2222:
223:
701:
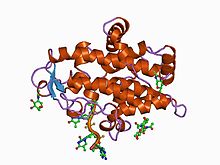
Aspergillus nuclease S1 is a monomeric protein of a molecular weight of 38 kilodalton. It requires Zn as a cofactor and is relatively stable against denaturing agents like urea, SDS, or formaldehyde. The optimum pH for its activity lies between 4-4.5. Aspergillus nuclease S1 is known to be inhibited
459:
Alternative names include endonuclease S1 (Aspergillus), single-stranded-nucleate endonuclease, deoxyribonuclease S1, deoxyribonuclease S1, Aspergillus nuclease S1, Neurospora crassa single-strand specific endonuclease, S1 nuclease, single-strand endodeoxyribonuclease, single-stranded DNA specific
530:
These requirements and distinguishing features are responsible for function efficacy. It is an enzyme and these four features are needed for enzyme functionality. The three zinc ions are vital for catalysis. The first two zincs activate the attacking water in hydrolysis whilst the third zinc ion
415:
Although its primary substrate is single-stranded, it can also occasionally introduce single-stranded breaks in double-stranded DNA or RNA, or DNA-RNA hybrids. The enzyme hydrolyses single stranded region in duplex DNA such as loops or gaps. It also cleaves a strand opposite a nick on the
734:. In molecular biology, it is used in removing single stranded tails from DNA molecules to create blunt ended molecules and opening hairpin loops generated during synthesis of double stranded cDNA.
859:
Romier C, Dominguez R, Lahm A, Dahl O, Suck D (September 1998). "Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs".
902:
Yang X, Pu F, Ren J, Qu X (July 2011). "DNA-templated ensemble for label-free and real-time fluorescence turn-on detection of enzymatic/oxidative cleavage of single-stranded DNA".
773:
Balabanova LA, Gafurov YM, Pivkin MV, Terentyeva NA, Likhatskaya GN, Rasskazov VA (February 2012). "An extracellular S1-type nuclease of marine fungus
Penicillium melinii".
722:
located in the cavity in the active site and its backbone supports the action one of the zinc ions. Such mechanisms are essential to the catalytic function of the enzyme.
1703:
1649:
347:
177:
824:
Podzimek T, Matoušek J, Lipovová P, Poučková P, Spiwok V, Santrůček J (February 2011). "Biochemical properties of three plant nucleases with anticancer potential".
447:
extracellular, that is, outside of the cell. Their function and distinguishing features mean they have potential in being exploited in the field of
1311:
1472:
702:
somewhat by 50 μM ATP and nearly completely by 1 mM ATP. 50% inhibition has been shown at 85 μM dAMP and 1 μM dATP but uninhibited by cAMP.
196:
1129:
1568:
872:
1279:
1296:
189:
1543:
639:
460:
endonuclease, single-strand-specific endodeoxyribonuclease, single strand-specific DNase and
Aspergillus oryzae S1 nuclease.
283:
156:
560:
367:
1457:
1558:
1552:
1353:
1103:
1401:
1941:
1512:
1301:
749:
443:
and are thought to be associated in programmed cell death and also in tissue differentiation. Furthermore, they are
1619:
1507:
150:
1428:
1338:
1291:
132:
355:
2097:
1850:
1791:
137:
1895:
1462:
1452:
1855:
1747:
1372:
201:
17:
125:
2212:
1441:
1437:
1433:
1349:
1182:
351:
60:
2262:
2198:
2185:
2172:
2159:
2146:
2133:
2120:
2082:
1609:
1165:
731:
303:
296:
308:
2257:
2092:
2046:
1989:
1357:
1206:
1120:
383:
153:
43:
752:, non-homologous family with similar DNA/RNA activity but accepts double-stranded substrate better
77:
1994:
1226:
1096:
484:
1782:
1421:
411:
Endonucleolytic cleavage to 5'-phosphomononucleotide and 5'-phosphooligonucleotide end-products
2242:
2015:
1934:
1717:
1614:
1406:
1367:
1231:
1153:
2087:
1900:
1735:
1730:
1663:
1323:
1148:
782:
431:
334:
113:
679:
600:
8:
2051:
1755:
1725:
1529:
1524:
1448:
1384:
1170:
397:
89:
55:
786:
386:
48:
2252:
2247:
1984:
1888:
1740:
1221:
1211:
1089:
1050:
1033:
884:
806:
743:
519:
421:
1004:
987:
963:
938:
1689:
1634:
1602:
1477:
1196:
1055:
1009:
968:
919:
876:
841:
798:
405:
342:
288:
144:
888:
810:
251:
2030:
2025:
1999:
1927:
1765:
1345:
1250:
1245:
1201:
1045:
999:
958:
950:
911:
868:
837:
833:
790:
330:
264:
2077:
2061:
1974:
1867:
1681:
1597:
1592:
1587:
1500:
1495:
1255:
1133:
1077:
513:
276:
1081:
689:
610:
2226:
2115:
2056:
1840:
1835:
1830:
1216:
1191:
1187:
1160:
1143:
469:
172:
794:
2236:
2020:
1979:
1581:
1490:
1240:
448:
1969:
1708:
1639:
1362:
1283:
1059:
954:
923:
873:
10.1002/(sici)1097-0134(19980901)32:4<414::aid-prot2>3.0.co;2-g
845:
802:
476:
468:
Most nucleases with EC 3.1.30.1 activity are homologous to each other in a
390:
880:
292:
2193:
2128:
1964:
1654:
1548:
1411:
1377:
1315:
1013:
711:
660:
581:
491:
436:
972:
939:"Stability of the unique anticodon loop conformation of E.coli tRNAfMet"
101:
1860:
915:
719:
655:
576:
509:
440:
2167:
2141:
1773:
1572:
1485:
1112:
715:
444:
401:
2221:
1818:
1813:
1808:
1629:
1264:
1116:
1073:
523:
271:
730:
Aspergillus nuclease S1 is used in the laboratory as a reagent in
1825:
1803:
1798:
1416:
772:
505:
120:
823:
2180:
1950:
1845:
1778:
1671:
1269:
1177:
393:
362:
222:
184:
96:
84:
72:
2154:
1787:
1538:
1534:
532:
718:
groups from 3' nucleotides. Additionally, the side chain of
400:(ssDNA) and RNA into oligo- or mononucleotides. This enzyme
1519:
1396:
1389:
1333:
1328:
1069:
324:
258:
246:
108:
1919:
988:"Specificity of the S1 nuclease from Aspergillus oryzae"
710:
This zinc-dependent nuclease protein domain produces 5'
858:
498:
1068:
This article incorporates text from the public domain
416:
complementary strand. It has no sequence specificity.
2210:
985:
986:Wiegand RC, Godson GN, Radding CM (November 1975).
1407:Fructose 6-P,2-kinase:fructose 2,6-bisphosphatase
1111:
475:Members of this family, including P1 and S1, are
2234:
435:. Members of the S1/P1 family are found in both
768:
766:
746:, similar activity but probably not homologous
227:P1 nuclease in complex with a substrate analog
1935:
1097:
479:with very distinguishing features, they are:
1031:
901:
817:
763:
1942:
1928:
1104:
1090:
852:
221:
1049:
1003:
962:
936:
419:Well-known versions include S1 found in
1297:Ubiquitin carboxy-terminal hydrolase L1
14:
2235:
1923:
1877:either deoxy- or ribo-
1085:
1458:Protein serine/threonine phosphatase
1032:Desai NA, Shankar V (January 2003).
1559:Cyclic nucleotide phosphodiesterase
1553:Clostridium perfringens alpha toxin
1354:Tartrate-resistant acid phosphatase
992:The Journal of Biological Chemistry
24:
1402:Pyruvate dehydrogenase phosphatase
1051:10.1111/j.1574-6976.2003.tb00626.x
1034:"Single-strand-specific nucleases"
1024:
483:a requirement for three zinc ions
25:
2274:
1302:4-hydroxybenzoyl-CoA thioesterase
937:Wrede P, Rich A (November 1979).
750:DNA/RNA non-specific endonuclease
2220:
1620:N-acetylglucosamine-6-sulfatase
1508:Sphingomyelin phosphodiesterase
454:
1429:Inositol-phosphate phosphatase
1292:Palmitoyl protein thioesterase
979:
930:
895:
838:10.1016/j.plantsci.2010.10.006
472:family called Nuclease S1/P1.
13:
1:
1792:RNA-induced silencing complex
1005:10.1016/S0021-9258(19)40751-5
756:
538:
319:Available protein structures:
1896:Serratia marcescens nuclease
1463:Dual-specificity phosphatase
1453:Protein tyrosine phosphatase
705:
463:
7:
1949:
1373:Fructose 1,6-bisphosphatase
737:
10:
2279:
1067:
732:nuclease protection assays
2106:
2098:Michaelis–Menten kinetics
2070:
2039:
2008:
1957:
1876:
1764:
1716:
1702:
1680:
1662:
1648:
1628:
1610:Galactosamine-6 sulfatase
1567:
1471:
1310:
1278:
1166:6-phosphogluconolactonase
1128:
1038:FEMS Microbiology Reviews
795:10.1007/s10126-011-9392-5
685:
675:
670:
666:
654:
646:
634:
629:
624:
606:
596:
591:
587:
575:
567:
555:
550:
545:
361:
341:
323:
318:
314:
302:
282:
270:
257:
245:
237:
232:
220:
215:
195:
183:
171:
166:
162:
143:
131:
119:
107:
95:
83:
71:
66:
54:
42:
37:
32:
1990:Diffusion-limited enzyme
1358:Purple acid phosphatases
904:Chemical Communications
725:
531:stabilizes the leaving
425:(yellow koji mold) and
1783:Microprocessor complex
1422:Beta-propeller phytase
943:Nucleic Acids Research
2083:Eadie–Hofstee diagram
2016:Allosteric regulation
1718:Endodeoxyribonuclease
1615:Iduronate-2-sulfatase
1368:Glucose 6-phosphatase
1154:Butyrylcholinesterase
2093:Lineweaver–Burk plot
1901:Micrococcal nuclease
1736:Deoxyribonuclease IV
1731:Deoxyribonuclease II
1664:Exodeoxyribonuclease
1324:Alkaline phosphatase
1149:Acetylcholinesterase
955:10.1093/nar/7.6.1457
775:Marine Biotechnology
640:Penicillium citrinum
432:Penicillium citrinum
1756:UvrABC endonuclease
1726:Deoxyribonuclease I
1449:Protein phosphatase
1385:Protein phosphatase
1183:Bile salt-dependent
1171:PAF acetylhydrolase
787:2012MarBt..14...87B
497:requires an acidic
398:single-stranded DNA
2052:Enzyme superfamily
1985:Enzyme promiscuity
1889:Mung bean nuclease
1748:Restriction enzyme
1741:Restriction enzyme
916:10.1039/c1cc12216a
744:Mung bean nuclease
561:Aspergillus oryzae
520:Disulphide bridges
490:containing common
422:Aspergillus oryzae
2208:
2207:
1917:
1916:
1913:
1912:
1909:
1908:
1698:
1697:
1690:Oligonucleotidase
1635:deoxyribonuclease
1603:Steroid sulfatase
1478:Phosphodiesterase
1207:Hormone-sensitive
699:
698:
695:
694:
620:
619:
616:
615:
406:chemical reaction
377:
376:
373:
372:
368:structure summary
211:
210:
207:
206:
126:metabolic pathway
16:(Redirected from
2270:
2263:Protein families
2225:
2224:
2216:
2088:Hanes–Woolf plot
2031:Enzyme activator
2026:Enzyme inhibitor
2000:Enzyme catalysis
1944:
1937:
1930:
1921:
1920:
1766:Endoribonuclease
1752:
1746:
1714:
1713:
1660:
1659:
1646:
1645:
1346:Acid phosphatase
1227:Monoacylglycerol
1137:ester hydrolases
1106:
1099:
1092:
1083:
1082:
1063:
1053:
1018:
1017:
1007:
983:
977:
976:
966:
934:
928:
927:
899:
893:
892:
856:
850:
849:
821:
815:
814:
770:
668:
667:
642:
622:
621:
589:
588:
563:
543:
542:
316:
315:
225:
213:
212:
164:
163:
30:
29:
27:Class of enzymes
21:
2278:
2277:
2273:
2272:
2271:
2269:
2268:
2267:
2258:Protein domains
2233:
2232:
2231:
2219:
2211:
2209:
2204:
2116:Oxidoreductases
2102:
2078:Enzyme kinetics
2066:
2062:List of enzymes
2035:
2004:
1975:Catalytic triad
1953:
1948:
1918:
1905:
1872:
1760:
1750:
1744:
1707:
1694:
1682:Exoribonuclease
1676:
1653:
1637:
1633:
1624:
1598:Arylsulfatase L
1593:Arylsulfatase B
1588:Arylsulfatase A
1563:
1476:
1467:
1306:
1274:
1136:
1124:
1110:
1080:
1066:
1027:
1025:Further reading
1022:
1021:
998:(22): 8848–55.
984:
980:
935:
931:
900:
896:
857:
853:
822:
818:
771:
764:
759:
740:
728:
708:
638:
559:
541:
514:N-glycosylation
512:asparagine via
504:contains three
466:
457:
228:
28:
23:
22:
15:
12:
11:
5:
2276:
2266:
2265:
2260:
2255:
2250:
2245:
2230:
2229:
2206:
2205:
2203:
2202:
2189:
2176:
2163:
2150:
2137:
2124:
2110:
2108:
2104:
2103:
2101:
2100:
2095:
2090:
2085:
2080:
2074:
2072:
2068:
2067:
2065:
2064:
2059:
2054:
2049:
2043:
2041:
2040:Classification
2037:
2036:
2034:
2033:
2028:
2023:
2018:
2012:
2010:
2006:
2005:
2003:
2002:
1997:
1992:
1987:
1982:
1977:
1972:
1967:
1961:
1959:
1955:
1954:
1947:
1946:
1939:
1932:
1924:
1915:
1914:
1911:
1910:
1907:
1906:
1904:
1903:
1898:
1893:
1892:
1891:
1880:
1878:
1874:
1873:
1871:
1870:
1865:
1864:
1863:
1858:
1853:
1848:
1838:
1833:
1828:
1823:
1822:
1821:
1816:
1811:
1806:
1796:
1795:
1794:
1785:
1770:
1768:
1762:
1761:
1759:
1758:
1753:
1738:
1733:
1728:
1722:
1720:
1711:
1700:
1699:
1696:
1695:
1693:
1692:
1686:
1684:
1678:
1677:
1675:
1674:
1668:
1666:
1657:
1643:
1626:
1625:
1623:
1622:
1617:
1612:
1607:
1606:
1605:
1600:
1595:
1590:
1577:
1575:
1565:
1564:
1562:
1561:
1556:
1546:
1541:
1532:
1527:
1522:
1517:
1516:
1515:
1505:
1504:
1503:
1498:
1488:
1482:
1480:
1469:
1468:
1466:
1465:
1460:
1455:
1446:
1445:
1444:
1426:
1425:
1424:
1414:
1409:
1404:
1399:
1394:
1393:
1392:
1382:
1381:
1380:
1370:
1365:
1360:
1343:
1342:
1341:
1336:
1331:
1320:
1318:
1308:
1307:
1305:
1304:
1299:
1294:
1288:
1286:
1276:
1275:
1273:
1272:
1267:
1261:
1260:
1259:
1258:
1253:
1248:
1237:
1236:
1235:
1234:
1232:Diacylglycerol
1229:
1224:
1219:
1214:
1209:
1204:
1199:
1194:
1185:
1174:
1173:
1168:
1163:
1161:Pectinesterase
1158:
1157:
1156:
1151:
1144:Cholinesterase
1140:
1138:
1126:
1125:
1109:
1108:
1101:
1094:
1086:
1065:
1064:
1028:
1026:
1023:
1020:
1019:
978:
949:(6): 1457–67.
929:
910:(28): 8133–5.
894:
851:
816:
761:
760:
758:
755:
754:
753:
747:
739:
736:
727:
724:
707:
704:
697:
696:
693:
692:
687:
683:
682:
677:
673:
672:
664:
663:
658:
652:
651:
648:
644:
643:
636:
632:
631:
627:
626:
618:
617:
614:
613:
608:
604:
603:
598:
594:
593:
585:
584:
579:
573:
572:
569:
565:
564:
557:
553:
552:
548:
547:
540:
537:
528:
527:
516:
502:
501:for catalysis.
495:
488:
470:protein domain
465:
462:
456:
453:
413:
412:
404:the following
375:
374:
371:
370:
365:
359:
358:
345:
339:
338:
328:
321:
320:
312:
311:
306:
300:
299:
286:
280:
279:
274:
268:
267:
262:
255:
254:
249:
243:
242:
241:S1-P1_nuclease
239:
235:
234:
230:
229:
226:
218:
217:
216:S1-P1 nuclease
209:
208:
205:
204:
199:
193:
192:
187:
181:
180:
175:
169:
168:
160:
159:
148:
141:
140:
135:
129:
128:
123:
117:
116:
111:
105:
104:
99:
93:
92:
87:
81:
80:
75:
69:
68:
64:
63:
58:
52:
51:
46:
40:
39:
35:
34:
26:
9:
6:
4:
3:
2:
2275:
2264:
2261:
2259:
2256:
2254:
2251:
2249:
2246:
2244:
2241:
2240:
2238:
2228:
2223:
2218:
2217:
2214:
2200:
2196:
2195:
2190:
2187:
2183:
2182:
2177:
2174:
2170:
2169:
2164:
2161:
2157:
2156:
2151:
2148:
2144:
2143:
2138:
2135:
2131:
2130:
2125:
2122:
2118:
2117:
2112:
2111:
2109:
2105:
2099:
2096:
2094:
2091:
2089:
2086:
2084:
2081:
2079:
2076:
2075:
2073:
2069:
2063:
2060:
2058:
2057:Enzyme family
2055:
2053:
2050:
2048:
2045:
2044:
2042:
2038:
2032:
2029:
2027:
2024:
2022:
2021:Cooperativity
2019:
2017:
2014:
2013:
2011:
2007:
2001:
1998:
1996:
1993:
1991:
1988:
1986:
1983:
1981:
1980:Oxyanion hole
1978:
1976:
1973:
1971:
1968:
1966:
1963:
1962:
1960:
1956:
1952:
1945:
1940:
1938:
1933:
1931:
1926:
1925:
1922:
1902:
1899:
1897:
1894:
1890:
1887:
1886:
1885:
1882:
1881:
1879:
1875:
1869:
1866:
1862:
1859:
1857:
1854:
1852:
1849:
1847:
1844:
1843:
1842:
1839:
1837:
1834:
1832:
1829:
1827:
1824:
1820:
1817:
1815:
1812:
1810:
1807:
1805:
1802:
1801:
1800:
1797:
1793:
1789:
1786:
1784:
1780:
1777:
1776:
1775:
1772:
1771:
1769:
1767:
1763:
1757:
1754:
1749:
1742:
1739:
1737:
1734:
1732:
1729:
1727:
1724:
1723:
1721:
1719:
1715:
1712:
1710:
1705:
1701:
1691:
1688:
1687:
1685:
1683:
1679:
1673:
1670:
1669:
1667:
1665:
1661:
1658:
1656:
1651:
1647:
1644:
1641:
1636:
1631:
1627:
1621:
1618:
1616:
1613:
1611:
1608:
1604:
1601:
1599:
1596:
1594:
1591:
1589:
1586:
1585:
1584:
1583:
1582:arylsulfatase
1579:
1578:
1576:
1574:
1570:
1566:
1560:
1557:
1554:
1550:
1547:
1545:
1542:
1540:
1536:
1533:
1531:
1528:
1526:
1523:
1521:
1518:
1514:
1511:
1510:
1509:
1506:
1502:
1499:
1497:
1494:
1493:
1492:
1491:Phospholipase
1489:
1487:
1484:
1483:
1481:
1479:
1474:
1470:
1464:
1461:
1459:
1456:
1454:
1450:
1447:
1443:
1439:
1435:
1432:
1431:
1430:
1427:
1423:
1420:
1419:
1418:
1415:
1413:
1410:
1408:
1405:
1403:
1400:
1398:
1395:
1391:
1388:
1387:
1386:
1383:
1379:
1376:
1375:
1374:
1371:
1369:
1366:
1364:
1361:
1359:
1355:
1351:
1347:
1344:
1340:
1337:
1335:
1332:
1330:
1327:
1326:
1325:
1322:
1321:
1319:
1317:
1313:
1309:
1303:
1300:
1298:
1295:
1293:
1290:
1289:
1287:
1285:
1281:
1277:
1271:
1268:
1266:
1263:
1262:
1257:
1254:
1252:
1249:
1247:
1244:
1243:
1242:
1241:Phospholipase
1239:
1238:
1233:
1230:
1228:
1225:
1223:
1220:
1218:
1215:
1213:
1210:
1208:
1205:
1203:
1200:
1198:
1195:
1193:
1189:
1186:
1184:
1181:
1180:
1179:
1176:
1175:
1172:
1169:
1167:
1164:
1162:
1159:
1155:
1152:
1150:
1147:
1146:
1145:
1142:
1141:
1139:
1135:
1131:
1127:
1122:
1118:
1114:
1107:
1102:
1100:
1095:
1093:
1088:
1087:
1084:
1079:
1075:
1071:
1061:
1057:
1052:
1047:
1044:(5): 457–91.
1043:
1039:
1035:
1030:
1029:
1015:
1011:
1006:
1001:
997:
993:
989:
982:
974:
970:
965:
960:
956:
952:
948:
944:
940:
933:
925:
921:
917:
913:
909:
905:
898:
890:
886:
882:
878:
874:
870:
867:(4): 414–24.
866:
862:
855:
847:
843:
839:
835:
832:(2): 343–51.
831:
827:
826:Plant Science
820:
812:
808:
804:
800:
796:
792:
788:
784:
780:
776:
769:
767:
762:
751:
748:
745:
742:
741:
735:
733:
723:
721:
717:
713:
703:
691:
688:
684:
681:
678:
674:
669:
665:
662:
659:
657:
653:
649:
645:
641:
637:
633:
628:
623:
612:
609:
605:
602:
599:
595:
590:
586:
583:
580:
578:
574:
570:
566:
562:
558:
554:
549:
544:
536:
534:
525:
521:
517:
515:
511:
508:bound to the
507:
503:
500:
496:
493:
489:
486:
482:
481:
480:
478:
477:glycoproteins
473:
471:
461:
452:
450:
449:biotechnology
446:
442:
438:
434:
433:
428:
424:
423:
417:
410:
409:
408:
407:
403:
399:
395:
392:
388:
385:
381:
369:
366:
364:
360:
357:
353:
349:
346:
344:
340:
336:
332:
329:
326:
322:
317:
313:
310:
307:
305:
301:
298:
294:
290:
287:
285:
281:
278:
275:
273:
269:
266:
263:
260:
256:
253:
250:
248:
244:
240:
236:
231:
224:
219:
214:
203:
200:
198:
194:
191:
188:
186:
182:
179:
176:
174:
170:
165:
161:
158:
155:
152:
149:
146:
142:
139:
136:
134:
130:
127:
124:
122:
118:
115:
112:
110:
106:
103:
102:NiceZyme view
100:
98:
94:
91:
88:
86:
82:
79:
76:
74:
70:
65:
62:
59:
57:
53:
50:
47:
45:
41:
36:
31:
19:
2243:Zinc enzymes
2194:Translocases
2191:
2178:
2165:
2152:
2139:
2129:Transferases
2126:
2113:
1970:Binding site
1883:
1751:}}
1745:{{
1709:Endonuclease
1640:ribonuclease
1580:
1363:Nucleotidase
1284:Thioesterase
1041:
1037:
995:
991:
981:
946:
942:
932:
907:
903:
897:
864:
860:
854:
829:
825:
819:
781:(1): 87–95.
778:
774:
729:
714:and cleaves
709:
700:
529:
474:
467:
458:
455:Nomenclature
430:
426:
420:
418:
414:
396:that splits
391:endonuclease
379:
378:
90:BRENDA entry
1965:Active site
1884:Nuclease S1
1655:Exonuclease
1549:Lecithinase
1378:Calcineurin
1316:Phosphatase
1222:Lipoprotein
1212:Endothelial
712:nucleotides
680:Swiss-model
630:Identifiers
625:Nuclease P1
601:Swiss-model
551:Identifiers
546:Nuclease S1
492:active site
437:prokaryotes
427:Nuclease P1
380:Nuclease S1
233:Identifiers
78:IntEnz view
61:37288-25-8
38:Identifiers
33:Nuclease S1
18:P1 nuclease
2237:Categories
2168:Isomerases
2142:Hydrolases
2009:Regulation
1197:Pancreatic
1134:Carboxylic
757:References
720:tryptophan
676:Structures
671:Search for
597:Structures
592:Search for
539:Properties
510:amino acid
494:motifs and
441:eukaryotes
331:structures
147:structures
114:KEGG entry
2253:Nucleases
2248:EC 3.1.30
2047:EC number
1774:RNase III
1632:(includes
1573:Sulfatase
1486:Autotaxin
1350:Prostatic
1202:Lysosomal
1117:esterases
1113:Hydrolase
1078:IPR003154
716:phosphate
706:Mechanism
526:residues.
485:cofactors
464:Structure
429:found in
402:catalyses
277:IPR003154
67:Databases
2071:Kinetics
1995:Cofactor
1958:Activity
1868:RNase T1
1630:Nuclease
1265:Cutinase
1074:InterPro
1060:12586391
924:21629944
889:24233161
861:Proteins
846:21421379
811:17856850
803:21647618
738:See also
690:InterPro
635:Organism
611:InterPro
556:Organism
533:oxyanion
524:cysteine
522:between
445:secreted
389:) is an
387:3.1.30.1
348:RCSB PDB
272:InterPro
202:proteins
190:articles
178:articles
151:RCSB PDB
49:3.1.30.1
2227:Biology
2181:Ligases
1951:Enzymes
1841:RNase E
1836:RNase Z
1831:RNase A
1826:RNase P
1799:RNase H
1417:Phytase
1217:Hepatic
1192:Lingual
1188:Gastric
881:9726413
783:Bibcode
686:Domains
656:UniProt
607:Domains
577:UniProt
506:glycans
309:cd11010
252:PF02265
138:profile
121:MetaCyc
56:CAS no.
2213:Portal
2155:Lyases
1779:Drosha
1704:3.1.21
1672:RecBCD
1650:3.1.11
1270:PETase
1178:Lipase
1058:
1014:171268
1012:
971:
964:342320
961:
922:
887:
879:
844:
809:
801:
661:P24289
647:Symbol
582:P24021
568:Symbol
394:enzyme
363:PDBsum
337:
327:
297:SUPFAM
265:CL0368
238:Symbol
185:PubMed
167:Search
157:PDBsum
97:ExPASy
85:BRENDA
73:IntEnz
44:EC no.
2107:Types
1788:Dicer
1743:;see
1569:3.1.6
1539:PDE4B
1535:PDE4A
1473:3.1.4
1442:IMPA3
1438:IMPA2
1434:IMPA1
1312:3.1.3
1280:3.1.2
1130:3.1.1
973:41223
885:S2CID
807:S2CID
293:SCOPe
284:SCOP2
133:PRIAM
2199:list
2192:EC7
2186:list
2179:EC6
2173:list
2166:EC5
2160:list
2153:EC4
2147:list
2140:EC3
2134:list
2127:EC2
2121:list
2114:EC1
1706:-31:
1652:-16:
1638:and
1544:PDE5
1530:PDE3
1525:PDE2
1520:PDE1
1412:PTEN
1397:OCRL
1390:PP2A
1339:ALPP
1334:ALPL
1329:ALPI
1123:3.1)
1072:and
1070:Pfam
1056:PMID
1010:PMID
969:PMID
920:PMID
877:PMID
842:PMID
799:PMID
726:Uses
650:NuP1
571:NucS
518:two
439:and
356:PDBj
352:PDBe
335:ECOD
325:Pfam
289:1ak0
261:clan
259:Pfam
247:Pfam
197:NCBI
154:PDBe
109:KEGG
1861:4/5
1046:doi
1000:doi
996:250
959:PMC
951:doi
912:doi
869:doi
834:doi
830:180
791:doi
343:PDB
304:CDD
173:PMC
145:PDB
2239::
1819:2C
1814:2B
1809:2A
1790::
1781::
1571::
1451::
1440:,
1436:,
1352:)/
1314::
1282::
1251:A2
1246:A1
1132::
1121:EC
1115::
1076::
1054:.
1042:26
1040:.
1036:.
1008:.
994:.
990:.
967:.
957:.
945:.
941:.
918:.
908:47
906:.
883:.
875:.
865:32
863:.
840:.
828:.
805:.
797:.
789:.
779:14
777:.
765:^
535:.
499:pH
451:.
384:EC
354:;
350:;
333:/
295:/
291:/
2215::
2201:)
2197:(
2188:)
2184:(
2175:)
2171:(
2162:)
2158:(
2149:)
2145:(
2136:)
2132:(
2123:)
2119:(
1943:e
1936:t
1929:v
1856:3
1851:2
1846:1
1804:1
1642:)
1555:)
1551:(
1537:/
1513:1
1501:D
1496:C
1475::
1356:/
1348:(
1256:B
1190:/
1119:(
1105:e
1098:t
1091:v
1062:.
1048::
1016:.
1002::
975:.
953::
947:7
926:.
914::
891:.
871::
848:.
836::
813:.
793::
785::
487:,
382:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.