493:) and quantify their absolute copy numbers in tumors and biological fluids, thus answering the basic questions about the absolute copy number of proteins in a single cell, which will be essential in digital modelling of mammalian cells and human body, and the relative levels of genetically abnormal proteins in tumors, and proving useful for diagnostic applications. SRM has also been used as a method of triggering full product ion scans of peptides to either a) confirm the specificity of the SRM transition, or b) detect specific
20:
497:
which are below the limit of detection of standard MS analyses. In 2017, SRM has been developed to be a highly sensitive and reproducible mass spectrometry-based protein targeted detection platform (entitled "SAFE-SRM"), and it has been demonstrated that the SRM-based new pipeline has major
137:
where the precursor ion ABCD is selected by the first stage of mass spectrometry (MS1), dissociates into molecule AB and product ion CD, and the latter is selected by the second stage of mass spectrometry (MS2) and detected. The precursor and product ion pair is called a SRM "transition".
234:
where ABCD is selected by MS1, dissociates into molecule AB and ion CD. The ion is selected in the second mass spectrometry stage MS2 then undergoes further fragmentation to form ion D which is selected in the third mass spectrometry stage MS3 and detected.
498:
advantages in clinical proteomics applications over traditional SRM pipelines, and it has demonstrated a dramatically improved diagnostic performance over that from antibody-based protein biomarker diagnostic methods, such as
1043:
229:
371:
307:
132:
690:
Kondrat, R. W.; McClusky, G. A.; Cooks, R. G. (1978). "Multiple reaction monitoring in mass spectrometry/mass spectrometry for direct analysis of complex mixtures".
1101:
1116:
1050:
27:
that undergo fragmentation followed by product ion selection in the MS2 stage. Additional stages of selection and fragmentation can be performed.
423:
to yield product ions whose signal abundances are indicative of the abundance of the peptide in the sample. This experiment can be performed on
376:
where ABCD is selected by MS1 and dissociates by two pathways, forming either AB or CD. The ions are selected sequentially by MS2 and detected.
564:
384:) is the application of SRM with parallel detection of all transitions in a single analysis using a high resolution mass spectrometer.
424:
245:) is the application of selected reaction monitoring to multiple product ions from one or more precursor ions, for example
154:
905:
1036:
494:
537:
148:) is the serial application of three or more stages of mass spectrometry to SRM, represented in a simple case by
791:(2012). "Selected reaction monitoring–based proteomics: workflows, potential, pitfalls and future directions".
649:
Murray, Kermit K.; Boyd, Robert K.; Eberlin, Marcos N.; Langley, G. John; Li, Liang; Naito, Yasuhide (2013).
1159:
727:"Parallel Reaction Monitoring for High Resolution and High Mass Accuracy Quantitative, Targeted Proteomics"
315:
251:
76:
1180:
1123:
1096:
1070:
511:
420:
1128:
440:
47:
1059:
921:"Multiple Reaction Monitoring to Identify Sites of Protein Phosphorylation with High Sensitivity"
516:
405:
393:
901:
59:
24:
451:. Much work goes into ensuring that transitions are selected that have maximum specificity.
973:
855:
552:
8:
1185:
1133:
444:
977:
859:
556:
1149:
996:
961:
878:
843:
824:
761:
726:
623:
590:
725:
Peterson, A. C.; Russell, J. D.; Bailey, D. J.; Westphall, M. S.; Coon, J. J. (2012).
1106:
1080:
1001:
942:
883:
828:
816:
808:
766:
748:
707:
672:
628:
610:
475:
471:
455:
397:
55:
902:"Selected Reaction Monitoring Mass Spectrometry for Absolute Protein Quantification"
991:
981:
932:
873:
863:
800:
756:
738:
699:
662:
618:
602:
560:
489:
SRM has been used to identify the proteins encoded by wild-type and mutant genes (
1111:
788:
651:"Definitions of terms relating to mass spectrometry (IUPAC Recommendations 2013)"
586:
490:
937:
920:
479:
1174:
812:
784:
752:
711:
676:
667:
650:
614:
582:
986:
868:
743:
439:
is cycled through the product ions which are detected upon exiting the last
1005:
946:
887:
820:
770:
632:
1028:
565:
10.1002/(SICI)1096-9888(199602)31:2<129::AID-JMS305>3.0.CO;2-T
962:"Selected reaction monitoring approach for validating peptide biomarkers"
467:
703:
606:
1075:
804:
408:
401:
591:"Selected reaction monitoring for quantitative proteomics: a tutorial"
23:
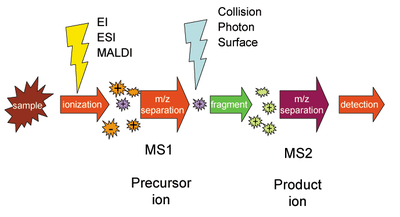
In selected reaction monitoring, the mass selection stage MS1 selects
463:
459:
1025:; quantify proteins in complex proteome digests by mass spectrometry
478:
that can provide the absolute quantification (i.e., copy number per
1102:
Stable isotope labeling by/with amino acids in cell culture (SILAC)
419:
population of mostly the intended species. This population is then
483:
412:
62:
is selected in the second mass spectrometer stage for detection.
54:
of a particular mass is selected in the first stage of a tandem
724:
499:
482:) of the native, light peptide, and by extension, its parent
1117:
Isobaric tags for relative and absolute quantitation (iTRAQ)
1022:
580:
416:
51:
960:
Wang Q, Zhang M, Tomita T, et al. (December 2017).
842:
Wang Q, Chaerkady R, Wu J, et al. (February 2011).
648:
19:
58:
and an ion product of a fragmentation reaction of the
447:. A precursor/product pair is often referred to as a
318:
254:
157:
79:
689:
415:
precursor is first isolated to obtain a substantial
919:Unwin RD, Griffiths JG, et al. (August 2005).
777:
718:
683:
576:
574:
365:
301:
223:
126:
1172:
783:
224:{\displaystyle ABCD^{+}\to AB+CD^{+}\to C+D^{+}}
959:
844:"Mutant proteins as cancer-specific biomarkers"
841:
571:
918:
535:
435:acts as a collision cell, and mass-resolving Q
1044:
70:A general case of SRM can be represented by
1058:
644:
642:
1051:
1037:
995:
985:
936:
877:
867:
760:
742:
666:
622:
639:
18:
1173:
1032:
366:{\displaystyle ABCD^{+}\to AB^{+}+CD}
302:{\displaystyle ABCD^{+}\to AB+CD^{+}}
127:{\displaystyle ABCD^{+}\to AB+CD^{+}}
538:"Tandem Mass Spectrometry: a Primer"
425:triple quadrupole mass spectrometers
925:Molecular & Cellular Proteomics
731:Molecular & Cellular Proteomics
13:
1124:Isotope-coded affinity tags (ICAT)
470:) peptides to a complex matrix as
14:
1197:
1016:
906:Journal of Visualized Experiments
474:, SRM can be used to construct a
1129:Metal-coded affinity tag (MeCAT)
495:post-translational modifications
953:
142:Consecutive reaction monitoring
16:Tandem mass spectrometry method
912:
894:
835:
529:
338:
274:
202:
177:
99:
1:
522:
392:SRM can be used for targeted
387:
1160:Data-independent acquisition
966:Proc. Natl. Acad. Sci. U.S.A
848:Proc. Natl. Acad. Sci. U.S.A
545:Journal of Mass Spectrometry
378:Parallel reaction monitoring
239:Multiple reaction monitoring
40:multiple reaction monitoring
32:Selected reaction monitoring
7:
505:
65:
10:
1202:
938:10.1074/mcp.M500113-MCP200
655:Pure and Applied Chemistry
458:with heavy-labeled (e.g.,
1155:Targeted proteomics / SRM
1142:
1097:Label-free quantification
1089:
1071:Protein mass spectrometry
1066:
595:Molecular Systems Biology
512:Protein mass spectrometry
431:isolates the precursor, q
668:10.1351/PAC-REC-06-04-06
427:, where mass-resolving Q
48:tandem mass spectrometry
1060:Quantitative proteomics
987:10.1073/pnas.1712731114
869:10.1073/pnas.1019203108
744:10.1074/mcp.O112.020131
536:E. de Hoffmann (1996).
517:Quantitative proteomics
472:concentration standards
394:quantitative proteomics
46:), is a method used in
1112:Tandem mass tags (TMT)
367:
303:
225:
128:
28:
368:
304:
226:
129:
22:
692:Analytical Chemistry
404:in, for example, an
316:
252:
155:
77:
1143:Acquisition methods
1134:N-terminal labeling
978:2017PNAS..11413519W
972:(51): 13519–13524.
860:2011PNAS..108.2444W
704:10.1021/ac50036a020
607:10.1038/msb.2008.61
557:1996JMSp...31..129D
445:electron multiplier
1150:Shotgun proteomics
805:10.1038/nmeth.2015
363:
299:
221:
124:
29:
1181:Mass spectrometry
1168:
1167:
1107:Isobaric labeling
1081:Mass spectrometry
737:(11): 1475–1488.
698:(14): 2017–2021.
476:calibration curve
456:isotopic labeling
398:mass spectrometry
56:mass spectrometer
1193:
1053:
1046:
1039:
1030:
1029:
1010:
1009:
999:
989:
957:
951:
950:
940:
916:
910:
909:
898:
892:
891:
881:
871:
839:
833:
832:
789:Aebersold, Ruedi
781:
775:
774:
764:
746:
722:
716:
715:
687:
681:
680:
670:
661:(7): 1515–1609.
646:
637:
636:
626:
587:Aebersold, Ruedi
585:; Domon, Bruno;
581:Lange, Vinzenz;
578:
569:
568:
542:
533:
372:
370:
369:
364:
353:
352:
337:
336:
308:
306:
305:
300:
298:
297:
273:
272:
230:
228:
227:
222:
220:
219:
201:
200:
176:
175:
133:
131:
130:
125:
123:
122:
98:
97:
1201:
1200:
1196:
1195:
1194:
1192:
1191:
1190:
1171:
1170:
1169:
1164:
1138:
1085:
1062:
1057:
1019:
1014:
1013:
958:
954:
917:
913:
900:
899:
895:
840:
836:
782:
778:
723:
719:
688:
684:
647:
640:
579:
572:
540:
534:
530:
525:
508:
491:mutant proteins
438:
434:
430:
390:
348:
344:
332:
328:
317:
314:
313:
293:
289:
268:
264:
253:
250:
249:
215:
211:
196:
192:
171:
167:
156:
153:
152:
118:
114:
93:
89:
78:
75:
74:
68:
38:), also called
17:
12:
11:
5:
1199:
1189:
1188:
1183:
1166:
1165:
1163:
1162:
1157:
1152:
1146:
1144:
1140:
1139:
1137:
1136:
1131:
1126:
1121:
1120:
1119:
1114:
1104:
1099:
1093:
1091:
1090:Quantification
1087:
1086:
1084:
1083:
1078:
1073:
1067:
1064:
1063:
1056:
1055:
1048:
1041:
1033:
1027:
1026:
1018:
1017:External links
1015:
1012:
1011:
952:
931:(8): 1134–44.
911:
893:
834:
799:(6): 555–566.
793:Nature Methods
785:Picotti, Paola
776:
717:
682:
638:
583:Picotti, Paola
570:
551:(2): 129–137.
527:
526:
524:
521:
520:
519:
514:
507:
504:
436:
432:
428:
389:
386:
374:
373:
362:
359:
356:
351:
347:
343:
340:
335:
331:
327:
324:
321:
310:
309:
296:
292:
288:
285:
282:
279:
276:
271:
267:
263:
260:
257:
232:
231:
218:
214:
210:
207:
204:
199:
195:
191:
188:
185:
182:
179:
174:
170:
166:
163:
160:
135:
134:
121:
117:
113:
110:
107:
104:
101:
96:
92:
88:
85:
82:
67:
64:
60:precursor ions
25:precursor ions
15:
9:
6:
4:
3:
2:
1198:
1187:
1184:
1182:
1179:
1178:
1176:
1161:
1158:
1156:
1153:
1151:
1148:
1147:
1145:
1141:
1135:
1132:
1130:
1127:
1125:
1122:
1118:
1115:
1113:
1110:
1109:
1108:
1105:
1103:
1100:
1098:
1095:
1094:
1092:
1088:
1082:
1079:
1077:
1074:
1072:
1069:
1068:
1065:
1061:
1054:
1049:
1047:
1042:
1040:
1035:
1034:
1031:
1024:
1021:
1020:
1007:
1003:
998:
993:
988:
983:
979:
975:
971:
967:
963:
956:
948:
944:
939:
934:
930:
926:
922:
915:
907:
903:
897:
889:
885:
880:
875:
870:
865:
861:
857:
854:(6): 2444–9.
853:
849:
845:
838:
830:
826:
822:
818:
814:
810:
806:
802:
798:
794:
790:
786:
780:
772:
768:
763:
758:
754:
750:
745:
740:
736:
732:
728:
721:
713:
709:
705:
701:
697:
693:
686:
678:
674:
669:
664:
660:
656:
652:
645:
643:
634:
630:
625:
620:
616:
612:
608:
604:
600:
596:
592:
588:
584:
577:
575:
566:
562:
558:
554:
550:
546:
539:
532:
528:
518:
515:
513:
510:
509:
503:
501:
496:
492:
487:
485:
481:
477:
473:
469:
465:
461:
457:
452:
450:
446:
442:
426:
422:
418:
414:
410:
407:
403:
399:
395:
385:
383:
379:
360:
357:
354:
349:
345:
341:
333:
329:
325:
322:
319:
312:
311:
294:
290:
286:
283:
280:
277:
269:
265:
261:
258:
255:
248:
247:
246:
244:
240:
236:
216:
212:
208:
205:
197:
193:
189:
186:
183:
180:
172:
168:
164:
161:
158:
151:
150:
149:
147:
143:
139:
119:
115:
111:
108:
105:
102:
94:
90:
86:
83:
80:
73:
72:
71:
63:
61:
57:
53:
49:
45:
41:
37:
33:
26:
21:
1154:
969:
965:
955:
928:
924:
914:
896:
851:
847:
837:
796:
792:
779:
734:
730:
720:
695:
691:
685:
658:
654:
598:
594:
548:
544:
531:
488:
453:
448:
406:electrospray
400:. Following
391:
381:
377:
375:
242:
238:
237:
233:
145:
141:
140:
136:
69:
50:in which an
43:
39:
35:
31:
30:
1186:Proteomics
1175:Categories
1076:Proteomics
523:References
449:transition
441:quadrupole
421:fragmented
402:ionization
388:Proteomics
829:205420574
813:1548-7091
753:1535-9476
712:0003-2700
677:1365-3075
615:1744-4292
339:→
275:→
203:→
178:→
100:→
1023:SRMatlas
1006:29203663
947:15923565
888:21248225
821:22669653
771:22865924
633:18854821
589:(2008).
506:See also
66:Variants
997:5754789
974:Bibcode
879:3038743
856:Bibcode
762:3494192
624:2583086
601:: 222.
553:Bibcode
484:protein
413:peptide
1004:
994:
945:
886:
876:
827:
819:
811:
769:
759:
751:
710:
675:
631:
621:
613:
454:Using
443:by an
409:source
825:S2CID
541:(PDF)
500:ELISA
466:, or
1002:PMID
943:PMID
884:PMID
817:PMID
809:ISSN
767:PMID
749:ISSN
708:ISSN
673:ISSN
629:PMID
611:ISSN
480:cell
411:, a
992:PMC
982:doi
970:114
933:doi
874:PMC
864:doi
852:108
801:doi
757:PMC
739:doi
700:doi
663:doi
619:PMC
603:doi
561:doi
417:ion
396:by
382:PRM
243:MRM
146:CRM
52:ion
44:MRM
36:SRM
1177::
1000:.
990:.
980:.
968:.
964:.
941:.
927:.
923:.
904:.
882:.
872:.
862:.
850:.
846:.
823:.
815:.
807:.
795:.
787:;
765:.
755:.
747:.
735:11
733:.
729:.
706:.
696:50
694:.
671:.
659:85
657:.
653:.
641:^
627:.
617:.
609:.
597:.
593:.
573:^
559:.
549:31
547:.
543:.
502:.
486:.
462:,
1052:e
1045:t
1038:v
1008:.
984::
976::
949:.
935::
929:4
908:.
890:.
866::
858::
831:.
803::
797:9
773:.
741::
714:.
702::
679:.
665::
635:.
605::
599:4
567:.
563::
555::
468:N
464:C
460:D
437:3
433:2
429:1
380:(
361:D
358:C
355:+
350:+
346:B
342:A
334:+
330:D
326:C
323:B
320:A
295:+
291:D
287:C
284:+
281:B
278:A
270:+
266:D
262:C
259:B
256:A
241:(
217:+
213:D
209:+
206:C
198:+
194:D
190:C
187:+
184:B
181:A
173:+
169:D
165:C
162:B
159:A
144:(
120:+
116:D
112:C
109:+
106:B
103:A
95:+
91:D
87:C
84:B
81:A
42:(
34:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.