458:
1743:
450:
26:
597:
concentrations and concentrations of NADH that are saturating, NADH inhibits the peroxidase activity of the NADH peroxidase by converting the enzyme to an unstable intermediate. NAD behaves as an activator by reversing the equilibria that lead to the unstable intermediate, thus converting the enzyme
686:"Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis"
908:
Crane EJ, Parsonage D, Poole LB, Claiborne A (October 1995). "Analysis of the kinetic mechanism of enterococcal NADH peroxidase reveals catalytic roles for NADH complexes with both oxidized and two-electron-reduced enzyme forms".
292:
873:
Crane EJ, Yeh JI, Luba J, Claiborne A (August 2000). "Analysis of the kinetic and redox properties of the NADH peroxidase R303M mutant: correlation with the crystal structure".
981:"The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of a flavin peroxidase for reduced diphosphopyridine nucleotide"
201:
446:
H. Glu-14 participates in forming the tight dimer interface that limits solvent accessibility, important for maintaining the oxidation state of the sulfenic acid.
1072:
Hansson L, Häggström MH (1984). "Effects of growth conditions on the activities of superoxide dismutase and NADH-oxidase/NADH-peroxidase inStreptococcus lactis".
649:
The actual function of NADH peroxidases and oxidases in plants is still unclear, but they could act in early signaling of oxidative stress through producing H
944:
Crane EJ, Parsonage D, Claiborne A (February 1996). "The active-site histidine-10 of enterococcal NADH peroxidase is not essential for catalytic activity".
738:"Heterogeneity among the flavin-containing NADH peroxidases of group D streptococci. Analysis of the enzyme from Streptococcus faecalis ATCC 9790"
1391:
1261:
434:
His10 of the NADH peroxidase is located near the N-terminus of the R1 helix within the FAD-binding site. One of the oxygen atoms of Cys42-SO
220:
442:
and to Cys42 N terminus. The His10 functions in part to stabilize the unusual Cys42-SOH redox center. Arg303 also stabilizes the Cys42-SO
1107:
Gordon J, Holman RA, McLeod JW (October 1953). "Further observations on the production of hydrogen peroxide by anaerobic bacteria".
213:
1235:
849:
164:
1183:
Chen SX, Schopfer P (March 1999). "Hydroxyl-radical production in physiological reactions. A novel function of peroxidase".
457:
1783:
1462:
357:
334:
1768:
684:
La
Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A (December 2007).
505:
intermediate has been observed, however, and the precise details of Cys42-SOH reduction have not been elucidated.
1773:
158:
497:
by the Cys42-thiolate, yielding E•NADH; and (4) rate-limiting hydride transfer from bound NADH, regenerating EH
140:
1618:
145:
630:. Second, the enzyme presents an additional mechanism for regeneration of the NAD essential to the strictly
449:
311:
25:
631:
373:
277:
225:
481:
The kinetic mechanism of the wild-type peroxidase involves (1) NADH reduction of E(FAD, Cys42-SOH) to EH
133:
1733:
1021:
1719:
1706:
1693:
1680:
1667:
1654:
1641:
1603:
1228:
68:
1613:
1567:
1510:
1374:
1252:
330:
246:
161:
51:
85:
1515:
1379:
1289:
475:
369:
1778:
1349:
1299:
461:
Four residues essential for active site functionality in NADH Peroxidase, Adapted from PDB 2NPX
833:
825:
1536:
1455:
1294:
1279:
471:
428:
431:
with respect to chain fold and location as well as conformation of the prosthetic group FAD
1608:
1221:
478:
of the peroxide bond to catalyze the two-electron reduction of hydrogen peroxide to water.
474:
is unique in that it utilizes the Cys42 thiol/sulfenic acid (-SH/-SOH) redox couple in the
121:
8:
1572:
1213:
1160:
1143:
615:
353:
97:
63:
56:
1763:
1505:
1364:
1165:
1089:
798:
781:
715:
997:
980:
841:
754:
737:
176:
1200:
1196:
1124:
1054:
1040:
1002:
961:
926:
890:
855:
845:
803:
759:
707:
702:
685:
342:
261:
152:
32:
1169:
1144:"Effect of Aluminium on Oxidative Stress Related Enzymes Activities in Barley Roots"
1093:
719:
668:
formation by NADH peroxidase and oxidase in cell wall loosening and reconstruction.
1551:
1546:
1520:
1448:
1274:
1192:
1155:
1116:
1081:
1044:
1036:
992:
953:
918:
882:
837:
793:
749:
697:
627:
1598:
1582:
1495:
1409:
1404:
1359:
1354:
388:
453:
Alignment of NADH, FAD and
Cysteine 42 in NADH Peroxidase, Adapted from PDB 2NPX
180:
1747:
1636:
1577:
1244:
384:
196:
1757:
1541:
1500:
1395:
171:
1120:
1490:
1204:
1128:
1006:
894:
859:
711:
485:(FAD, Cys42-SH) in an initial priming step; (2) rapid binding of NADH to EH
338:
965:
930:
807:
763:
387:, specifically those acting on a peroxide as acceptor (peroxidases). The
30:
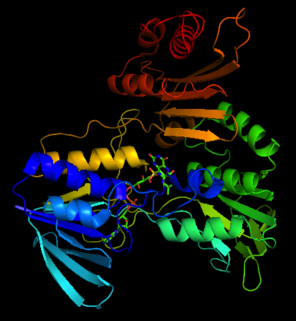
The structure of NADH peroxidase from
Enterococcus faecalis. Adapted from
1714:
1649:
1485:
1312:
1058:
1049:
922:
249:
109:
1369:
1248:
1085:
315:
238:
957:
886:
36:
1688:
1662:
826:"Crystal Structures of Oxidized and Reduced Forms of NADH Peroxidase"
643:
439:
257:
1742:
1284:
618:
conditions and represents an enzymatic defense available against H
683:
634:
of this organism. The enzyme may also protect against exogenous H
128:
1701:
1471:
1141:
253:
208:
104:
92:
80:
1675:
1429:
1424:
1419:
1414:
361:
907:
1342:
1337:
1332:
1327:
1322:
1317:
1307:
302:
The presumed function of NADH peroxidase is to inactivate H
116:
1440:
1243:
614:
NADH eliminates potentially toxic hydrogen peroxide under
943:
1142:Šimonovičová M, Tamás L, Huttová J, Mistrík I (2004).
1731:
872:
779:
280:
782:"NADH binding site and catalysis of NADH peroxidase"
735:
427:The crystal structure of NADH peroxidase resembles
1106:
286:
1135:
832:. Methods in Enzymology. Vol. 353. pp.
780:Stehle T, Claiborne A, Schulz GE (January 1993).
1755:
1071:
598:to the kinetically active complex that reduces H
326:causes damage to essential cellular components.
660:An alternative role may include regulation of H
1456:
1229:
1022:"DPNH peroxidase: effector activities of DPN"
775:
773:
736:Miller H, Poole LB, Claiborne A (June 1990).
731:
729:
314:during glycerol metabolism or dismutation of
1182:
1065:
823:
413:nicotinamide adenine dinucleotide peroxidase
1100:
677:
1463:
1449:
1236:
1222:
819:
817:
770:
726:
581:Inhibitors include Ag, Cl, Co, Cu, Hg, NaN
310:generated within the cell, for example by
1176:
1159:
1048:
1013:
996:
972:
937:
901:
866:
797:
753:
701:
1019:
978:
456:
448:
814:
438:H is hydrogen-bonded both to the His10
405:diphosphopyridine nucleotide peroxidase
1756:
830:Redox Cell Biology and Genetics Part B
609:
1444:
1217:
465:
393:NADH:hydrogen-peroxide oxidoreductase
383:This enzyme belongs to the family of
395:. Other names in common use include
287:{\displaystyle \rightleftharpoons }
13:
1161:10.1023/B:BIOP.0000033454.95515.8a
799:10.1111/j.1432-1033.1993.tb19889.x
14:
1795:
1741:
1197:10.1046/j.1432-1327.1999.00199.x
703:10.1111/j.1365-2958.2007.05987.x
380:intermediate has been observed.
24:
281:
1:
1029:Biochem. Biophys. Res. Commun
998:10.1016/S0021-9258(18)64952-X
842:10.1016/S0076-6879(02)53035-4
755:10.1016/S0021-9258(19)38750-2
671:
1041:10.1016/0006-291X(77)91267-0
824:Yeh JI, Claiborne A (2002).
642:and contribute to bacterial
422:
312:glycerol-3-phosphate oxidase
7:
1470:
1020:Dolin MI (September 1977).
10:
1800:
1784:Enzymes of known structure
376:, however no discrete FADH
1627:
1619:Michaelis–Menten kinetics
1591:
1560:
1529:
1478:
1390:
1260:
470:The NADH peroxidase from
219:
207:
195:
190:
186:
170:
151:
139:
127:
115:
103:
91:
79:
74:
62:
50:
45:
23:
18:
1511:Diffusion-limited enzyme
1375:Iodothyronine deiodinase
391:of this enzyme class is
1769:NADPH-dependent enzymes
1380:Iodotyrosine deiodinase
1290:Cytochrome c peroxidase
1121:10.1002/path.1700660224
979:Dolin MI (March 1957).
632:fermentative metabolism
1774:NADH-dependent enzymes
1350:Horseradish peroxidase
1300:Glutathione peroxidase
462:
454:
288:
1604:Eadie–Hofstee diagram
1537:Allosteric regulation
1295:Eosinophil peroxidase
1280:Fatty-acid peroxidase
558:E•NADH + H → EH
472:Enterococcus faecalis
460:
452:
429:glutathione reductase
289:
1614:Lineweaver–Burk plot
1074:Current Microbiology
509:E + NADH → (EH
489:; (3) reduction of H
476:heterolytic cleavage
278:
923:10.1021/bi00043a016
610:Biological Function
333:of this enzyme are
1573:Enzyme superfamily
1506:Enzyme promiscuity
1365:Thyroid peroxidase
1148:Biologia Plantarum
1109:J Pathol Bacteriol
1086:10.1007/BF01626563
551:→ E•NADH + H
513:'•NAD)* → EH
501:. No discrete FADH
466:Reaction mechanism
463:
455:
352:, whereas its two
284:
1729:
1728:
1438:
1437:
958:10.1021/bi952347y
887:10.1021/bi000553m
851:978-0-12-182256-9
589:. At suboptimal H
532:+ NADH → EH
368:. It employs one
262:chemical reaction
235:
234:
231:
230:
134:metabolic pathway
1791:
1746:
1745:
1737:
1609:Hanes–Woolf plot
1552:Enzyme activator
1547:Enzyme inhibitor
1521:Enzyme catalysis
1465:
1458:
1451:
1442:
1441:
1275:NADPH peroxidase
1238:
1231:
1224:
1215:
1214:
1209:
1208:
1180:
1174:
1173:
1163:
1139:
1133:
1132:
1104:
1098:
1097:
1069:
1063:
1062:
1052:
1026:
1017:
1011:
1010:
1000:
976:
970:
969:
941:
935:
934:
917:(43): 14114–24.
905:
899:
898:
881:(34): 10353–64.
870:
864:
863:
821:
812:
811:
801:
777:
768:
767:
757:
733:
724:
723:
705:
681:
628:oxidative stress
517:'•NAD → EH
417:NADH2 peroxidase
293:
291:
290:
285:
188:
187:
39:
28:
16:
15:
1799:
1798:
1794:
1793:
1792:
1790:
1789:
1788:
1754:
1753:
1752:
1740:
1732:
1730:
1725:
1637:Oxidoreductases
1623:
1599:Enzyme kinetics
1587:
1583:List of enzymes
1556:
1525:
1496:Catalytic triad
1474:
1469:
1439:
1434:
1386:
1360:Myeloperoxidase
1355:Lactoperoxidase
1270:NADH peroxidase
1256:
1245:Oxidoreductases
1242:
1212:
1185:Eur. J. Biochem
1181:
1177:
1140:
1136:
1105:
1101:
1070:
1066:
1024:
1018:
1014:
977:
973:
942:
938:
906:
902:
871:
867:
852:
822:
815:
786:Eur. J. Biochem
778:
771:
748:(17): 9857–63.
734:
727:
682:
678:
674:
667:
663:
656:
652:
641:
637:
625:
621:
612:
605:
601:
596:
592:
588:
584:
576:
573:•NAD → EH
572:
565:
561:
554:
550:
546:
542:
535:
531:
524:
520:
516:
512:
504:
500:
496:
492:
488:
484:
468:
445:
437:
425:
409:NADH-peroxidase
397:DPNH peroxidase
389:systematic name
385:oxidoreductases
379:
365:
350:
346:
325:
321:
309:
305:
297:
279:
276:
275:
274:
270:
243:NADH peroxidase
41:
31:
19:NADH peroxidase
12:
11:
5:
1797:
1787:
1786:
1781:
1776:
1771:
1766:
1751:
1750:
1727:
1726:
1724:
1723:
1710:
1697:
1684:
1671:
1658:
1645:
1631:
1629:
1625:
1624:
1622:
1621:
1616:
1611:
1606:
1601:
1595:
1593:
1589:
1588:
1586:
1585:
1580:
1575:
1570:
1564:
1562:
1561:Classification
1558:
1557:
1555:
1554:
1549:
1544:
1539:
1533:
1531:
1527:
1526:
1524:
1523:
1518:
1513:
1508:
1503:
1498:
1493:
1488:
1482:
1480:
1476:
1475:
1468:
1467:
1460:
1453:
1445:
1436:
1435:
1433:
1432:
1427:
1422:
1417:
1412:
1407:
1401:
1399:
1388:
1387:
1385:
1384:
1383:
1382:
1377:
1367:
1362:
1357:
1352:
1347:
1346:
1345:
1340:
1335:
1330:
1325:
1320:
1315:
1310:
1297:
1292:
1287:
1282:
1277:
1272:
1266:
1264:
1258:
1257:
1241:
1240:
1233:
1226:
1218:
1211:
1210:
1175:
1154:(2): 261–266.
1134:
1099:
1080:(6): 345–351.
1064:
1035:(1): 393–400.
1012:
971:
936:
900:
865:
850:
813:
792:(1–2): 221–6.
769:
725:
696:(5): 1148–63.
690:Mol. Microbiol
675:
673:
670:
665:
661:
654:
650:
639:
635:
623:
619:
616:aerobic growth
611:
608:
603:
599:
594:
590:
586:
582:
579:
578:
574:
570:
567:
563:
559:
556:
552:
548:
544:
540:
537:
533:
529:
526:
522:
518:
514:
510:
502:
498:
494:
490:
486:
482:
467:
464:
443:
435:
424:
421:
401:NAD peroxidase
377:
363:
348:
344:
323:
319:
318:, before the H
307:
303:
300:
299:
295:
283:
272:
268:
233:
232:
229:
228:
223:
217:
216:
211:
205:
204:
199:
193:
192:
184:
183:
174:
168:
167:
156:
149:
148:
143:
137:
136:
131:
125:
124:
119:
113:
112:
107:
101:
100:
95:
89:
88:
83:
77:
76:
72:
71:
66:
60:
59:
54:
48:
47:
43:
42:
29:
21:
20:
9:
6:
4:
3:
2:
1796:
1785:
1782:
1780:
1779:Flavoproteins
1777:
1775:
1772:
1770:
1767:
1765:
1762:
1761:
1759:
1749:
1744:
1739:
1738:
1735:
1721:
1717:
1716:
1711:
1708:
1704:
1703:
1698:
1695:
1691:
1690:
1685:
1682:
1678:
1677:
1672:
1669:
1665:
1664:
1659:
1656:
1652:
1651:
1646:
1643:
1639:
1638:
1633:
1632:
1630:
1626:
1620:
1617:
1615:
1612:
1610:
1607:
1605:
1602:
1600:
1597:
1596:
1594:
1590:
1584:
1581:
1579:
1578:Enzyme family
1576:
1574:
1571:
1569:
1566:
1565:
1563:
1559:
1553:
1550:
1548:
1545:
1543:
1542:Cooperativity
1540:
1538:
1535:
1534:
1532:
1528:
1522:
1519:
1517:
1514:
1512:
1509:
1507:
1504:
1502:
1501:Oxyanion hole
1499:
1497:
1494:
1492:
1489:
1487:
1484:
1483:
1481:
1477:
1473:
1466:
1461:
1459:
1454:
1452:
1447:
1446:
1443:
1431:
1428:
1426:
1423:
1421:
1418:
1416:
1413:
1411:
1408:
1406:
1403:
1402:
1400:
1397:
1396:peroxiredoxin
1393:
1389:
1381:
1378:
1376:
1373:
1372:
1371:
1368:
1366:
1363:
1361:
1358:
1356:
1353:
1351:
1348:
1344:
1341:
1339:
1336:
1334:
1331:
1329:
1326:
1324:
1321:
1319:
1316:
1314:
1311:
1309:
1306:
1303:
1302:
1301:
1298:
1296:
1293:
1291:
1288:
1286:
1283:
1281:
1278:
1276:
1273:
1271:
1268:
1267:
1265:
1263:
1259:
1254:
1250:
1246:
1239:
1234:
1232:
1227:
1225:
1220:
1219:
1216:
1206:
1202:
1198:
1194:
1191:(3): 726–35.
1190:
1186:
1179:
1171:
1167:
1162:
1157:
1153:
1149:
1145:
1138:
1130:
1126:
1122:
1118:
1115:(2): 527–37.
1114:
1110:
1103:
1095:
1091:
1087:
1083:
1079:
1075:
1068:
1060:
1056:
1051:
1050:2027.42/22844
1046:
1042:
1038:
1034:
1030:
1023:
1016:
1008:
1004:
999:
994:
991:(1): 557–73.
990:
986:
985:J. Biol. Chem
982:
975:
967:
963:
959:
955:
952:(7): 2380–7.
951:
947:
940:
932:
928:
924:
920:
916:
912:
904:
896:
892:
888:
884:
880:
876:
869:
861:
857:
853:
847:
843:
839:
835:
831:
827:
820:
818:
809:
805:
800:
795:
791:
787:
783:
776:
774:
765:
761:
756:
751:
747:
743:
742:J. Biol. Chem
739:
732:
730:
721:
717:
713:
709:
704:
699:
695:
691:
687:
680:
676:
669:
658:
647:
645:
633:
629:
617:
607:
568:
557:
538:
527:
508:
507:
506:
479:
477:
473:
459:
451:
447:
441:
432:
430:
420:
418:
414:
410:
406:
402:
398:
394:
390:
386:
381:
375:
371:
367:
359:
355:
351:
340:
336:
332:
327:
317:
313:
266:
265:
264:
263:
259:
255:
251:
248:
244:
240:
227:
224:
222:
218:
215:
212:
210:
206:
203:
200:
198:
194:
189:
185:
182:
178:
175:
173:
172:Gene Ontology
169:
166:
163:
160:
157:
154:
150:
147:
144:
142:
138:
135:
132:
130:
126:
123:
120:
118:
114:
111:
110:NiceZyme view
108:
106:
102:
99:
96:
94:
90:
87:
84:
82:
78:
73:
70:
67:
65:
61:
58:
55:
53:
49:
44:
38:
34:
27:
22:
17:
1715:Translocases
1712:
1699:
1686:
1673:
1660:
1650:Transferases
1647:
1634:
1491:Binding site
1304:
1269:
1188:
1184:
1178:
1151:
1147:
1137:
1112:
1108:
1102:
1077:
1073:
1067:
1032:
1028:
1015:
988:
984:
974:
949:
946:Biochemistry
945:
939:
914:
911:Biochemistry
910:
903:
878:
875:Biochemistry
874:
868:
829:
789:
785:
745:
741:
693:
689:
679:
659:
648:
613:
585:, Pb, and SO
580:
480:
469:
433:
426:
416:
412:
408:
404:
400:
396:
392:
382:
328:
301:
267:NADH + H + H
242:
236:
98:BRENDA entry
1486:Active site
1262:1.11.1.1-14
1249:peroxidases
86:IntEnz view
46:Identifiers
1758:Categories
1689:Isomerases
1663:Hydrolases
1530:Regulation
1370:Deiodinase
672:References
626:-mediated
543:•NADH* + H
331:substrates
316:superoxide
239:enzymology
155:structures
122:KEGG entry
69:9032-24-0
1764:EC 1.11.1
1568:EC number
1392:1.11.1.15
644:virulence
521:+ NAD + H
440:imidazole
423:Structure
294:NAD + 2 H
282:⇌
258:catalyzes
75:Databases
1592:Kinetics
1516:Cofactor
1479:Activity
1285:Catalase
1205:10103001
1170:34802416
1129:13118459
1094:27660179
1007:13416259
895:10956025
860:12078517
720:40046805
712:17971082
562:•NAD + H
370:cofactor
354:products
252:) is an
250:1.11.1.1
226:proteins
214:articles
202:articles
159:RCSB PDB
57:1.11.1.1
40:.
1748:Biology
1702:Ligases
1472:Enzymes
966:8652580
931:7578008
808:8425532
764:2161844
181:QuickGO
146:profile
129:MetaCyc
64:CAS no.
1734:Portal
1676:Lyases
1203:
1168:
1127:
1092:
1059:199166
1057:
1005:
964:
929:
893:
858:
848:
806:
762:
718:
710:
536:•NADH*
415:, and
341:, and
329:The 3
254:enzyme
209:PubMed
191:Search
177:AmiGO
165:PDBsum
105:ExPASy
93:BRENDA
81:IntEnz
52:EC no.
1628:Types
1255:1.11)
1166:S2CID
1090:S2CID
1025:(PDF)
834:44–54
716:S2CID
577:+ NAD
256:that
141:PRIAM
1720:list
1713:EC7
1707:list
1700:EC6
1694:list
1687:EC5
1681:list
1674:EC4
1668:list
1661:EC3
1655:list
1648:EC2
1642:list
1635:EC1
1201:PMID
1125:PMID
1055:PMID
1003:PMID
962:PMID
927:PMID
891:PMID
856:PMID
846:ISBN
804:PMID
760:PMID
708:PMID
360:and
356:are
335:NADH
260:the
241:, a
221:NCBI
162:PDBe
117:KEGG
37:2NPX
1305:GPX
1193:doi
1189:260
1156:doi
1117:doi
1082:doi
1045:hdl
1037:doi
993:doi
989:225
954:doi
919:doi
883:doi
838:doi
794:doi
790:211
750:doi
746:265
698:doi
374:FAD
358:NAD
237:In
197:PMC
153:PDB
33:PDB
1760::
1253:EC
1247::
1199:.
1187:.
1164:.
1152:48
1150:.
1146:.
1123:.
1113:66
1111:.
1088:.
1078:10
1076:.
1053:.
1043:.
1033:78
1031:.
1027:.
1001:.
987:.
983:.
960:.
950:35
948:.
925:.
915:34
913:.
889:.
879:39
877:.
854:.
844:.
836:.
828:.
816:^
802:.
788:.
784:.
772:^
758:.
744:.
740:.
728:^
714:.
706:.
694:66
692:.
688:.
657:.
646:.
606:.
569:EH
539:EH
528:EH
419:.
411:,
407:,
403:,
399:,
372:,
337:,
247:EC
179:/
35::
1736::
1722:)
1718:(
1709:)
1705:(
1696:)
1692:(
1683:)
1679:(
1670:)
1666:(
1657:)
1653:(
1644:)
1640:(
1464:e
1457:t
1450:v
1430:6
1425:5
1420:4
1415:3
1410:2
1405:1
1398:)
1394:(
1343:8
1338:7
1333:6
1328:5
1323:4
1318:3
1313:2
1308:1
1251:(
1237:e
1230:t
1223:v
1207:.
1195::
1172:.
1158::
1131:.
1119::
1096:.
1084::
1061:.
1047::
1039::
1009:.
995::
968:.
956::
933:.
921::
897:.
885::
862:.
840::
810:.
796::
766:.
752::
722:.
700::
666:2
664:O
662:2
655:2
653:O
651:2
640:2
638:O
636:2
624:2
622:O
620:2
604:2
602:O
600:2
595:2
593:O
591:2
587:4
583:3
575:2
571:2
566:O
564:2
560:2
555:O
553:2
549:2
547:O
545:2
541:2
534:2
530:2
525:O
523:2
519:2
515:2
511:2
503:2
499:2
495:2
493:O
491:2
487:2
483:2
444:3
436:3
378:2
366:O
364:2
362:H
349:2
347:O
345:2
343:H
339:H
324:2
322:O
320:2
308:2
306:O
304:2
298:O
296:2
273:2
271:O
269:2
245:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.