692:
42:
298:
that are believed to be conduits for regulated water transport. Mitochondrial matrix has a pH of about 7.8, which is higher than the pH of the intermembrane space of the mitochondria, which is around 7.0–7.4. Mitochondrial DNA was discovered by Nash and Margit in 1963. One to many double stranded
909:
The concentration of oxaloacetate in particular is kept low, so any fluctuations in this concentrations serve to drive the citric acid cycle forward. The production of ATP also serves as a means of regulation by acting as an inhibitor for isocitrate dehydrogenase, pyruvate dehydrogenase, the
786:. The energy is used in order to rotate ATP synthase which facilitates the passage of a proton, producing ATP. A pH difference between the matrix and intermembrane space creates an electrochemical gradient by which ATP synthase can pass a proton into the matrix favorably.
695:
ATP synthesis as seen from the perspective of the matrix. Conditions produced by the relationships between the catabolic pathways (citric acid cycle and oxidative phosphorylation) and structural makeup (lipid bilayer and electron transport chain) of matrix facilitate ATP
781:
by the energy of the electrons going through the electron transport chain. Four electrons are finally accepted by oxygen in the matrix to complete the electron transport chain. The protons return to the mitochondrial matrix through the protein
1566:
Denton, Richard M.; Randle, Philip J.; Bridges, Barbara J.; Cooper, Ronald H.; Kerbey, Alan L.; Pask, Helen T.; Severson, David L.; Stansbie, David; Whitehouse, Susan (1975-10-01). "Regulation of mammalian pyruvate dehydrogenase".
290:, compartment has a higher aqueous:protein content of around 3.8 μL/mg protein relative to that occurring in mitochondrial matrix where such levels typically are near 0.8 μL/mg protein. It is not known how mitochondria maintain
922:
The mitochondria contains its own set of DNA used to produce proteins found in the electron transport chain. The mitochondrial DNA only codes for about thirteen proteins that are used in processing mitochondrial transcripts,
1138:
Porcelli, Anna Maria; Ghelli, Anna; Zanna, Claudia; Pinton, Paolo; Rizzuto, Rosario; Rugolo, Michela (2005-01-28). "pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant".
1364:
Anderson, S.; Bankier, A. T.; Barrell, B. G.; de Bruijn, M. H. L.; Coulson, A. R.; Drouin, J.; Eperon, I. C.; Nierlich, D. P.; Roe, B. A. (1981-04-09). "Sequence and organization of the human mitochondrial genome".
822:. After these initial steps the urea cycle continues in the inner membrane space until ornithine once again enters the matrix through a transport channel to continue the first to steps within matrix.
885:
which increases the reaction rate in the cycle. Concentration of intermediates and coenzymes in the matrix also increase or decrease the rate of ATP production due to
761:
are produced in the matrix or transported in through porin and transport proteins in order to undergo oxidation through oxidative phosphorylation. NADH and FADH
200:. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm. The mitochondrial matrix contains the
1531:; Gehring, Heinz; Christen, Philipp (1984-04-15). "Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure".
869:
Regulation within the matrix is primarily controlled by ion concentration, metabolite concentration and energy charge. Availability of ions such as
1743:
598:
that facilitates the production of ATP through the pumping of protons. The gradient also provides control of the concentration of ions such as
2224:
1179:
Dimroth, P.; Kaim, G.; Matthey, U. (2000-01-01). "Crucial role of the membrane potential for ATP synthesis by F(1)F(o) ATP synthases".
1282:
King, John V.; Liang, Wenguang G.; Scherpelz, Kathryn P.; Schilling, Alexander B.; Meredith, Stephen C.; Tang, Wei-Jen (2014-07-08).
2219:
2310:
1892:
1978:
794:
The first two steps of the urea cycle take place within the mitochondrial matrix of liver and kidney cells. In the first step
1348:
1119:
1038:
1787:
1736:
2078:
2013:
1004:
2023:
1848:
268:
2349:
2147:
1782:
803:
515:
391:
1419:
Iuchi, S.; Lin, E. C. C. (1993-07-01). "Adaptation of
Escherichia coli to redox environments by gene expression".
882:
499:
2516:
2344:
2008:
1729:
861:. These amino acids are then used either within the matrix or transported into the cytosol to produce proteins.
2339:
2125:
2083:
906:
503:
1929:
1871:
1761:
657:
Following glycolysis, the citric acid cycle is activated by the production of acetyl-CoA. The oxidation of
197:
108:
53:
243:
The composition of the matrix based on its structures and contents produce an environment that allows the
2541:
1983:
774:
755:
674:
272:
1339:
Alberts, Bruce; Johnson, Alexander; Lewis, julian; Roberts, Keith; Peters, Walter; Raff, Martin (1994).
2157:
2152:
1956:
1822:
1054:
Mitchell, Peter; Moyle, Jennifer (1967-01-14). "Chemiosmotic
Hypothesis of Oxidative Phosphorylation".
1033:. San Francisco: W.H. Freeman. pp. 509–527, 569–579, 614–616, 638–641, 732–735, 739–748, 770–773.
807:
519:
691:
1882:
276:
260:
229:
1472:"Mutations and polymorphisms in the human ornithine transcarbamylase gene: Mutation update addendum"
602:
driven by the mitochondrial membrane potential. The membrane only allows nonpolar molecules such as
2073:
1951:
898:
878:
735:
710:
685:
595:
559:
495:
252:
2120:
1907:
894:
830:
727:
714:
681:
2115:
1772:
874:
527:
483:
460:
280:
221:
98:
482:
Enzymes from processes that take place in the matrix. The citric acid cycle is facilitated by
2536:
2489:
2229:
2105:
1941:
1919:
886:
464:
2330:
2236:
2093:
1988:
1374:
1063:
940:
722:
555:
523:
511:
311:
content, and in humans is maternally derived. Mitochondria of mammals have 55s ribosomes.
8:
811:
799:
778:
468:
299:
mainly circular DNA is present in mitochondrial matrix. Mitochondrial DNA is 1% of total
287:
73:
1378:
1284:"Molecular basis of substrate recognition and degradation by human presequence protease"
1067:
2260:
2028:
1902:
1707:
1652:
1619:
1600:
1509:
1452:
1432:
1398:
1316:
1283:
1087:
1259:
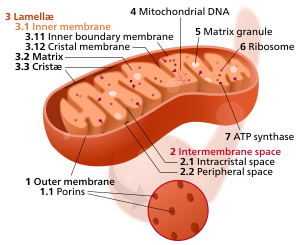
1226:
353:
2250:
2053:
1834:
1699:
1676:
1657:
1639:
1592:
1584:
1548:
1544:
1501:
1493:
1444:
1436:
1390:
1344:
1321:
1303:
1264:
1246:
1204:
1196:
1156:
1115:
1079:
1034:
1000:
957:
924:
870:
625:
329:
264:
256:
225:
201:
158:
1711:
1604:
1456:
531:
2063:
1856:
1691:
1647:
1631:
1576:
1540:
1513:
1483:
1428:
1402:
1382:
1311:
1295:
1254:
1238:
1188:
1148:
1091:
1071:
952:
911:
902:
890:
487:
220:. The enzymes in the matrix facilitate reactions responsible for the production of
1968:
1861:
1528:
936:
633:
629:
63:
1635:
294:
balance across the inner mitochondrial membrane, although the membrane contains
1152:
838:
666:
624:
to enter the matrix. Molecules enter and exit the mitochondrial matrix through
603:
237:
217:
1299:
1112:
Distribution of metabolites between mitochondria and cytosol of perfused liver
742:
through the oxidation of carbons in two cycles. The oxidation of NADH and FADH
2530:
1752:
1643:
1588:
1497:
1440:
1307:
1250:
1200:
962:
928:
910:
electron transport chain protein complexes, and ATP synthase. ADP acts as an
558:
that contains the complexes of oxidative phosphorylation. which contains the
189:
31:
1488:
1471:
636:. These attributed characteristics allow for control over concentrations of
2385:
2360:
2018:
1897:
1810:
1661:
1325:
1268:
1208:
1160:
932:
837:
can be converted into amino acids within the matrix through the process of
834:
783:
658:
567:
535:
418:
395:
376:
357:
177:
22:
1703:
1552:
1527:
Kirsch, Jack F.; Eichele, Gregor; Ford, Geoffrey C.; Vincent, Michael G.;
1505:
1448:
1394:
1083:
1721:
1596:
1192:
802:
through the investment of two ATP molecules. This step is facilitated by
579:
2137:
1799:
1695:
1580:
846:
841:. These reactions are facilitated by transaminases in order to produce
819:
731:
670:
641:
583:
575:
380:
349:
341:
325:
248:
213:
1242:
680:. The production of acetyl-CoA begins the citric acid cycle while the
594:
The electron transport chain is responsible for establishing a pH and
2068:
1386:
1075:
850:
842:
815:
765:
undergo oxidation in the electron transport chain by transferring an
571:
491:
430:
426:
384:
365:
295:
244:
41:
873:
various functions of the citric acid cycle. in the matrix activates
644:
necessary for regulation and determines the rate of ATP production.
148:
2214:
2088:
858:
766:
718:
539:
507:
361:
337:
333:
308:
233:
205:
170:
2374:
2369:
2289:
854:
795:
706:
589:
566:
of the inner membrane and consists of four protein complexes and
345:
304:
291:
1110:
Soboll, S; Scholz, R; Freisl, M; Elbers, R; Heldt, H.W. (1976).
2479:
2474:
2444:
2439:
2319:
2299:
2294:
2284:
2279:
2274:
2269:
1363:
999:. New York City: John Wiley & Sons, Inc. pp. 582–584.
701:
610:
563:
543:
451:
369:
209:
849:
from oxaloacetate. Transamination of α-ketoglutarate produces
2499:
2494:
2484:
2469:
2464:
2459:
2454:
2449:
2434:
2429:
2424:
2419:
2414:
2409:
2404:
2399:
2394:
2190:
2177:
770:
422:
406:
632:. Molecules are then able to leave the mitochondria through
1281:
402:
1338:
1565:
1137:
637:
617:
599:
443:
434:
414:
410:
300:
1526:
1109:
997:
1620:"Mitochondrial Protein Synthesis, Import, and Assembly"
1225:
Karmen, A.; Wroblewski, F.; Ladue, J. S. (1955-01-01).
730:
which is on the inner membrane and is part of protein
1224:
995:
Voet, Donald; Voet, Judith; Pratt, Charlotte (2013).
433:). Additional metabolites present in the matrix are
16:
Space within the inner membrane of the mitochondrion
1141:
661:
by pyruvate dehydrogenase in the matrix produces CO
255:and enzymes in the matrix play a large role in the
1178:
1028:
704:for the citric acid cycle are in the matrix (e.g.
401:. The electron transport chain oxidizes coenzymes
1470:Tuchman, Mendel; Plante, Robert J. (1995-01-01).
2528:
582:(coenzyme Q: cytochrome c oxidoreductase), and
409:. Protein synthesis makes use of mitochondrial
616:and small non charged polar molecules such as
590:Inner membrane control over matrix composition
1737:
1469:
1053:
994:
889:and cataplerotic effects. NADH can act as an
421:. Regulation of processes makes use of ions (
328:involved in processes within the matrix. The
749:
738:. The cycle produces coenzymes NADH and FADH
673:pathway that produces acetyl-CoA, NADH, and
549:
746:produces GTP from succinyl-CoA synthetase.
2225:Mitochondrial permeability transition pore
1751:
1744:
1730:
1674:
534:. Amino acid production is facilitated by
275:through oxidation that will be reduced in
1651:
1487:
1315:
1258:
1029:Stryer, L; Berg, J; Tymoczko, JL (2002).
2220:Mitochondrial membrane transport protein
1418:
690:
324:The matrix is host to a wide variety of
578:(succinate:coenzyme Q oxidoreductase),
538:. Amino acid metabolism is mediated by
2529:
1979:Cholesterol side-chain cleavage enzyme
1227:"Transaminase activity in human blood"
1114:. New york: Elsevier. pp. 29–40.
669:of fatty acids serves as an alternate
1725:
1414:
1412:
1231:The Journal of Clinical Investigation
47:Components of a typical mitochondrion
1343:. New york: Garland Publishing Inc.
1220:
1218:
1174:
1172:
1170:
1133:
1131:
1105:
1103:
1101:
1024:
1022:
1020:
1018:
1016:
990:
988:
986:
984:
982:
980:
978:
917:
652:
1893:Coenzyme Q – cytochrome c reductase
1677:"Protein synthesis in mitochondria"
1617:
1569:Molecular and Cellular Biochemistry
1181:The Journal of Experimental Biology
514:. The urea cycle is facilitated by
251:pathways to proceed favorably. The
13:
2079:Oxoglutarate dehydrogenase complex
2014:Glycerol-3-phosphate dehydrogenase
1433:10.1111/j.1365-2958.1993.tb01664.x
1409:
574:(NADH:coenzyme Q oxidoreductase),
14:
2553:
2024:Carnitine palmitoyltransferase II
1675:Grivell, L.A.; Pel, H.J. (1994).
1332:
1215:
1167:
1128:
1098:
1013:
975:
943:of the electron transport chain.
825:
806:. The second step facilitated by
2148:Carbamoyl phosphate synthetase I
1788:Long-chain-fatty-acid—CoA ligase
1783:Carnitine palmitoyltransferase I
804:carbamoyl phosphate synthetase I
516:carbamoyl phosphate synthetase I
40:
2009:Glutamate aspartate transporter
1668:
1611:
1559:
1520:
1463:
1357:
2126:Pyruvate dehydrogenase complex
2084:Succinyl coenzyme A synthetase
1275:
1047:
777:. Protons are pulled into the
319:
314:
1:
1618:Fox, Thomas D. (2012-12-01).
1341:Molecular Biology of the Cell
968:
883:α-ketoglutarate dehydrogenase
864:
789:
715:α-ketoglutarate dehydrogenase
500:α-ketoglutarate dehydrogenase
238:beta oxidation of fatty acids
1930:Dihydroorotate dehydrogenase
1545:10.1016/0022-2836(84)90333-4
1533:Journal of Molecular Biology
647:
7:
1984:Steroid 11-beta-hydroxylase
1636:10.1534/genetics.112.141267
946:
212:, small organic molecules,
10:
2558:
2158:N-Acetylglutamate synthase
2153:Ornithine transcarbamylase
1957:Glycerol phosphate shuttle
1823:monoamine neurotransmitter
1153:10.1016/j.bbrc.2004.11.105
808:ornithine transcarbamylase
520:ornithine transcarbamylase
477:
2512:
2383:
2358:
2328:
2308:
2258:
2249:
2210:
2203:
2186:
2166:
2134:
2102:
2050:
2041:
1997:
1965:
1938:
1916:
1883:oxidative phosphorylation
1879:
1870:
1847:
1819:
1796:
1769:
1760:
1300:10.1016/j.str.2014.05.003
750:Oxidative phosphorylation
684:produced are used in the
550:Inner membrane components
303:of a cell. It is rich in
277:oxidative phosphorylation
261:oxidative phosphorylation
230:oxidative phosphorylation
39:
30:
21:
2074:Isocitrate dehydrogenase
1952:Malate-aspartate shuttle
899:isocitrate dehydrogenase
879:isocitrate dehydrogenase
736:electron transport chain
711:isocitrate dehydrogenase
686:electron transport chain
665:, acetyl-CoA, and NADH.
596:electrochemical gradient
586:(cytochrome c oxidase).
560:electron transport chain
554:The inner membrane is a
496:isocitrate dehydrogenase
253:electron transport chain
196:is the space within the
2121:Glutamate dehydrogenase
1908:Succinate dehydrogenase
1489:10.1002/humu.1380050404
907:pyruvate dehydrogenase.
728:succinate dehydrogenase
504:succinyl-CoA synthetase
117:Inner boundary membrane
2517:mitochondrial diseases
2116:Aspartate transaminase
1773:fatty acid degradation
1421:Molecular Microbiology
875:pyruvate dehydrogenase
697:
570:. These complexes are
528:acyl-CoA dehydrogenase
484:pyruvate dehydrogenase
2230:Mitochondrial carrier
2106:anaplerotic reactions
1942:mitochondrial shuttle
1920:pyrimidine metabolism
694:
562:that is found on the
2237:Translocator protein
2094:Malate dehydrogenase
1989:Aldosterone synthase
1193:10.1242/jeb.203.1.51
723:malate dehydrogenase
556:phospholipid bilayer
544:presequence protease
524:pyruvate carboxylase
512:malate dehydrogenase
1849:Intermembrane space
1529:Jansonius, Johan N.
1379:1981Natur.290..457A
1068:1967Natur.213..137M
812:carbamoyl phosphate
800:carbamoyl phosphate
779:intermembrane space
522:. β-Oxidation uses
392:carbamoyl phosphate
288:intermembrane space
74:Intermembrane space
2542:Matrices (biology)
2204:Other/to be sorted
2169:alcohol metabolism
2029:Uncoupling protein
1903:NADH dehydrogenase
1696:10.1007/bf00986960
1581:10.1007/BF01731731
925:ribosomal proteins
798:is converted into
698:
626:transport proteins
83:Intracristal space
2524:
2523:
2508:
2507:
2251:Mitochondrial DNA
2245:
2244:
2199:
2198:
2054:citric acid cycle
2037:
2036:
1843:
1842:
1835:Monoamine oxidase
1373:(5806): 457–465.
1350:978-0-8153-3218-3
1243:10.1172/JCI103055
1121:978-0-444-10925-5
1062:(5072): 137–139.
1040:978-0-7167-4684-3
958:Mitochondrial DNA
941:protein complexes
918:Protein synthesis
653:Citric acid cycle
398:
387:
372:
330:citric acid cycle
265:citric acid cycle
257:citric acid cycle
226:citric acid cycle
202:mitochondrial DNA
186:
185:
159:Mitochondrial DNA
2549:
2388:
2363:
2333:
2313:
2263:
2256:
2255:
2208:
2207:
2171:
2141:
2109:
2064:Citrate synthase
2057:
2048:
2047:
2002:
1972:
1945:
1923:
1886:
1877:
1876:
1857:Adenylate kinase
1828:
1804:
1776:
1767:
1766:
1746:
1739:
1732:
1723:
1722:
1716:
1715:
1681:
1672:
1666:
1665:
1655:
1630:(4): 1203–1234.
1615:
1609:
1608:
1563:
1557:
1556:
1524:
1518:
1517:
1491:
1467:
1461:
1460:
1416:
1407:
1406:
1387:10.1038/290457a0
1361:
1355:
1354:
1336:
1330:
1329:
1319:
1279:
1273:
1272:
1262:
1222:
1213:
1212:
1176:
1165:
1164:
1135:
1126:
1125:
1107:
1096:
1095:
1076:10.1038/213137a0
1051:
1045:
1044:
1026:
1011:
1010:
992:
953:Matrix (biology)
937:protein subunits
903:citrate synthase
707:citrate synthase
630:ion transporters
488:citrate synthase
396:
385:
370:
141:
124:Cristal membrane
90:Peripheral space
44:
34:
25:
19:
18:
2557:
2556:
2552:
2551:
2550:
2548:
2547:
2546:
2527:
2526:
2525:
2520:
2504:
2384:
2379:
2359:
2354:
2329:
2324:
2309:
2304:
2259:
2241:
2195:
2182:
2167:
2162:
2135:
2130:
2103:
2098:
2051:
2033:
1998:
1993:
1969:steroidogenesis
1966:
1961:
1939:
1934:
1917:
1912:
1880:
1866:
1862:Creatine kinase
1839:
1825:
1820:
1815:
1797:
1792:
1770:
1756:
1750:
1720:
1719:
1679:
1673:
1669:
1616:
1612:
1564:
1560:
1525:
1521:
1468:
1464:
1417:
1410:
1362:
1358:
1351:
1337:
1333:
1294:(7): 996–1007.
1280:
1276:
1223:
1216:
1187:(Pt 1): 51–59.
1177:
1168:
1136:
1129:
1122:
1108:
1099:
1052:
1048:
1041:
1027:
1014:
1007:
993:
976:
971:
949:
920:
895:α-ketoglutarate
867:
831:α-Ketoglutarate
828:
792:
764:
759:
752:
745:
741:
678:
664:
655:
650:
621:
614:
607:
592:
552:
480:
472:
459:
455:
447:
442:
438:
354:α-ketoglutarate
322:
317:
286:The cytosolic,
232:, oxidation of
216:cofactors, and
182:
181:
173:
166:
161:
151:
142:
136:
125:
118:
111:
101:
91:
84:
76:
66:
56:
32:
23:
17:
12:
11:
5:
2555:
2545:
2544:
2539:
2522:
2521:
2513:
2510:
2509:
2506:
2505:
2503:
2502:
2497:
2492:
2487:
2482:
2477:
2472:
2467:
2462:
2457:
2452:
2447:
2442:
2437:
2432:
2427:
2422:
2417:
2412:
2407:
2402:
2397:
2391:
2389:
2381:
2380:
2378:
2377:
2372:
2366:
2364:
2356:
2355:
2353:
2352:
2347:
2342:
2336:
2334:
2326:
2325:
2323:
2322:
2316:
2314:
2306:
2305:
2303:
2302:
2297:
2292:
2287:
2282:
2277:
2272:
2266:
2264:
2253:
2247:
2246:
2243:
2242:
2240:
2239:
2234:
2233:
2232:
2227:
2217:
2211:
2205:
2201:
2200:
2197:
2196:
2194:
2193:
2187:
2184:
2183:
2181:
2180:
2174:
2172:
2164:
2163:
2161:
2160:
2155:
2150:
2144:
2142:
2132:
2131:
2129:
2128:
2123:
2118:
2112:
2110:
2100:
2099:
2097:
2096:
2091:
2086:
2081:
2076:
2071:
2066:
2060:
2058:
2045:
2039:
2038:
2035:
2034:
2032:
2031:
2026:
2021:
2016:
2011:
2005:
2003:
1995:
1994:
1992:
1991:
1986:
1981:
1975:
1973:
1963:
1962:
1960:
1959:
1954:
1948:
1946:
1936:
1935:
1933:
1932:
1926:
1924:
1914:
1913:
1911:
1910:
1905:
1900:
1895:
1889:
1887:
1874:
1872:Inner membrane
1868:
1867:
1865:
1864:
1859:
1853:
1851:
1845:
1844:
1841:
1840:
1838:
1837:
1831:
1829:
1817:
1816:
1814:
1813:
1807:
1805:
1794:
1793:
1791:
1790:
1785:
1779:
1777:
1764:
1762:Outer membrane
1758:
1757:
1749:
1748:
1741:
1734:
1726:
1718:
1717:
1690:(3): 183–194.
1684:Mol. Biol. Rep
1667:
1610:
1558:
1539:(3): 497–525.
1519:
1482:(4): 293–295.
1476:Human Mutation
1462:
1408:
1356:
1349:
1331:
1274:
1237:(1): 126–131.
1214:
1166:
1147:(4): 799–804.
1127:
1120:
1097:
1046:
1039:
1012:
1006:978-1118129180
1005:
973:
972:
970:
967:
966:
965:
960:
955:
948:
945:
919:
916:
866:
863:
839:transamination
827:
826:Transamination
824:
791:
788:
769:to regenerate
762:
757:
751:
748:
743:
739:
676:
667:Beta oxidation
662:
654:
651:
649:
646:
619:
612:
605:
591:
588:
551:
548:
532:β-ketothiolase
479:
476:
470:
457:
453:
445:
440:
436:
321:
318:
316:
313:
224:, such as the
218:inorganic ions
198:inner membrane
184:
183:
165:Matrix granule
153:
152:
143:
128:
127:
126:
119:
109:Inner membrane
93:
92:
85:
68:
67:
54:Outer membrane
45:
37:
36:
28:
27:
15:
9:
6:
4:
3:
2:
2554:
2543:
2540:
2538:
2535:
2534:
2532:
2519:
2518:
2511:
2501:
2498:
2496:
2493:
2491:
2488:
2486:
2483:
2481:
2478:
2476:
2473:
2471:
2468:
2466:
2463:
2461:
2458:
2456:
2453:
2451:
2448:
2446:
2443:
2441:
2438:
2436:
2433:
2431:
2428:
2426:
2423:
2421:
2418:
2416:
2413:
2411:
2408:
2406:
2403:
2401:
2398:
2396:
2393:
2392:
2390:
2387:
2382:
2376:
2373:
2371:
2368:
2367:
2365:
2362:
2357:
2351:
2348:
2346:
2343:
2341:
2338:
2337:
2335:
2332:
2327:
2321:
2318:
2317:
2315:
2312:
2307:
2301:
2298:
2296:
2293:
2291:
2288:
2286:
2283:
2281:
2278:
2276:
2273:
2271:
2268:
2267:
2265:
2262:
2257:
2254:
2252:
2248:
2238:
2235:
2231:
2228:
2226:
2223:
2222:
2221:
2218:
2216:
2213:
2212:
2209:
2206:
2202:
2192:
2189:
2188:
2185:
2179:
2176:
2175:
2173:
2170:
2165:
2159:
2156:
2154:
2151:
2149:
2146:
2145:
2143:
2140:
2139:
2133:
2127:
2124:
2122:
2119:
2117:
2114:
2113:
2111:
2108:
2107:
2101:
2095:
2092:
2090:
2087:
2085:
2082:
2080:
2077:
2075:
2072:
2070:
2067:
2065:
2062:
2061:
2059:
2056:
2055:
2049:
2046:
2044:
2040:
2030:
2027:
2025:
2022:
2020:
2017:
2015:
2012:
2010:
2007:
2006:
2004:
2001:
1996:
1990:
1987:
1985:
1982:
1980:
1977:
1976:
1974:
1971:
1970:
1964:
1958:
1955:
1953:
1950:
1949:
1947:
1944:
1943:
1937:
1931:
1928:
1927:
1925:
1922:
1921:
1915:
1909:
1906:
1904:
1901:
1899:
1896:
1894:
1891:
1890:
1888:
1885:
1884:
1878:
1875:
1873:
1869:
1863:
1860:
1858:
1855:
1854:
1852:
1850:
1846:
1836:
1833:
1832:
1830:
1827:
1824:
1818:
1812:
1809:
1808:
1806:
1803:
1801:
1795:
1789:
1786:
1784:
1781:
1780:
1778:
1775:
1774:
1768:
1765:
1763:
1759:
1754:
1753:Mitochondrial
1747:
1742:
1740:
1735:
1733:
1728:
1727:
1724:
1713:
1709:
1705:
1701:
1697:
1693:
1689:
1685:
1678:
1671:
1663:
1659:
1654:
1649:
1645:
1641:
1637:
1633:
1629:
1625:
1621:
1614:
1606:
1602:
1598:
1594:
1590:
1586:
1582:
1578:
1574:
1570:
1562:
1554:
1550:
1546:
1542:
1538:
1534:
1530:
1523:
1515:
1511:
1507:
1503:
1499:
1495:
1490:
1485:
1481:
1477:
1473:
1466:
1458:
1454:
1450:
1446:
1442:
1438:
1434:
1430:
1426:
1422:
1415:
1413:
1404:
1400:
1396:
1392:
1388:
1384:
1380:
1376:
1372:
1368:
1360:
1352:
1346:
1342:
1335:
1327:
1323:
1318:
1313:
1309:
1305:
1301:
1297:
1293:
1289:
1285:
1278:
1270:
1266:
1261:
1256:
1252:
1248:
1244:
1240:
1236:
1232:
1228:
1221:
1219:
1210:
1206:
1202:
1198:
1194:
1190:
1186:
1182:
1175:
1173:
1171:
1162:
1158:
1154:
1150:
1146:
1142:
1134:
1132:
1123:
1117:
1113:
1106:
1104:
1102:
1093:
1089:
1085:
1081:
1077:
1073:
1069:
1065:
1061:
1057:
1050:
1042:
1036:
1032:
1025:
1023:
1021:
1019:
1017:
1008:
1002:
998:
991:
989:
987:
985:
983:
981:
979:
974:
964:
963:Mitochondrion
961:
959:
956:
954:
951:
950:
944:
942:
939:found in the
938:
934:
930:
929:ribosomal RNA
926:
915:
913:
908:
904:
900:
896:
892:
888:
884:
880:
876:
872:
862:
860:
856:
852:
848:
844:
840:
836:
832:
823:
821:
817:
813:
809:
805:
801:
797:
787:
785:
780:
776:
772:
768:
760:
747:
737:
733:
729:
726:) except for
725:
724:
720:
716:
712:
708:
703:
693:
689:
687:
683:
679:
672:
668:
660:
645:
643:
639:
635:
631:
627:
623:
615:
608:
601:
597:
587:
585:
581:
577:
573:
569:
565:
561:
557:
547:
545:
541:
537:
536:transaminases
533:
529:
525:
521:
517:
513:
509:
505:
501:
497:
493:
489:
485:
475:
473:
466:
462:
456:
449:
439:
432:
428:
424:
420:
416:
412:
408:
404:
400:
393:
389:
383:makes use of
382:
378:
374:
367:
363:
359:
355:
351:
347:
343:
339:
335:
331:
327:
312:
310:
306:
302:
297:
293:
289:
284:
282:
278:
274:
270:
266:
262:
258:
254:
250:
246:
241:
239:
235:
231:
227:
223:
219:
215:
211:
207:
203:
199:
195:
191:
190:mitochondrion
180:
179:
176:
172:
169:
164:
160:
157:
150:
147:
144:
140:
135:
132:
129:
123:
120:
116:
113:
112:
110:
107:
104:
103:
102:
100:
97:
89:
86:
82:
79:
78:
77:
75:
72:
65:
62:
59:
58:
57:
55:
52:
48:
43:
38:
35:
33:mitochondrion
29:
26:
20:
2537:Cell anatomy
2514:
2361:ATP synthase
2168:
2136:
2104:
2052:
2042:
2019:ATP synthase
1999:
1967:
1940:
1918:
1898:Cytochrome c
1881:
1821:
1811:Kynureninase
1798:
1771:
1687:
1683:
1670:
1627:
1623:
1613:
1575:(1): 27–53.
1572:
1568:
1561:
1536:
1532:
1522:
1479:
1475:
1465:
1424:
1420:
1370:
1366:
1359:
1340:
1334:
1291:
1287:
1277:
1234:
1230:
1184:
1180:
1144:
1140:
1111:
1059:
1055:
1049:
1031:Biochemistry
1030:
996:
933:transfer RNA
921:
868:
835:oxaloacetate
829:
793:
784:ATP synthase
753:
705:
699:
656:
593:
568:ATP synthase
553:
481:
377:oxaloacetate
358:succinyl-CoA
323:
285:
242:
193:
187:
178:ATP synthase
174:
167:
162:
155:
154:
145:
139:You are here
138:
133:
130:
121:
114:
105:
95:
94:
87:
80:
70:
69:
60:
50:
49:
46:
24:Cell biology
2311:Complex III
1427:(1): 9–15.
887:anaplerotic
700:All of the
642:metabolites
580:complex III
399:-citrulline
326:metabolites
320:Metabolites
315:Composition
279:to produce
2531:Categories
2331:Complex IV
2138:urea cycle
1826:metabolism
1802:metabolism
1800:tryptophan
969:References
871:Ca control
865:Regulation
847:asparagine
820:citrulline
790:Urea cycle
732:complex II
696:synthesis.
682:co-enzymes
584:complex IV
576:complex II
542:, such as
388:-ornithine
381:urea cycle
350:isocitrate
342:acetyl-CoA
296:aquaporins
236:, and the
214:nucleotide
208:, soluble
2515:see also
2261:Complex I
2069:Aconitase
1644:0016-6731
1589:0300-8177
1498:1098-1004
1441:1365-2958
1308:1878-4186
1288:Structure
1251:0021-9738
1201:0022-0949
912:activator
891:inhibitor
851:glutamate
843:aspartate
816:ornithine
810:converts
767:electrons
754:NADH and
671:catabolic
648:Processes
572:complex I
540:proteases
492:aconitase
366:succinate
332:involves
267:produces
249:catabolic
206:ribosomes
137: ◄
2215:Frataxin
2089:Fumarase
1755:proteins
1712:21200502
1662:23212899
1624:Genetics
1605:27367543
1457:39165641
1326:24931469
1269:13221663
1209:10600673
1161:15607740
947:See also
859:arginine
719:fumarase
659:pyruvate
508:fumarase
362:fumarate
338:pyruvate
334:acyl-CoA
309:cytosine
245:anabolic
234:pyruvate
171:Ribosome
2375:MT-ATP8
2370:MT-ATP6
2290:MT-ND4L
1704:7969106
1653:3512135
1553:6143829
1514:2951786
1506:7627182
1449:8412675
1403:4355527
1395:7219534
1375:Bibcode
1317:4128088
1092:4149605
1084:4291593
1064:Bibcode
855:proline
796:ammonia
734:in the
702:enzymes
564:cristae
478:Enzymes
373:-malate
346:citrate
305:guanine
292:osmotic
210:enzymes
188:In the
99:Lamella
2480:MT-TS2
2475:MT-TS1
2445:MT-TL2
2440:MT-TL1
2350:MT-CO3
2345:MT-CO2
2340:MT-CO1
2320:MT-CYB
2300:MT-ND6
2295:MT-ND5
2285:MT-ND4
2280:MT-ND3
2275:MT-ND2
2270:MT-ND1
2043:Matrix
1710:
1702:
1660:
1650:
1642:
1603:
1597:171557
1595:
1587:
1551:
1512:
1504:
1496:
1455:
1447:
1439:
1401:
1393:
1367:Nature
1347:
1324:
1314:
1306:
1267:
1260:438594
1257:
1249:
1207:
1199:
1159:
1118:
1090:
1082:
1056:Nature
1037:
1003:
935:, and
905:, and
881:, and
857:, and
721:, and
530:, and
510:, and
467:, and
417:, and
394:, and
379:. The
375:, and
263:. The
194:matrix
192:, the
149:Cristæ
134:Matrix
2500:MT-TY
2495:MT-TW
2490:MT-TV
2485:MT-TT
2470:MT-TR
2465:MT-TQ
2460:MT-TP
2455:MT-TN
2450:MT-TM
2435:MT-TK
2430:MT-TI
2425:MT-TH
2420:MT-TG
2415:MT-TF
2410:MT-TE
2405:MT-TD
2400:MT-TC
2395:MT-TA
2191:PMPCB
2178:ALDH2
2000:other
1708:S2CID
1680:(PDF)
1601:S2CID
1510:S2CID
1453:S2CID
1399:S2CID
1088:S2CID
818:into
634:porin
407:FADH2
273:FADH2
64:Porin
2386:tRNA
1700:PMID
1658:PMID
1640:ISSN
1593:PMID
1585:ISSN
1549:PMID
1502:PMID
1494:ISSN
1445:PMID
1437:ISSN
1391:PMID
1345:ISBN
1322:PMID
1304:ISSN
1265:PMID
1247:ISSN
1205:PMID
1197:ISSN
1157:PMID
1116:ISBN
1080:PMID
1035:ISBN
1001:ISBN
893:for
845:and
833:and
814:and
773:and
756:FADH
675:FADH
640:and
638:ions
628:and
609:and
518:and
419:tRNA
405:and
403:NADH
307:and
271:and
269:NADH
259:and
247:and
122:3.12
115:3.11
1692:doi
1648:PMC
1632:doi
1628:192
1577:doi
1541:doi
1537:174
1484:doi
1429:doi
1383:doi
1371:290
1312:PMC
1296:doi
1255:PMC
1239:doi
1189:doi
1185:203
1149:doi
1145:326
1072:doi
1060:213
775:FAD
771:NAD
465:ADP
461:ATP
415:RNA
411:DNA
301:DNA
281:ATP
222:ATP
146:3.3
131:3.2
106:3.1
88:2.2
81:2.1
61:1.1
2533::
1706:.
1698:.
1688:19
1686:.
1682:.
1656:.
1646:.
1638:.
1626:.
1622:.
1599:.
1591:.
1583:.
1571:.
1547:.
1535:.
1508:.
1500:.
1492:.
1478:.
1474:.
1451:.
1443:.
1435:.
1423:.
1411:^
1397:.
1389:.
1381:.
1369:.
1320:.
1310:.
1302:.
1292:22
1290:.
1286:.
1263:.
1253:.
1245:.
1235:34
1233:.
1229:.
1217:^
1203:.
1195:.
1183:.
1169:^
1155:.
1143:.
1130:^
1100:^
1086:.
1078:.
1070:.
1058:.
1015:^
977:^
931:,
927:,
914:.
901:,
897:,
877:,
853:,
717:,
713:,
709:,
604:CO
600:Ca
546:.
526:,
506:,
502:,
498:,
494:,
490:,
486:,
474:.
463:,
450:,
435:CO
431:Mg
423:Ca
413:,
390:,
368:,
364:,
360:,
356:,
352:,
348:,
344:,
340:,
336:,
283:.
240:.
228:,
204:,
1745:e
1738:t
1731:v
1714:.
1694::
1664:.
1634::
1607:.
1579::
1573:9
1555:.
1543::
1516:.
1486::
1480:5
1459:.
1431::
1425:9
1405:.
1385::
1377::
1353:.
1328:.
1298::
1271:.
1241::
1211:.
1191::
1163:.
1151::
1124:.
1094:.
1074::
1066::
1043:.
1009:.
763:2
758:2
744:2
740:2
688:.
677:2
663:2
622:O
620:2
618:H
613:2
611:O
606:2
471:i
469:P
458:,
454:2
452:O
448:O
446:2
444:H
441:,
437:2
429:/
427:K
425:/
397:L
386:L
371:L
175:7
168:6
163:5
156:4
96:3
71:2
51:1
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.