NK cells
2454:B10 cell
2324:12593103
2316:12086750
2273:32202240
2232:28466096
2175:31789952
2140:27798187
2089:30720585
2040:26229701
1997:19111164
1956:15053338
1918:22841983
1883:17977483
1852:73512236
1844:30843881
1785:31610223
1744:16258538
1668:20860480
1619:43479181
1611:14605368
1565:36305874
1478:31776337
1421:27585956
1372:21045195
1223:23216612
1131:25211552
1052:34696417
1001:32709926
952:15030305
917:19647627
799:See also
649:DNA-PKcs
579:perforin
477:molecule
418:pathogen
281:undergo
278:antigens
140:affinity
3179:T cells
3153:Opsonin
3132:NK cell
3120:Humoral
3000:Central
2967:Allergy
2916:Isotype
2816:Epitope
2787:Antigen
2476:T cells
2405:B1 cell
2397:B cells
2223:7079893
2131:5098842
2080:6527475
2032:8765018
1835:6483516
1735:2908083
1659:3045535
1591:Bibcode
1583:Science
1556:9814191
1469:6881447
1448:Bibcode
1412:5241346
1363:3035066
1323:4325396
1315:9624004
1295:Bibcode
1272:9623994
1250:Bibcode
1139:5655814
1043:8537190
1020:Viruses
992:7608190
682:During
667:Unlike
647:), and
641:lamin A
595:caspase
498:either
325:may be
221:T cells
217:protein
198:in the
127:called
105:antigen
84:) is a
3125:B cell
3113:T cell
2858:B cell
2821:Linear
2809:Hapten
2420:Memory
2415:Plasma
2322:
2314:
2279:
2271:
2230:
2220:
2181:
2173:
2138:
2128:
2087:
2077:
2038:
2030:
1995:
1985:
1954:
1916:
1881:
1850:
1842:
1832:
1791:
1783:
1742:
1732:
1666:
1656:
1617:
1609:
1563:
1553:
1510:
1476:
1466:
1419:
1409:
1370:
1360:
1321:
1313:
1287:Nature
1270:
1242:Nature
1221:
1172:
1162:
1137:
1129:
1085:
1075:
1050:
1040:
999:
989:
950:
915:
876:
866:
793:myelin
700:CXCL13
692:CXCR5+
585:, and
454:T cell
451:Signal
204:thymus
192:stress
185:iodine
181:thymus
94:cancer
2532:Naïve
2440:Pre-B
2435:Naïve
2320:S2CID
2277:S2CID
2179:S2CID
2036:S2CID
1848:S2CID
1789:S2CID
1615:S2CID
1539:(1).
1342:Blood
1319:S2CID
1135:S2CID
874:S2CID
704:IL-21
696:HBsAg
673:viral
441:for T
276:self
160:IFN-γ
156:TNF-α
2764:and
2492:αβ (
2449:cell
2312:PMID
2269:PMID
2228:PMID
2171:PMID
2136:PMID
2085:PMID
2059:Pain
2028:PMID
1993:PMID
1983:ISBN
1952:PMID
1914:PMID
1879:PMID
1840:PMID
1781:PMID
1740:PMID
1664:PMID
1607:PMID
1561:PMID
1508:ISBN
1474:PMID
1417:PMID
1368:PMID
1311:PMID
1268:PMID
1219:PMID
1188:link
1170:OCLC
1160:ISBN
1127:PMID
1083:OCLC
1073:ISBN
1048:PMID
997:PMID
948:PMID
913:PMID
864:ISBN
733:and
675:and
504:CD86
500:CD80
493:CD28
403:host
392:IL-2
384:APCs
246:and
158:and
138:The
18:CD8+
3086:HLA
3082:MHC
2447:reg
2304:doi
2259:doi
2218:PMC
2210:doi
2163:doi
2126:PMC
2116:doi
2075:PMC
2067:doi
2063:160
2020:doi
1975:doi
1944:doi
1906:doi
1830:PMC
1820:doi
1771:doi
1730:PMC
1722:doi
1691:doi
1654:PMC
1646:doi
1599:doi
1587:302
1551:PMC
1541:doi
1537:220
1500:doi
1464:PMC
1456:doi
1407:PMC
1399:doi
1358:PMC
1350:doi
1346:117
1303:doi
1291:393
1258:doi
1246:393
1211:doi
1117:doi
1038:PMC
1028:doi
987:PMC
979:doi
940:doi
905:doi
856:doi
747:HIV
623:Fas
502:or
482:CD8
469:TCR
457:APC
294:an
272:MHC
270:to
248:CD8
244:CD4
212:TCR
129:CD8
80:or
66:CTL
3175::
2846::
2594:γδ
2568:VM
2560:RM
2552:EM
2544:CM
2524:/
2522:17
2516:/
2508:/
2505:FH
2501:/
2318:.
2310:.
2300:25
2298:.
2275:.
2267:.
2255:38
2253:.
2249:.
2226:.
2216:.
2206:39
2204:.
2200:.
2177:.
2169:.
2159:25
2157:.
2134:.
2124:.
2112:36
2110:.
2106:.
2083:.
2073:.
2061:.
2057:.
2034:.
2026:.
2016:26
2014:.
1991:.
1981:.
1950:.
1938:.
1926:^
1912:.
1902:12
1900:.
1875:25
1873:.
1869:.
1846:.
1838:.
1828:.
1814:.
1810:.
1787:.
1779:.
1767:72
1765:.
1761:.
1738:.
1728:.
1718:11
1716:.
1712:.
1685:.
1662:.
1652:.
1640:.
1636:.
1613:.
1605:.
1597:.
1585:.
1573:^
1559:.
1549:.
1535:.
1531:.
1506:.
1486:^
1472:.
1462:.
1454:.
1444:10
1442:.
1438:.
1415:.
1405:.
1395:74
1393:.
1389:.
1366:.
1356:.
1344:.
1340:.
1317:.
1309:.
1301:.
1289:.
1266:.
1256:.
1244:.
1240:.
1217:.
1207:32
1205:.
1184:}}
1180:{{
1168:.
1133:.
1125:.
1113:44
1111:.
1107:.
1095:^
1081:.
1046:.
1036:.
1024:13
1022:.
1018:.
995:.
985:.
975:17
973:.
969:.
946:.
936:23
934:.
911:.
901:25
899:.
880:.
872:.
862:.
729:,
581:,
565:.
523:.
394:.
135:.
76:,
72:,
68:,
64:,
60:,
49:A
3084:/
2842:/
2751:e
2744:t
2737:v
2566:T
2558:T
2550:T
2542:T
2528:)
2520:h
2518:T
2514:3
2512:h
2510:T
2503:T
2445:B
2381:e
2374:t
2367:v
2326:.
2306::
2283:.
2261::
2234:.
2212::
2185:.
2165::
2142:.
2118::
2091:.
2069::
2042:.
2022::
1999:.
1977::
1958:.
1946::
1940:2
1920:.
1908::
1885:.
1854:.
1822::
1816:4
1795:.
1773::
1746:.
1724::
1697:.
1693::
1687:1
1670:.
1648::
1642:5
1621:.
1601::
1593::
1567:.
1543::
1516:.
1502::
1480:.
1458::
1450::
1423:.
1401::
1374:.
1352::
1325:.
1305::
1297::
1274:.
1260::
1252::
1225:.
1213::
1190:)
1176:.
1141:.
1119::
1089:.
1054:.
1030::
1003:.
981::
954:.
942::
919:.
907::
858::
636:C
632:H
614:C
610:C
603:C
575:C
551:C
517:C
443:C
360:)
354:(
349:)
345:(
341:.
331:.
289:.
148:C
144:C
57:C
55:T
45:.
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.