20:
5327:
3016:
107:
5313:
1585:
626:
720:
5351:
5339:
1597:
observed test statistic, which is 3.13, versus an expected value of 2.06. The blue point corresponds to the fifth smallest test statistic, which is -1.75, versus an expected value of -1.96. The graph suggests that it is unlikely that all the null hypotheses are true, and that most or all instances of a true alternative hypothesis result from deviations in the positive direction.
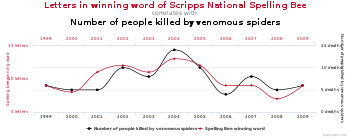
27:(uncorrected multiple comparisons) showing a correlation between the number of letters in a spelling bee's winning word and the number of people in the United States killed by venomous spiders. Given a large enough pool of variables for the same time period, it is possible to find a pair of graphs that show a
1540:
studies, there has been a serious problem with non-replication — a result being strongly statistically significant in one study but failing to be replicated in a follow-up study. Such non-replication can have many causes, but it is widely considered that failure to fully account for the consequences
1596:
under the null hypothesis. The departure of the upper tail of the distribution from the expected trend along the diagonal is due to the presence of substantially more large test statistic values than would be expected if all null hypotheses were true. The red point corresponds to the fourth largest
122:
Multiple comparisons arise when a statistical analysis involves multiple simultaneous statistical tests, each of which has a potential to produce a "discovery". A stated confidence level generally applies only to each test considered individually, but often it is desirable to have a confidence level
1615:
are positively correlated, which commonly occurs in practice. . On the other hand, the approach remains valid even in the presence of correlation among the test statistics, as long as the
Poisson distribution can be shown to provide a good approximation for the number of significant results. This
1610:
For example, if 1000 independent tests are performed, each at level α = 0.05, we expect 0.05 × 1000 = 50 significant tests to occur when all null hypotheses are true. Based on the
Poisson distribution with mean 50, the probability of observing more than 61 significant tests is less than
1606:
as a model for the number of significant results at a given level α that would be found when all null hypotheses are true. If the observed number of positives is substantially greater than what should be expected, this suggests that there are likely to be some true positives among the significant
127:
Suppose the treatment is a new way of teaching writing to students, and the control is the standard way of teaching writing. Students in the two groups can be compared in terms of grammar, spelling, organization, content, and so on. As more attributes are compared, it becomes increasingly likely
172:
level will contain the true value of the parameter in 95% of samples. However, if one considers 100 confidence intervals simultaneously, each with 95% coverage probability, the expected number of non-covering intervals is 5. If the intervals are statistically independent from each other, the
144:
In both examples, as the number of comparisons increases, it becomes more likely that the groups being compared will appear to differ in terms of at least one attribute. Our confidence that a result will generalize to independent data should generally be weaker if it is observed as part of an
1611:
0.05, so if more than 61 significant results are observed, it is very likely that some of them correspond to situations where the alternative hypothesis holds. A drawback of this approach is that it overstates the evidence that some of the alternative hypotheses are true when the
148:
For example, if one test is performed at the 5% level and the corresponding null hypothesis is true, there is only a 5% risk of incorrectly rejecting the null hypothesis. However, if 100 tests are each conducted at the 5% level and all corresponding null hypotheses are true, the
1601:
A basic question faced at the outset of analyzing a large set of testing results is whether there is evidence that any of the alternative hypotheses are true. One simple meta-test that can be applied when it is assumed that the tests are independent of each other is to use the
1569:(FDR) is often preferred. The FDR, loosely defined as the expected proportion of false positives among all significant tests, allows researchers to identify a set of "candidate positives" that can be more rigorously evaluated in a follow-up study.
1564:
remains the most accepted parameter for ascribing significance levels to statistical tests. Alternatively, if a study is viewed as exploratory, or if significant results can be easily re-tested in an independent study, control of the
854:
139:
in terms of the reduction of any one of a number of disease symptoms. As more symptoms are considered, it becomes increasingly likely that the drug will appear to be an improvement over existing drugs in terms of at least one
948:
1311:
1479:
1139:
1233:
1024:
680:
62:
The larger the number of inferences made, the more likely erroneous inferences become. Several statistical techniques have been developed to address this problem, for example, by requiring a
161:) is 5. If the tests are statistically independent from each other (i.e. are performed on independent samples), the probability of at least one incorrect rejection is approximately 99.4%.
118:
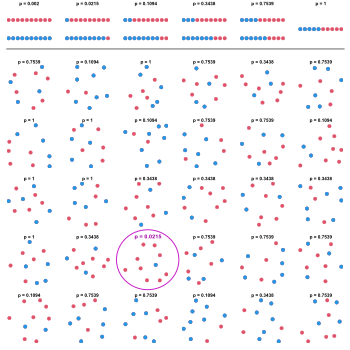
Although the 30 samples were all simulated under the null, one of the resulting p-values is small enough to produce a false rejection at the typical level 0.05 in the absence of correction.
1616:
scenario arises, for instance, when mining significant frequent itemsets from transactional datasets. Furthermore, a careful two stage analysis can bound the FDR at a pre-specified level.
1572:
The practice of trying many unadjusted comparisons in the hope of finding a significant one is a known problem, whether applied unintentionally or deliberately, is sometimes called "
116:
of the null hypothesis that blue and red are equally probable is performed. The first row shows the possible p-values as a function of the number of blue and red dots in the sample.
123:
for the whole family of simultaneous tests. Failure to compensate for multiple comparisons can have important real-world consequences, as illustrated by the following examples:
218:, we reject the null hypothesis if the test is declared significant. We do not reject the null hypothesis if the test is non-significant. Summing each type of outcome over all
886:
86:. Over the ensuing decades, many procedures were developed to address the problem. In 1996, the first international conference on multiple comparison procedures took place in
1528:. A different set of techniques have been developed for "large-scale multiple testing", in which thousands or even greater numbers of tests are performed. For example, in
1046:
457:
375:
1371:
567:
532:
418:
341:
1345:
777:
308:
1553:, often leading to the testing of large numbers of hypotheses with no prior basis for expecting many of the hypotheses to be true. In this situation, very high
1536:, expression levels of tens of thousands of genes can be measured, and genotypes for millions of genetic markers can be measured. Particularly in the field of
1506:
1159:
700:
894:
2073:
2368:
2305:
1238:
176:
Techniques have been developed to prevent the inflation of false positive rates and non-coverage rates that occur with multiple statistical tests.
2169:
746:
refers to making statistical tests more stringent in order to counteract the problem of multiple testing. The best known such adjustment is the
2492:
Phipson, B.; Smyth, G. K. (2010). "Permutation P-values Should Never Be Zero: Calculating Exact P-values when
Permutations are Randomly Drawn".
1376:
4448:
1969:
4953:
2590:
1054:
5103:
2071:
Benjamini, Yoav; Hochberg, Yosef (1995). "Controlling the false discovery rate: a practical and powerful approach to multiple testing".
4727:
3368:
2915:
2447:
Farcomeni, A. (2008). "A Review of Modern
Multiple Hypothesis Testing, with particular attention to the false discovery proportion".
2167:
Efron, Bradley; Tibshirani, Robert; Storey, John D.; Tusher, Virginia (2001). "Empirical Bayes analysis of a microarray experiment".
4501:
1752:
888:
increases as the number of comparisons increases. If we do not assume that the comparisons are independent, then we can still say:
5377:
4940:
1164:
960:
1800:
1524:
Traditional methods for multiple comparisons adjustments focus on correcting for modest numbers of comparisons, often in an
3363:
3063:
652:
3967:
3115:
2939:
1724:
2390:; Vandin, F (June 2012). "An Efficient Rigorous Approach for Identifying Statistically Significant Frequent Itemsets".
1141:. A marginally less conservative correction can be obtained by solving the equation for the family-wise error rate of
602:
4750:
4642:
2990:
2828:
2737:
2583:
1898:
78:
The problem of multiple comparisons received increased attention in the 1950s with the work of statisticians such as
5355:
4928:
4802:
2270:
1740:
184:
The following table defines the possible outcomes when testing multiple null hypotheses. Suppose we have a number
612:
4986:
4647:
4392:
3763:
3353:
3003:
2711:
2697:
1321:, which uniformly delivers more power than the simple Bonferroni correction, by testing only the lowest p-value (
608:
5037:
4249:
4056:
3945:
3903:
3008:
1696:
215:
3977:
5382:
5280:
4239:
3142:
145:
analysis that involves multiple comparisons, rather than an analysis that involves only a single comparison.
4831:
4780:
4765:
4755:
4624:
4496:
4463:
4289:
4244:
4074:
2909:
2874:
2806:
2662:
2576:
5343:
5175:
4976:
4900:
4201:
3955:
3624:
3088:
3030:
2979:
2891:
732:
5060:
5032:
5027:
4775:
4534:
4440:
4420:
4328:
4039:
3857:
3340:
3212:
2787:
1816:
Benjamini, Y. (2010). "Simultaneous and selective inference: Current successes and future challenges".
1757:
1729:
1318:
862:
4792:
4560:
4281:
4206:
4135:
4064:
3984:
3972:
3842:
3830:
3823:
3531:
3252:
1691:
1635:
than the normal quantiles, this suggests that some of the significant results may be true positives.
1550:
498:
489:
479:
469:
66:
for individual comparisons, so as to compensate for the number of inferences being made. Methods for
638:
128:
that the treatment and control groups will appear to differ on at least one attribute due to random
5275:
5042:
4905:
4590:
4555:
4519:
4304:
3746:
3655:
3614:
3526:
3217:
3056:
2965:
2960:
2925:
2809:
2727:
2682:
2677:
1890:
1708:
1048:. The most conservative method, which is free of dependence and distributional assumptions, is the
5184:
4797:
4737:
4674:
4312:
4296:
4034:
3896:
3886:
3736:
3650:
2933:
1916:"Adjusting for multiple testing when reporting research results: the Bonferroni vs Holm methods"
5222:
5152:
4945:
4882:
4637:
4524:
3521:
3418:
3325:
3204:
3103:
2920:
2896:
2840:
2782:
1659:
1632:
1561:
1546:
767:
751:
460:
112:
30 samples of 10 dots of random color (blue or red) are observed. On each sample, a two-tailed
67:
19:
5247:
5189:
5132:
4958:
4851:
4760:
4486:
4370:
4229:
4221:
4111:
4103:
3918:
3814:
3792:
3751:
3716:
3683:
3629:
3604:
3559:
3498:
3458:
3260:
3083:
3025:
2869:
2769:
2743:
2722:
2702:
2639:
2622:
2599:
2362:
2299:
1767:
1734:
1713:
1513:
1509:
1314:
1049:
1031:
954:
747:
429:
347:
63:
52:
1882:
1350:
849:{\displaystyle {\bar {\alpha }}=1-\left(1-\alpha _{\{{\text{per comparison}}\}}\right)^{m}.}
750:, but other methods have been developed. Such methods are typically designed to control the
5170:
4745:
4694:
4670:
4632:
4550:
4529:
4481:
4360:
4338:
4307:
4216:
4093:
4044:
3962:
3935:
3891:
3847:
3609:
3385:
3265:
2885:
2823:
2817:
2383:
2113:
1988:
1669:
1644:
1603:
1566:
1525:
755:
545:
534:
is the number of rejected null hypotheses (also called "discoveries", either true or false)
505:
396:
319:
173:
probability that at least one interval does not contain the population parameter is 99.4%.
169:
165:
28:
8:
5317:
5242:
5165:
4846:
4610:
4603:
4565:
4473:
4453:
4425:
4158:
4024:
4019:
4009:
4001:
3819:
3780:
3670:
3660:
3569:
3348:
3304:
3222:
3147:
3049:
2985:
2950:
2859:
1883:
1681:
1675:
1664:
1554:
1541:
of making multiple comparisons is one of the causes. It has been argued that advances in
1537:
1485:
1324:
287:
70:
give the probability of false positives resulting from the multiple comparisons problem.
2117:
2000:
1992:
1964:
5331:
5142:
4996:
4892:
4841:
4717:
4614:
4598:
4575:
4352:
4086:
4069:
4029:
3940:
3835:
3797:
3768:
3728:
3688:
3634:
3551:
3237:
3232:
3020:
2627:
2527:
2501:
2480:
2399:
2345:
2320:
2246:
2213:
2194:
2186:
2131:
2082:
2048:
2023:
2004:
1978:
1940:
1915:
1841:
1491:
1144:
685:
2565:
comic about the multiple comparisons problem, using jelly beans and acne as an example
2166:
2144:
2101:
1560:
For large-scale testing problems where the goal is to provide definitive results, the
91:
5326:
5237:
5207:
5199:
5019:
5010:
4935:
4866:
4722:
4707:
4682:
4570:
4511:
4377:
4365:
3991:
3908:
3852:
3775:
3619:
3541:
3320:
3194:
3015:
2777:
2764:
2754:
2667:
2644:
2636:
2632:
2607:
2519:
2472:
2350:
2290:
2251:
2233:
2149:
2053:
2035:
2008:
1945:
1894:
1833:
1796:
1686:
56:
2531:
2484:
5262:
5217:
4981:
4968:
4861:
4836:
4770:
4702:
4580:
4188:
4081:
4014:
3927:
3874:
3693:
3564:
3358:
3242:
3157:
3124:
2955:
2649:
2511:
2464:
2456:
2409:
2340:
2332:
2285:
2241:
2225:
2198:
2178:
2139:
2121:
2043:
1996:
1935:
1927:
1845:
1825:
1719:
2552:
943:{\displaystyle {\bar {\alpha }}\leq m\cdot \alpha _{\{{\text{per comparison}}\}},}
634:
5179:
4923:
4785:
4712:
4387:
4261:
4234:
4211:
4180:
3807:
3802:
3756:
3486:
3137:
2971:
2901:
2854:
2436:
1762:
590:
421:
150:
4669:
5128:
5123:
3586:
3516:
3162:
2657:
2182:
1965:"The look-elsewhere effect from a unified Bayesian and frequentist perspective"
1624:
1620:
1612:
1593:
1533:
154:
129:
2540:
Resampling-based
Multiple Testing: Examples and Methods for p-Value Adjustment
2336:
1028:
There are different ways to assure that the family-wise error rate is at most
859:
Hence, unless the tests are perfectly positively dependent (i.e., identical),
5371:
5285:
5252:
5115:
5076:
4887:
4856:
4320:
4274:
3879:
3581:
3408:
3172:
3167:
2795:
2672:
2460:
2237:
2039:
1772:
1306:{\displaystyle \alpha _{\{{\text{per comparison}}\}}=1-{(1-{\alpha })}^{1/m}}
113:
95:
24:
2413:
2126:
1508:
from the prior-to-posterior volume ratio. Continuous generalizations of the
106:
5227:
5160:
5137:
5052:
4382:
3678:
3576:
3511:
3453:
3438:
3375:
3330:
2945:
2523:
2515:
2476:
2354:
2255:
2153:
2057:
1837:
1829:
1628:
1589:
158:
136:
2553:
A gallery of examples of implausible correlations sourced by data dredging
1949:
1931:
1474:{\displaystyle \alpha _{\mathrm {\{per\ comparison\}} }={\alpha }/(m-i+1)}
5270:
5232:
4915:
4816:
4678:
4491:
4458:
3950:
3867:
3862:
3506:
3463:
3443:
3423:
3413:
3182:
2801:
2732:
2717:
2687:
2229:
1592:
for a simulated set of test statistics that have been standardized to be
1542:
2568:
2468:
1579:
4116:
3596:
3296:
3227:
3177:
3152:
3072:
2617:
2190:
2135:
2086:
1878:
649:
Probability that at least one null hypothesis is wrongly rejected, for
90:. This is an active research area with work being done by, for example
36:
633:
Graphs are unavailable due to technical issues. There is more info on
83:
4269:
4121:
3741:
3536:
3448:
3433:
3428:
3393:
2749:
2387:
1631:
of the test statistics. If the observed quantiles are markedly more
1584:
1573:
2381:
1134:{\displaystyle \alpha _{\mathrm {\{per\ comparison\}} }={\alpha }/m}
3785:
3403:
3280:
3275:
3270:
2506:
2022:
Qu, Hui-Qi; Tien, Matthew; Polychronakos, Constantin (2010-10-01).
1983:
1529:
87:
2404:
5290:
4991:
1859:
1619:
Another common approach that can be used in situations where the
5212:
4193:
4167:
4147:
3398:
3189:
1557:
are expected unless multiple comparisons adjustments are made.
179:
59:
a subset of parameters selected based on the observed values.
3041:
79:
1703:
General methods of alpha adjustment for multiple comparisons
1347:) against the strictest criterion, and the higher p-values (
3132:
2561:
2494:
2024:"Statistical significance in genetic association studies"
2557:
2441:
Multiple
Testing Procedures with Application to Genomics
2099:
2070:
1228:{\displaystyle \alpha _{\mathrm {\{per\ comparison\}} }}
1019:{\displaystyle 0.2649=1-(1-.05)^{6}\leq .05\times 6=0.3}
1876:
1549:
have made it far easier to generate large datasets for
2318:
2268:
1580:
Assessing whether any alternative hypotheses are true
1494:
1379:
1353:
1327:
1241:
1167:
1147:
1057:
1034:
963:
897:
865:
780:
688:
655:
548:
508:
432:
399:
350:
322:
290:
4954:
Autoregressive conditional heteroskedasticity (ARCH)
675:{\displaystyle \alpha _{\text{per comparison}}=0.05}
603:
Family-wise error rate § Controlling procedures
2547:
Multiple comparisons and multiple testing using SAS
2021:
682:, as a function of the number of independent tests
110:Production of a small p-value by multiple testing.
4416:
2102:"Statistical significance for genome-wide studies"
2074:Journal of the Royal Statistical Society, Series B
1500:
1473:
1365:
1339:
1305:
1227:
1153:
1133:
1040:
1018:
942:
880:
848:
694:
674:
613:False discovery rate § Controlling procedures
561:
526:
451:
412:
369:
335:
302:
609:False coverage rate § Controlling procedures
164:The multiple comparisons problem also applies to
5369:
4502:Multivariate adaptive regression splines (MARS)
2170:Journal of the American Statistical Association
1519:
1970:Journal of Cosmology and Astroparticle Physics
1373:) against progressively less strict criteria.
707:
225: yields the following random variables:
3057:
2584:
1913:
2545:P. Westfall, R. Tobias, R. Wolfinger (2011)
2491:
2367:: CS1 maint: multiple names: authors list (
2304:: CS1 maint: multiple names: authors list (
2214:"How does multiple testing correction work?"
1431:
1386:
1255:
1247:
1219:
1174:
1109:
1064:
932:
924:
827:
819:
1962:
765:independent comparisons are performed, the
180:Classification of multiple hypothesis tests
3102:
3064:
3050:
2591:
2577:
2429:F. Bretz, T. Hothorn, P. Westfall (2010),
1877:Kutner, Michael; Nachtsheim, Christopher;
168:. A single confidence interval with a 95%
16:Statistical interpretation with many tests
3715:
2598:
2505:
2446:
2403:
2344:
2289:
2245:
2143:
2125:
2047:
1982:
1939:
1815:
1793:Simultaneous Statistical Inference 2nd Ed
596:
2271:"Deming, data and observational studies"
1956:
1753:Testing hypotheses suggested by the data
1583:
1484:For continuous problems, one can employ
105:
18:
2538:P. H. Westfall and S. S. Young (1993),
2449:Statistical Methods in Medical Research
2100:Storey, JD; Tibshirani, Robert (2003).
1963:Bayer, Adrian E.; Seljak, Uroš (2020).
728:This section may need to be cleaned up.
153:of incorrect rejections (also known as
5370:
5028:Kaplan–Meier estimator (product limit)
2262:
1790:
573:is an observable random variable, and
135:Suppose we consider the efficacy of a
23:An example of coincidence produced by
5101:
4668:
4415:
3714:
3484:
3101:
3045:
2572:
2321:"Data dredging, bias, or confounding"
2312:
2211:
391:is the total number hypotheses tested
5338:
5038:Accelerated failure time (AFT) model
713:
619:
5350:
4633:Analysis of variance (ANOVA, anova)
3485:
2940:Generalized randomized block design
2028:Clinical and Investigative Medicine
51:occurs when one considers a set of
13:
4728:Cochran–Mantel–Haenszel statistics
3354:Pearson product-moment correlation
2423:
1914:Aickin, M; Gensler, H (May 1996).
1532:, when using technologies such as
1428:
1425:
1422:
1419:
1416:
1413:
1410:
1407:
1404:
1401:
1395:
1392:
1389:
1216:
1213:
1210:
1207:
1204:
1201:
1198:
1195:
1192:
1189:
1183:
1180:
1177:
1106:
1103:
1100:
1097:
1094:
1091:
1088:
1085:
1082:
1079:
1073:
1070:
1067:
14:
5394:
2991:Sequential probability ratio test
1885:Applied Linear Statistical Models
472:(also called "false discoveries")
272:Test is declared non-significant
240:Alternative hypothesis is true (H
5349:
5337:
5325:
5312:
5311:
5102:
3014:
2916:Polynomial and rational modeling
2291:10.1111/j.1740-9713.2011.00506.x
2212:Noble, William S. (2009-12-01).
1725:Duncan's new multiple range test
881:{\displaystyle {\bar {\alpha }}}
718:
624:
482:(also called "true discoveries")
188:of null hypotheses, denoted by:
4987:Least-squares spectral analysis
2439:and M. J. van der Laan (2008),
2375:
2319:Smith, G. D., Shah, E. (2002).
2269:Young, S. S., Karr, A. (2011).
2205:
2160:
490:false negatives (Type II error)
64:stricter significance threshold
5378:Statistical hypothesis testing
3968:Mean-unbiased minimum-variance
3071:
2683:Replication versus subsampling
2386:; Pietracaprina, A; Pucci, G;
2093:
2064:
2015:
1907:
1889:. McGraw-Hill Irwin. pp.
1870:
1852:
1809:
1784:
1697:Statistical hypothesis testing
1468:
1450:
1285:
1271:
989:
976:
904:
872:
787:
470:false positives (Type I error)
1:
5281:Geographic information system
4497:Simultaneous equations models
2001:10.1088/1475-7516/2020/10/009
1778:
1741:Benjamini–Hochberg procedure
252:Test is declared significant
101:
4464:Coefficient of determination
4075:Uniformly most powerful test
2910:Response surface methodology
2818:Analysis of variance (Anova)
2431:Multiple Comparisons Using R
1795:. Springer Verlag New York.
1520:Large-scale multiple testing
1161:independent comparisons for
312:
270:
250:
7:
5033:Proportional hazards models
4977:Spectral density estimation
4959:Vector autoregression (VAR)
4393:Maximum posterior estimator
3625:Randomized controlled trial
2980:Randomized controlled trial
1638:
1317:. Another procedure is the
744:Multiple testing correction
733:Multiple testing correction
708:Multiple testing correction
10:
5399:
4793:Multivariate distributions
3213:Average absolute deviation
2183:10.1198/016214501753382129
1758:Texas sharpshooter fallacy
606:
600:
569:are true null hypotheses,
542:hypothesis tests of which
233:Null hypothesis is true (H
73:
5307:
5261:
5198:
5151:
5114:
5110:
5097:
5069:
5051:
5018:
5009:
4967:
4914:
4875:
4824:
4815:
4781:Structural equation model
4736:
4693:
4689:
4664:
4623:
4589:
4543:
4510:
4472:
4439:
4435:
4411:
4351:
4260:
4179:
4143:
4134:
4117:Score/Lagrange multiplier
4102:
4055:
4000:
3926:
3917:
3727:
3723:
3710:
3669:
3643:
3595:
3550:
3532:Sample size determination
3497:
3493:
3480:
3384:
3339:
3313:
3295:
3251:
3203:
3123:
3114:
3110:
3097:
3079:
2999:
2868:
2763:
2696:
2606:
2337:10.1136/bmj.325.7378.1437
1692:Experimentwise error rate
5276:Environmental statistics
4798:Elliptical distributions
4591:Generalized linear model
4520:Simple linear regression
4290:Hodges–Lehmann estimator
3747:Probability distribution
3656:Stochastic approximation
3218:Coefficient of variation
2966:Repeated measures design
2678:Restricted randomization
2549:, 2nd edn, SAS Institute
2461:10.1177/0962280206079046
1709:Closed testing procedure
1313:, which is known as the
730:It has been merged from
49:multiple testing problem
4936:Cross-correlation (XCF)
4544:Non-standard predictors
3978:Lehmann–Scheffé theorem
3651:Adaptive clinical trial
2414:10.1145/2220357.2220359
2127:10.1073/pnas.1530509100
1623:can be standardized to
1041:{\displaystyle \alpha }
452:{\displaystyle m-m_{0}}
370:{\displaystyle m-m_{0}}
5332:Mathematics portal
5153:Engineering statistics
5061:Nelson–Aalen estimator
4638:Analysis of covariance
4525:Ordinary least squares
4449:Pearson product-moment
3853:Statistical functional
3764:Empirical distribution
3597:Controlled experiments
3326:Frequency distribution
3104:Descriptive statistics
3021:Mathematics portal
2783:Ordinary least squares
2516:10.2202/1544-6115.1585
1881:; Li, William (2005).
1830:10.1002/bimj.200900299
1730:Holm–Bonferroni method
1660:Family-wise error rate
1598:
1562:family-wise error rate
1547:information technology
1502:
1475:
1367:
1366:{\displaystyle i>1}
1341:
1319:Holm–Bonferroni method
1307:
1229:
1155:
1135:
1042:
1020:
944:
882:
850:
768:family-wise error rate
752:family-wise error rate
696:
676:
597:Controlling procedures
563:
528:
461:alternative hypotheses
459:is the number of true
453:
424:, an unknown parameter
420:is the number of true
414:
371:
337:
304:
119:
68:family-wise error rate
53:statistical inferences
32:
5248:Population statistics
5190:System identification
4924:Autocorrelation (ACF)
4852:Exponential smoothing
4766:Discriminant analysis
4761:Canonical correlation
4625:Partition of variance
4487:Regression validation
4331:(Jonckheere–Terpstra)
4230:Likelihood-ratio test
3919:Frequentist inference
3831:Location–scale family
3752:Sampling distribution
3717:Statistical inference
3684:Cross-sectional study
3671:Observational studies
3630:Randomized experiment
3459:Stem-and-leaf display
3261:Central limit theorem
2618:Scientific experiment
2600:Design of experiments
1932:10.2105/ajph.86.5.726
1791:Miller, R.G. (1981).
1768:Look-elsewhere effect
1735:Harmonic mean p-value
1714:Bonferroni correction
1587:
1503:
1476:
1368:
1342:
1308:
1230:
1156:
1136:
1050:Bonferroni correction
1043:
1021:
945:
883:
851:
748:Bonferroni correction
697:
677:
601:Further information:
564:
562:{\displaystyle m_{0}}
529:
527:{\displaystyle R=V+S}
454:
415:
413:{\displaystyle m_{0}}
372:
338:
336:{\displaystyle m_{0}}
305:
109:
22:
5383:Multiple comparisons
5171:Probabilistic design
4756:Principal components
4599:Exponential families
4551:Nonlinear regression
4530:General linear model
4492:Mixed effects models
4482:Errors and residuals
4459:Confounding variable
4361:Bayesian probability
4339:Van der Waerden test
4329:Ordered alternative
4094:Multiple comparisons
3973:Rao–Blackwellization
3936:Estimating equations
3892:Statistical distance
3610:Factorial experiment
3143:Arithmetic-Geometric
2892:Fractional factorial
2230:10.1038/nbt1209-1135
2218:Nature Biotechnology
1670:False discovery rate
1629:normal quantile plot
1604:Poisson distribution
1590:normal quantile plot
1567:false discovery rate
1555:false positive rates
1551:exploratory analysis
1526:analysis of variance
1492:
1377:
1351:
1325:
1239:
1165:
1145:
1055:
1032:
961:
895:
863:
778:
771:(FWER), is given by
756:false discovery rate
686:
653:
546:
506:
430:
397:
348:
320:
288:
170:coverage probability
166:confidence intervals
41:multiple comparisons
29:spurious correlation
5243:Official statistics
5166:Methods engineering
4847:Seasonal adjustment
4615:Poisson regressions
4535:Bayesian regression
4474:Regression analysis
4454:Partial correlation
4426:Regression analysis
4025:Prediction interval
4020:Likelihood interval
4010:Confidence interval
4002:Interval estimation
3963:Unbiased estimators
3781:Model specification
3661:Up-and-down designs
3349:Partial correlation
3305:Index of dispersion
3223:Interquartile range
3026:Statistical outline
2986:Sequential analysis
2951:Graeco-Latin square
2860:Multiple comparison
2807:Hierarchical model:
2331:(7378): 1437–1438.
2118:2003PNAS..100.9440S
1993:2020JCAP...10..009B
1818:Biometrical Journal
1682:Interval estimation
1676:False coverage rate
1665:False positive rate
1538:genetic association
1340:{\displaystyle i=1}
953:which follows from
303:{\displaystyle m-R}
5263:Spatial statistics
5143:Medical statistics
5043:First hitting time
4997:Whittle likelihood
4648:Degrees of freedom
4643:Multivariate ANOVA
4576:Heteroscedasticity
4388:Bayesian estimator
4353:Bayesian inference
4202:Kolmogorov–Smirnov
4087:Randomization test
4057:Testing hypotheses
4030:Tolerance interval
3941:Maximum likelihood
3836:Exponential family
3769:Density estimation
3729:Statistical theory
3689:Natural experiment
3635:Scientific control
3552:Survey methodology
3238:Standard deviation
3031:Statistical topics
2623:Statistical design
2392:Journal of the ACM
2177:(456): 1151–1160.
1920:Am J Public Health
1864:mcp-conference.org
1599:
1516:are presented in.
1498:
1471:
1363:
1337:
1303:
1225:
1151:
1131:
1038:
1016:
955:Boole's inequality
940:
878:
846:
692:
672:
559:
524:
449:
410:
367:
333:
300:
120:
55:simultaneously or
33:
5365:
5364:
5303:
5302:
5299:
5298:
5238:National accounts
5208:Actuarial science
5200:Social statistics
5093:
5092:
5089:
5088:
5085:
5084:
5020:Survival function
5005:
5004:
4867:Granger causality
4708:Contingency table
4683:Survival analysis
4660:
4659:
4656:
4655:
4512:Linear regression
4407:
4406:
4403:
4402:
4378:Credible interval
4347:
4346:
4130:
4129:
3946:Method of moments
3815:Parametric family
3776:Statistical model
3706:
3705:
3702:
3701:
3620:Random assignment
3542:Statistical power
3476:
3475:
3472:
3471:
3321:Contingency table
3291:
3290:
3158:Generalized/power
3039:
3038:
2926:Central composite
2824:Cochran's theorem
2778:Linear regression
2755:Nuisance variable
2668:Random assignment
2645:Experimental unit
2398:(3): 12:1–12:22.
2224:(12): 1135–1137.
2112:(16): 9440–9445.
1802:978-0-387-90548-8
1687:Post-hoc analysis
1501:{\displaystyle m}
1488:logic to compute
1400:
1253:
1188:
1154:{\displaystyle m}
1078:
930:
907:
875:
825:
790:
741:
740:
695:{\displaystyle m}
663:
646:
645:
589:are unobservable
497:is the number of
488:is the number of
478:is the number of
468:is the number of
385:
384:
203:, ...,
5390:
5353:
5352:
5341:
5340:
5330:
5329:
5315:
5314:
5218:Crime statistics
5112:
5111:
5099:
5098:
5016:
5015:
4982:Fourier analysis
4969:Frequency domain
4949:
4896:
4862:Structural break
4822:
4821:
4771:Cluster analysis
4718:Log-linear model
4691:
4690:
4666:
4665:
4607:
4581:Homoscedasticity
4437:
4436:
4413:
4412:
4332:
4324:
4316:
4315:(Kruskal–Wallis)
4300:
4285:
4240:Cross validation
4225:
4207:Anderson–Darling
4154:
4141:
4140:
4112:Likelihood-ratio
4104:Parametric tests
4082:Permutation test
4065:1- & 2-tails
3956:Minimum distance
3928:Point estimation
3924:
3923:
3875:Optimal decision
3826:
3725:
3724:
3712:
3711:
3694:Quasi-experiment
3644:Adaptive designs
3495:
3494:
3482:
3481:
3359:Rank correlation
3121:
3120:
3112:
3111:
3099:
3098:
3066:
3059:
3052:
3043:
3042:
3019:
3018:
2956:Orthogonal array
2593:
2586:
2579:
2570:
2569:
2535:
2509:
2488:
2418:
2417:
2407:
2379:
2373:
2372:
2366:
2358:
2348:
2316:
2310:
2309:
2303:
2295:
2293:
2275:
2266:
2260:
2259:
2249:
2209:
2203:
2202:
2164:
2158:
2157:
2147:
2129:
2097:
2091:
2090:
2068:
2062:
2061:
2051:
2034:(5): E266–E270.
2019:
2013:
2012:
1986:
1960:
1954:
1953:
1943:
1911:
1905:
1904:
1888:
1874:
1868:
1867:
1856:
1850:
1849:
1813:
1807:
1806:
1788:
1747:Related concepts
1720:Bonferroni bound
1514:Šidák correction
1507:
1505:
1504:
1499:
1480:
1478:
1477:
1472:
1449:
1444:
1436:
1435:
1434:
1398:
1372:
1370:
1369:
1364:
1346:
1344:
1343:
1338:
1315:Šidák correction
1312:
1310:
1309:
1304:
1302:
1301:
1297:
1288:
1284:
1259:
1258:
1254:
1251:
1234:
1232:
1231:
1226:
1224:
1223:
1222:
1186:
1160:
1158:
1157:
1152:
1140:
1138:
1137:
1132:
1127:
1122:
1114:
1113:
1112:
1076:
1047:
1045:
1044:
1039:
1025:
1023:
1022:
1017:
997:
996:
949:
947:
946:
941:
936:
935:
931:
928:
909:
908:
900:
887:
885:
884:
879:
877:
876:
868:
855:
853:
852:
847:
842:
841:
836:
832:
831:
830:
826:
823:
792:
791:
783:
722:
721:
714:
701:
699:
698:
693:
681:
679:
678:
673:
665:
664:
661:
628:
627:
620:
591:random variables
588:
584:
580:
576:
572:
568:
566:
565:
560:
558:
557:
541:
533:
531:
530:
525:
496:
487:
477:
467:
458:
456:
455:
450:
448:
447:
419:
417:
416:
411:
409:
408:
390:
381:
376:
374:
373:
368:
366:
365:
342:
340:
339:
334:
332:
331:
309:
307:
306:
301:
282:
277:
267:
262:
257:
228:
227:
216:statistical test
213:
5398:
5397:
5393:
5392:
5391:
5389:
5388:
5387:
5368:
5367:
5366:
5361:
5324:
5295:
5257:
5194:
5180:quality control
5147:
5129:Clinical trials
5106:
5081:
5065:
5053:Hazard function
5047:
5001:
4963:
4947:
4910:
4906:Breusch–Godfrey
4894:
4871:
4811:
4786:Factor analysis
4732:
4713:Graphical model
4685:
4652:
4619:
4605:
4585:
4539:
4506:
4468:
4431:
4430:
4399:
4343:
4330:
4322:
4314:
4298:
4283:
4262:Rank statistics
4256:
4235:Model selection
4223:
4181:Goodness of fit
4175:
4152:
4126:
4098:
4051:
3996:
3985:Median unbiased
3913:
3824:
3757:Order statistic
3719:
3698:
3665:
3639:
3591:
3546:
3489:
3487:Data collection
3468:
3380:
3335:
3309:
3287:
3247:
3199:
3116:Continuous data
3106:
3093:
3075:
3070:
3040:
3035:
3013:
2995:
2972:Crossover study
2963:
2961:Latin hypercube
2897:Plackett–Burman
2876:
2873:
2872:
2864:
2767:
2759:
2700:
2692:
2609:
2602:
2597:
2426:
2424:Further reading
2421:
2384:Mitzenmacher, M
2380:
2376:
2360:
2359:
2317:
2313:
2297:
2296:
2273:
2267:
2263:
2210:
2206:
2165:
2161:
2098:
2094:
2069:
2065:
2020:
2016:
1961:
1957:
1912:
1908:
1901:
1875:
1871:
1858:
1857:
1853:
1814:
1810:
1803:
1789:
1785:
1781:
1763:Model selection
1641:
1621:test statistics
1613:test statistics
1582:
1522:
1493:
1490:
1489:
1445:
1440:
1385:
1384:
1380:
1378:
1375:
1374:
1352:
1349:
1348:
1326:
1323:
1322:
1293:
1289:
1280:
1270:
1269:
1250:
1246:
1242:
1240:
1237:
1236:
1173:
1172:
1168:
1166:
1163:
1162:
1146:
1143:
1142:
1123:
1118:
1063:
1062:
1058:
1056:
1053:
1052:
1033:
1030:
1029:
992:
988:
962:
959:
958:
927:
923:
919:
899:
898:
896:
893:
892:
867:
866:
864:
861:
860:
837:
822:
818:
814:
807:
803:
802:
782:
781:
779:
776:
775:
737:
723:
719:
710:
705:
704:
703:
687:
684:
683:
660:
656:
654:
651:
650:
647:
642:
629:
625:
615:
605:
599:
586:
582:
578:
574:
570:
553:
549:
547:
544:
543:
539:
507:
504:
503:
494:
485:
475:
465:
443:
439:
431:
428:
427:
422:null hypotheses
404:
400:
398:
395:
394:
388:
379:
361:
357:
349:
346:
345:
327:
323:
321:
318:
317:
289:
286:
285:
280:
275:
265:
260:
255:
243:
236:
223:
211:
202:
195:
189:
182:
155:false positives
151:expected number
117:
111:
104:
92:Emmanuel Candès
76:
17:
12:
11:
5:
5396:
5386:
5385:
5380:
5363:
5362:
5360:
5359:
5347:
5335:
5321:
5308:
5305:
5304:
5301:
5300:
5297:
5296:
5294:
5293:
5288:
5283:
5278:
5273:
5267:
5265:
5259:
5258:
5256:
5255:
5250:
5245:
5240:
5235:
5230:
5225:
5220:
5215:
5210:
5204:
5202:
5196:
5195:
5193:
5192:
5187:
5182:
5173:
5168:
5163:
5157:
5155:
5149:
5148:
5146:
5145:
5140:
5135:
5126:
5124:Bioinformatics
5120:
5118:
5108:
5107:
5095:
5094:
5091:
5090:
5087:
5086:
5083:
5082:
5080:
5079:
5073:
5071:
5067:
5066:
5064:
5063:
5057:
5055:
5049:
5048:
5046:
5045:
5040:
5035:
5030:
5024:
5022:
5013:
5007:
5006:
5003:
5002:
5000:
4999:
4994:
4989:
4984:
4979:
4973:
4971:
4965:
4964:
4962:
4961:
4956:
4951:
4943:
4938:
4933:
4932:
4931:
4929:partial (PACF)
4920:
4918:
4912:
4911:
4909:
4908:
4903:
4898:
4890:
4885:
4879:
4877:
4876:Specific tests
4873:
4872:
4870:
4869:
4864:
4859:
4854:
4849:
4844:
4839:
4834:
4828:
4826:
4819:
4813:
4812:
4810:
4809:
4808:
4807:
4806:
4805:
4790:
4789:
4788:
4778:
4776:Classification
4773:
4768:
4763:
4758:
4753:
4748:
4742:
4740:
4734:
4733:
4731:
4730:
4725:
4723:McNemar's test
4720:
4715:
4710:
4705:
4699:
4697:
4687:
4686:
4662:
4661:
4658:
4657:
4654:
4653:
4651:
4650:
4645:
4640:
4635:
4629:
4627:
4621:
4620:
4618:
4617:
4601:
4595:
4593:
4587:
4586:
4584:
4583:
4578:
4573:
4568:
4563:
4561:Semiparametric
4558:
4553:
4547:
4545:
4541:
4540:
4538:
4537:
4532:
4527:
4522:
4516:
4514:
4508:
4507:
4505:
4504:
4499:
4494:
4489:
4484:
4478:
4476:
4470:
4469:
4467:
4466:
4461:
4456:
4451:
4445:
4443:
4433:
4432:
4429:
4428:
4423:
4417:
4409:
4408:
4405:
4404:
4401:
4400:
4398:
4397:
4396:
4395:
4385:
4380:
4375:
4374:
4373:
4368:
4357:
4355:
4349:
4348:
4345:
4344:
4342:
4341:
4336:
4335:
4334:
4326:
4318:
4302:
4299:(Mann–Whitney)
4294:
4293:
4292:
4279:
4278:
4277:
4266:
4264:
4258:
4257:
4255:
4254:
4253:
4252:
4247:
4242:
4232:
4227:
4224:(Shapiro–Wilk)
4219:
4214:
4209:
4204:
4199:
4191:
4185:
4183:
4177:
4176:
4174:
4173:
4165:
4156:
4144:
4138:
4136:Specific tests
4132:
4131:
4128:
4127:
4125:
4124:
4119:
4114:
4108:
4106:
4100:
4099:
4097:
4096:
4091:
4090:
4089:
4079:
4078:
4077:
4067:
4061:
4059:
4053:
4052:
4050:
4049:
4048:
4047:
4042:
4032:
4027:
4022:
4017:
4012:
4006:
4004:
3998:
3997:
3995:
3994:
3989:
3988:
3987:
3982:
3981:
3980:
3975:
3960:
3959:
3958:
3953:
3948:
3943:
3932:
3930:
3921:
3915:
3914:
3912:
3911:
3906:
3901:
3900:
3899:
3889:
3884:
3883:
3882:
3872:
3871:
3870:
3865:
3860:
3850:
3845:
3840:
3839:
3838:
3833:
3828:
3812:
3811:
3810:
3805:
3800:
3790:
3789:
3788:
3783:
3773:
3772:
3771:
3761:
3760:
3759:
3749:
3744:
3739:
3733:
3731:
3721:
3720:
3708:
3707:
3704:
3703:
3700:
3699:
3697:
3696:
3691:
3686:
3681:
3675:
3673:
3667:
3666:
3664:
3663:
3658:
3653:
3647:
3645:
3641:
3640:
3638:
3637:
3632:
3627:
3622:
3617:
3612:
3607:
3601:
3599:
3593:
3592:
3590:
3589:
3587:Standard error
3584:
3579:
3574:
3573:
3572:
3567:
3556:
3554:
3548:
3547:
3545:
3544:
3539:
3534:
3529:
3524:
3519:
3517:Optimal design
3514:
3509:
3503:
3501:
3491:
3490:
3478:
3477:
3474:
3473:
3470:
3469:
3467:
3466:
3461:
3456:
3451:
3446:
3441:
3436:
3431:
3426:
3421:
3416:
3411:
3406:
3401:
3396:
3390:
3388:
3382:
3381:
3379:
3378:
3373:
3372:
3371:
3366:
3356:
3351:
3345:
3343:
3337:
3336:
3334:
3333:
3328:
3323:
3317:
3315:
3314:Summary tables
3311:
3310:
3308:
3307:
3301:
3299:
3293:
3292:
3289:
3288:
3286:
3285:
3284:
3283:
3278:
3273:
3263:
3257:
3255:
3249:
3248:
3246:
3245:
3240:
3235:
3230:
3225:
3220:
3215:
3209:
3207:
3201:
3200:
3198:
3197:
3192:
3187:
3186:
3185:
3180:
3175:
3170:
3165:
3160:
3155:
3150:
3148:Contraharmonic
3145:
3140:
3129:
3127:
3118:
3108:
3107:
3095:
3094:
3092:
3091:
3086:
3080:
3077:
3076:
3069:
3068:
3061:
3054:
3046:
3037:
3036:
3034:
3033:
3028:
3023:
3011:
3006:
3000:
2997:
2996:
2994:
2993:
2988:
2983:
2975:
2974:
2969:
2958:
2953:
2948:
2943:
2937:
2929:
2928:
2923:
2918:
2913:
2905:
2904:
2899:
2894:
2889:
2881:
2879:
2866:
2865:
2863:
2862:
2857:
2851:
2850:
2838:
2826:
2821:
2813:
2812:
2804:
2799:
2791:
2790:
2785:
2780:
2774:
2772:
2761:
2760:
2758:
2757:
2752:
2747:
2740:
2735:
2730:
2725:
2720:
2715:
2707:
2705:
2694:
2693:
2691:
2690:
2685:
2680:
2675:
2670:
2665:
2658:Optimal design
2653:
2652:
2647:
2642:
2630:
2625:
2620:
2614:
2612:
2604:
2603:
2596:
2595:
2588:
2581:
2573:
2567:
2566:
2555:
2550:
2543:
2536:
2489:
2455:(4): 347–388.
2444:
2434:
2425:
2422:
2420:
2419:
2374:
2311:
2284:(3): 116–120.
2261:
2204:
2159:
2092:
2081:(1): 125–133.
2063:
2014:
1955:
1926:(5): 726–728.
1906:
1899:
1869:
1851:
1824:(6): 708–721.
1808:
1801:
1782:
1780:
1777:
1776:
1775:
1770:
1765:
1760:
1755:
1749:
1748:
1744:
1743:
1738:
1732:
1727:
1722:
1716:
1711:
1705:
1704:
1700:
1699:
1694:
1689:
1684:
1679:
1673:
1667:
1662:
1656:
1655:
1651:
1650:
1640:
1637:
1581:
1578:
1521:
1518:
1497:
1470:
1467:
1464:
1461:
1458:
1455:
1452:
1448:
1443:
1439:
1433:
1430:
1427:
1424:
1421:
1418:
1415:
1412:
1409:
1406:
1403:
1397:
1394:
1391:
1388:
1383:
1362:
1359:
1356:
1336:
1333:
1330:
1300:
1296:
1292:
1287:
1283:
1279:
1276:
1273:
1268:
1265:
1262:
1257:
1252:per comparison
1249:
1245:
1235:. This yields
1221:
1218:
1215:
1212:
1209:
1206:
1203:
1200:
1197:
1194:
1191:
1185:
1182:
1179:
1176:
1171:
1150:
1130:
1126:
1121:
1117:
1111:
1108:
1105:
1102:
1099:
1096:
1093:
1090:
1087:
1084:
1081:
1075:
1072:
1069:
1066:
1061:
1037:
1015:
1012:
1009:
1006:
1003:
1000:
995:
991:
987:
984:
981:
978:
975:
972:
969:
966:
951:
950:
939:
934:
929:per comparison
926:
922:
918:
915:
912:
906:
903:
874:
871:
857:
856:
845:
840:
835:
829:
824:per comparison
821:
817:
813:
810:
806:
801:
798:
795:
789:
786:
739:
738:
726:
724:
717:
709:
706:
691:
671:
668:
662:per comparison
659:
648:
644:
643:
632:
630:
623:
618:
617:
616:
598:
595:
556:
552:
536:
535:
523:
520:
517:
514:
511:
501:
499:true negatives
492:
483:
480:true positives
473:
463:
446:
442:
438:
435:
425:
407:
403:
392:
383:
382:
377:
364:
360:
356:
353:
343:
330:
326:
315:
311:
310:
299:
296:
293:
283:
278:
273:
269:
268:
263:
258:
253:
249:
248:
245:
241:
238:
234:
231:
221:
207:
200:
193:
181:
178:
142:
141:
133:
130:sampling error
103:
100:
75:
72:
15:
9:
6:
4:
3:
2:
5395:
5384:
5381:
5379:
5376:
5375:
5373:
5358:
5357:
5348:
5346:
5345:
5336:
5334:
5333:
5328:
5322:
5320:
5319:
5310:
5309:
5306:
5292:
5289:
5287:
5286:Geostatistics
5284:
5282:
5279:
5277:
5274:
5272:
5269:
5268:
5266:
5264:
5260:
5254:
5253:Psychometrics
5251:
5249:
5246:
5244:
5241:
5239:
5236:
5234:
5231:
5229:
5226:
5224:
5221:
5219:
5216:
5214:
5211:
5209:
5206:
5205:
5203:
5201:
5197:
5191:
5188:
5186:
5183:
5181:
5177:
5174:
5172:
5169:
5167:
5164:
5162:
5159:
5158:
5156:
5154:
5150:
5144:
5141:
5139:
5136:
5134:
5130:
5127:
5125:
5122:
5121:
5119:
5117:
5116:Biostatistics
5113:
5109:
5105:
5100:
5096:
5078:
5077:Log-rank test
5075:
5074:
5072:
5068:
5062:
5059:
5058:
5056:
5054:
5050:
5044:
5041:
5039:
5036:
5034:
5031:
5029:
5026:
5025:
5023:
5021:
5017:
5014:
5012:
5008:
4998:
4995:
4993:
4990:
4988:
4985:
4983:
4980:
4978:
4975:
4974:
4972:
4970:
4966:
4960:
4957:
4955:
4952:
4950:
4948:(Box–Jenkins)
4944:
4942:
4939:
4937:
4934:
4930:
4927:
4926:
4925:
4922:
4921:
4919:
4917:
4913:
4907:
4904:
4902:
4901:Durbin–Watson
4899:
4897:
4891:
4889:
4886:
4884:
4883:Dickey–Fuller
4881:
4880:
4878:
4874:
4868:
4865:
4863:
4860:
4858:
4857:Cointegration
4855:
4853:
4850:
4848:
4845:
4843:
4840:
4838:
4835:
4833:
4832:Decomposition
4830:
4829:
4827:
4823:
4820:
4818:
4814:
4804:
4801:
4800:
4799:
4796:
4795:
4794:
4791:
4787:
4784:
4783:
4782:
4779:
4777:
4774:
4772:
4769:
4767:
4764:
4762:
4759:
4757:
4754:
4752:
4749:
4747:
4744:
4743:
4741:
4739:
4735:
4729:
4726:
4724:
4721:
4719:
4716:
4714:
4711:
4709:
4706:
4704:
4703:Cohen's kappa
4701:
4700:
4698:
4696:
4692:
4688:
4684:
4680:
4676:
4672:
4667:
4663:
4649:
4646:
4644:
4641:
4639:
4636:
4634:
4631:
4630:
4628:
4626:
4622:
4616:
4612:
4608:
4602:
4600:
4597:
4596:
4594:
4592:
4588:
4582:
4579:
4577:
4574:
4572:
4569:
4567:
4564:
4562:
4559:
4557:
4556:Nonparametric
4554:
4552:
4549:
4548:
4546:
4542:
4536:
4533:
4531:
4528:
4526:
4523:
4521:
4518:
4517:
4515:
4513:
4509:
4503:
4500:
4498:
4495:
4493:
4490:
4488:
4485:
4483:
4480:
4479:
4477:
4475:
4471:
4465:
4462:
4460:
4457:
4455:
4452:
4450:
4447:
4446:
4444:
4442:
4438:
4434:
4427:
4424:
4422:
4419:
4418:
4414:
4410:
4394:
4391:
4390:
4389:
4386:
4384:
4381:
4379:
4376:
4372:
4369:
4367:
4364:
4363:
4362:
4359:
4358:
4356:
4354:
4350:
4340:
4337:
4333:
4327:
4325:
4319:
4317:
4311:
4310:
4309:
4306:
4305:Nonparametric
4303:
4301:
4295:
4291:
4288:
4287:
4286:
4280:
4276:
4275:Sample median
4273:
4272:
4271:
4268:
4267:
4265:
4263:
4259:
4251:
4248:
4246:
4243:
4241:
4238:
4237:
4236:
4233:
4231:
4228:
4226:
4220:
4218:
4215:
4213:
4210:
4208:
4205:
4203:
4200:
4198:
4196:
4192:
4190:
4187:
4186:
4184:
4182:
4178:
4172:
4170:
4166:
4164:
4162:
4157:
4155:
4150:
4146:
4145:
4142:
4139:
4137:
4133:
4123:
4120:
4118:
4115:
4113:
4110:
4109:
4107:
4105:
4101:
4095:
4092:
4088:
4085:
4084:
4083:
4080:
4076:
4073:
4072:
4071:
4068:
4066:
4063:
4062:
4060:
4058:
4054:
4046:
4043:
4041:
4038:
4037:
4036:
4033:
4031:
4028:
4026:
4023:
4021:
4018:
4016:
4013:
4011:
4008:
4007:
4005:
4003:
3999:
3993:
3990:
3986:
3983:
3979:
3976:
3974:
3971:
3970:
3969:
3966:
3965:
3964:
3961:
3957:
3954:
3952:
3949:
3947:
3944:
3942:
3939:
3938:
3937:
3934:
3933:
3931:
3929:
3925:
3922:
3920:
3916:
3910:
3907:
3905:
3902:
3898:
3895:
3894:
3893:
3890:
3888:
3885:
3881:
3880:loss function
3878:
3877:
3876:
3873:
3869:
3866:
3864:
3861:
3859:
3856:
3855:
3854:
3851:
3849:
3846:
3844:
3841:
3837:
3834:
3832:
3829:
3827:
3821:
3818:
3817:
3816:
3813:
3809:
3806:
3804:
3801:
3799:
3796:
3795:
3794:
3791:
3787:
3784:
3782:
3779:
3778:
3777:
3774:
3770:
3767:
3766:
3765:
3762:
3758:
3755:
3754:
3753:
3750:
3748:
3745:
3743:
3740:
3738:
3735:
3734:
3732:
3730:
3726:
3722:
3718:
3713:
3709:
3695:
3692:
3690:
3687:
3685:
3682:
3680:
3677:
3676:
3674:
3672:
3668:
3662:
3659:
3657:
3654:
3652:
3649:
3648:
3646:
3642:
3636:
3633:
3631:
3628:
3626:
3623:
3621:
3618:
3616:
3613:
3611:
3608:
3606:
3603:
3602:
3600:
3598:
3594:
3588:
3585:
3583:
3582:Questionnaire
3580:
3578:
3575:
3571:
3568:
3566:
3563:
3562:
3561:
3558:
3557:
3555:
3553:
3549:
3543:
3540:
3538:
3535:
3533:
3530:
3528:
3525:
3523:
3520:
3518:
3515:
3513:
3510:
3508:
3505:
3504:
3502:
3500:
3496:
3492:
3488:
3483:
3479:
3465:
3462:
3460:
3457:
3455:
3452:
3450:
3447:
3445:
3442:
3440:
3437:
3435:
3432:
3430:
3427:
3425:
3422:
3420:
3417:
3415:
3412:
3410:
3409:Control chart
3407:
3405:
3402:
3400:
3397:
3395:
3392:
3391:
3389:
3387:
3383:
3377:
3374:
3370:
3367:
3365:
3362:
3361:
3360:
3357:
3355:
3352:
3350:
3347:
3346:
3344:
3342:
3338:
3332:
3329:
3327:
3324:
3322:
3319:
3318:
3316:
3312:
3306:
3303:
3302:
3300:
3298:
3294:
3282:
3279:
3277:
3274:
3272:
3269:
3268:
3267:
3264:
3262:
3259:
3258:
3256:
3254:
3250:
3244:
3241:
3239:
3236:
3234:
3231:
3229:
3226:
3224:
3221:
3219:
3216:
3214:
3211:
3210:
3208:
3206:
3202:
3196:
3193:
3191:
3188:
3184:
3181:
3179:
3176:
3174:
3171:
3169:
3166:
3164:
3161:
3159:
3156:
3154:
3151:
3149:
3146:
3144:
3141:
3139:
3136:
3135:
3134:
3131:
3130:
3128:
3126:
3122:
3119:
3117:
3113:
3109:
3105:
3100:
3096:
3090:
3087:
3085:
3082:
3081:
3078:
3074:
3067:
3062:
3060:
3055:
3053:
3048:
3047:
3044:
3032:
3029:
3027:
3024:
3022:
3017:
3012:
3010:
3007:
3005:
3002:
3001:
2998:
2992:
2989:
2987:
2984:
2982:
2981:
2977:
2976:
2973:
2970:
2968:
2967:
2962:
2959:
2957:
2954:
2952:
2949:
2947:
2944:
2941:
2938:
2936:
2935:
2931:
2930:
2927:
2924:
2922:
2919:
2917:
2914:
2912:
2911:
2907:
2906:
2903:
2900:
2898:
2895:
2893:
2890:
2888:
2887:
2883:
2882:
2880:
2878:
2871:
2867:
2861:
2858:
2856:
2855:Compare means
2853:
2852:
2849:
2847:
2843:
2839:
2837:
2835:
2831:
2827:
2825:
2822:
2820:
2819:
2815:
2814:
2811:
2808:
2805:
2803:
2800:
2798:
2797:
2796:Random effect
2793:
2792:
2789:
2786:
2784:
2781:
2779:
2776:
2775:
2773:
2771:
2766:
2762:
2756:
2753:
2751:
2748:
2746:
2745:
2741:
2739:
2738:Orthogonality
2736:
2734:
2731:
2729:
2726:
2724:
2721:
2719:
2716:
2714:
2713:
2709:
2708:
2706:
2704:
2699:
2695:
2689:
2686:
2684:
2681:
2679:
2676:
2674:
2673:Randomization
2671:
2669:
2666:
2664:
2660:
2659:
2655:
2654:
2651:
2648:
2646:
2643:
2641:
2638:
2634:
2631:
2629:
2626:
2624:
2621:
2619:
2616:
2615:
2613:
2611:
2605:
2601:
2594:
2589:
2587:
2582:
2580:
2575:
2574:
2571:
2564:
2563:
2558:
2556:
2554:
2551:
2548:
2544:
2541:
2537:
2533:
2529:
2525:
2521:
2517:
2513:
2508:
2503:
2500:: Article39.
2499:
2495:
2490:
2486:
2482:
2478:
2474:
2470:
2466:
2462:
2458:
2454:
2450:
2445:
2442:
2438:
2435:
2432:
2428:
2427:
2415:
2411:
2406:
2401:
2397:
2393:
2389:
2385:
2378:
2370:
2364:
2356:
2352:
2347:
2342:
2338:
2334:
2330:
2326:
2322:
2315:
2307:
2301:
2292:
2287:
2283:
2279:
2272:
2265:
2257:
2253:
2248:
2243:
2239:
2235:
2231:
2227:
2223:
2219:
2215:
2208:
2200:
2196:
2192:
2188:
2184:
2180:
2176:
2172:
2171:
2163:
2155:
2151:
2146:
2141:
2137:
2133:
2128:
2123:
2119:
2115:
2111:
2107:
2103:
2096:
2088:
2084:
2080:
2076:
2075:
2067:
2059:
2055:
2050:
2045:
2041:
2037:
2033:
2029:
2025:
2018:
2010:
2006:
2002:
1998:
1994:
1990:
1985:
1980:
1976:
1972:
1971:
1966:
1959:
1951:
1947:
1942:
1937:
1933:
1929:
1925:
1921:
1917:
1910:
1902:
1900:9780072386882
1896:
1892:
1887:
1886:
1880:
1873:
1865:
1861:
1855:
1847:
1843:
1839:
1835:
1831:
1827:
1823:
1819:
1812:
1804:
1798:
1794:
1787:
1783:
1774:
1773:Data dredging
1771:
1769:
1766:
1764:
1761:
1759:
1756:
1754:
1751:
1750:
1746:
1745:
1742:
1739:
1736:
1733:
1731:
1728:
1726:
1723:
1721:
1717:
1715:
1712:
1710:
1707:
1706:
1702:
1701:
1698:
1695:
1693:
1690:
1688:
1685:
1683:
1680:
1677:
1674:
1671:
1668:
1666:
1663:
1661:
1658:
1657:
1653:
1652:
1649:
1647:
1643:
1642:
1636:
1634:
1630:
1627:is to make a
1626:
1622:
1617:
1614:
1608:
1605:
1595:
1591:
1586:
1577:
1575:
1570:
1568:
1563:
1558:
1556:
1552:
1548:
1544:
1539:
1535:
1531:
1527:
1517:
1515:
1511:
1495:
1487:
1482:
1465:
1462:
1459:
1456:
1453:
1446:
1441:
1437:
1381:
1360:
1357:
1354:
1334:
1331:
1328:
1320:
1316:
1298:
1294:
1290:
1281:
1277:
1274:
1266:
1263:
1260:
1243:
1169:
1148:
1128:
1124:
1119:
1115:
1059:
1051:
1035:
1026:
1013:
1010:
1007:
1004:
1001:
998:
993:
985:
982:
979:
973:
970:
967:
964:
956:
937:
920:
916:
913:
910:
901:
891:
890:
889:
869:
843:
838:
833:
815:
811:
808:
804:
799:
796:
793:
784:
774:
773:
772:
770:
769:
764:
759:
757:
753:
749:
745:
735:
734:
729:
725:
716:
715:
712:
689:
669:
666:
657:
640:
639:MediaWiki.org
636:
631:
622:
621:
614:
610:
604:
594:
592:
554:
550:
521:
518:
515:
512:
509:
502:
500:
493:
491:
484:
481:
474:
471:
464:
462:
444:
440:
436:
433:
426:
423:
405:
401:
393:
387:
386:
378:
362:
358:
354:
351:
344:
328:
324:
316:
313:
297:
294:
291:
284:
279:
274:
271:
264:
259:
254:
251:
246:
239:
232:
230:
229:
226:
224:
217:
210:
206:
199:
192:
187:
177:
174:
171:
167:
162:
160:
159:Type I errors
156:
152:
146:
138:
134:
131:
126:
125:
124:
115:
114:binomial test
108:
99:
97:
96:Vladimir Vovk
93:
89:
85:
81:
71:
69:
65:
60:
58:
54:
50:
46:
42:
38:
30:
26:
25:data dredging
21:
5354:
5342:
5323:
5316:
5228:Econometrics
5178: /
5161:Chemometrics
5138:Epidemiology
5131: /
5104:Applications
4946:ARIMA model
4893:Q-statistic
4842:Stationarity
4738:Multivariate
4681: /
4677: /
4675:Multivariate
4673: /
4613: /
4609: /
4383:Bayes factor
4282:Signed rank
4194:
4168:
4160:
4148:
3843:Completeness
3679:Cohort study
3577:Opinion poll
3512:Missing data
3499:Study design
3454:Scatter plot
3376:Scatter plot
3369:Spearman's ρ
3331:Grouped data
2978:
2964:
2946:Latin square
2932:
2908:
2884:
2845:
2841:
2834:multivariate
2833:
2829:
2816:
2794:
2742:
2710:
2656:
2560:
2546:
2539:
2497:
2493:
2469:11573/142139
2452:
2448:
2440:
2430:
2395:
2391:
2377:
2363:cite journal
2328:
2324:
2314:
2300:cite journal
2281:
2278:Significance
2277:
2264:
2221:
2217:
2207:
2174:
2168:
2162:
2109:
2105:
2095:
2078:
2072:
2066:
2031:
2027:
2017:
1974:
1968:
1958:
1923:
1919:
1909:
1884:
1872:
1863:
1854:
1821:
1817:
1811:
1792:
1786:
1654:Key concepts
1645:
1618:
1609:
1600:
1571:
1559:
1523:
1483:
1027:
952:
858:
766:
762:
760:
743:
742:
731:
727:
711:
537:
219:
208:
204:
197:
190:
185:
183:
175:
163:
147:
143:
121:
77:
61:
48:
45:multiplicity
44:
40:
34:
5356:WikiProject
5271:Cartography
5233:Jurimetrics
5185:Reliability
4916:Time domain
4895:(Ljung–Box)
4817:Time-series
4695:Categorical
4679:Time-series
4671:Categorical
4606:(Bernoulli)
4441:Correlation
4421:Correlation
4217:Jarque–Bera
4189:Chi-squared
3951:M-estimator
3904:Asymptotics
3848:Sufficiency
3615:Interaction
3527:Replication
3507:Effect size
3464:Violin plot
3444:Radar chart
3424:Forest plot
3414:Correlogram
3364:Kendall's τ
2921:Box–Behnken
2802:Mixed model
2733:Confounding
2728:Interaction
2718:Effect size
2688:Sample size
2433:, CRC Press
2382:Kirsch, A;
1977:(10): 009.
1879:Neter, John
1543:measurement
1534:microarrays
957:. Example:
635:Phabricator
5372:Categories
5223:Demography
4941:ARMA model
4746:Regression
4323:(Friedman)
4284:(Wilcoxon)
4222:Normality
4212:Lilliefors
4159:Student's
4035:Resampling
3909:Robustness
3897:divergence
3887:Efficiency
3825:(monotone)
3820:Likelihood
3737:Population
3570:Stratified
3522:Population
3341:Dependence
3297:Count data
3228:Percentile
3205:Dispersion
3138:Arithmetic
3073:Statistics
2877:randomized
2875:Completely
2846:covariance
2608:Scientific
2507:1603.05766
2443:, Springer
1984:2007.13821
1779:References
1510:Bonferroni
607:See also:
102:Definition
37:statistics
4604:Logistic
4371:posterior
4297:Rank sum
4045:Jackknife
4040:Bootstrap
3858:Bootstrap
3793:Parameter
3742:Statistic
3537:Statistic
3449:Run chart
3434:Pie chart
3429:Histogram
3419:Fan chart
3394:Bar chart
3276:L-moments
3163:Geometric
2886:Factorial
2770:inference
2750:Covariate
2712:Treatment
2698:Treatment
2437:S. Dudoit
2405:1002.1104
2238:1087-0156
2040:0147-958X
2009:220830693
1737:procedure
1633:dispersed
1607:results.
1574:p-hacking
1457:−
1442:α
1382:α
1282:α
1278:−
1267:−
1244:α
1170:α
1120:α
1060:α
1036:α
1005:×
999:≤
983:−
974:−
921:α
917:⋅
911:≤
905:¯
902:α
873:¯
870:α
816:α
812:−
800:−
788:¯
785:α
658:α
437:−
355:−
295:−
57:estimates
5318:Category
5011:Survival
4888:Johansen
4611:Binomial
4566:Isotonic
4153:(normal)
3798:location
3605:Blocking
3560:Sampling
3439:Q–Q plot
3404:Box plot
3386:Graphics
3281:Skewness
3271:Kurtosis
3243:Variance
3173:Heronian
3168:Harmonic
3009:Category
3004:Glossary
2810:Bayesian
2788:Bayesian
2744:Blocking
2723:Contrast
2703:blocking
2663:Bayesian
2650:Blinding
2640:validity
2637:external
2633:Internal
2532:10735784
2524:21044043
2485:12777404
2477:17698936
2388:Upfal, E
2355:12493654
2256:20010596
2154:12883005
2058:20926032
1838:21154895
1639:See also
1625:Z-scores
1594:Z-scores
1530:genomics
1486:Bayesian
214:Using a
140:symptom.
88:Tel Aviv
5344:Commons
5291:Kriging
5176:Process
5133:studies
4992:Wavelet
4825:General
3992:Plug-in
3786:L space
3565:Cluster
3266:Moments
3084:Outline
2902:Taguchi
2870:Designs
2628:Control
2542:, Wiley
2346:1124898
2247:2907892
2199:9076863
2191:3085878
2136:3144228
2114:Bibcode
2087:2346101
2049:3270946
1989:Bibcode
1950:8629727
1941:1380484
1846:8806192
754:or the
637:and on
196:,
84:Scheffé
74:History
5213:Census
4803:Normal
4751:Manova
4571:Robust
4321:2-way
4313:1-way
4151:-test
3822:
3399:Biplot
3190:Median
3183:Lehmer
3125:Center
2942:(GRBD)
2842:Ancova
2830:Manova
2765:Models
2610:method
2530:
2522:
2483:
2475:
2353:
2343:
2254:
2244:
2236:
2197:
2189:
2152:
2145:170937
2142:
2134:
2085:
2056:
2046:
2038:
2007:
1948:
1938:
1897:
1893:–745.
1860:"Home"
1844:
1836:
1799:
1718:Boole–
1648:-value
1399:
1187:
1077:
965:0.2649
611:, and
585:, and
314:Total
247:Total
132:alone.
39:, the
4837:Trend
4366:prior
4308:anova
4197:-test
4171:-test
4163:-test
4070:Power
4015:Pivot
3808:shape
3803:scale
3253:Shape
3233:Range
3178:Heinz
3153:Cubic
3089:Index
2934:Block
2528:S2CID
2502:arXiv
2481:S2CID
2400:arXiv
2274:(PDF)
2195:S2CID
2187:JSTOR
2132:JSTOR
2083:JSTOR
2005:S2CID
1979:arXiv
1842:S2CID
1678:(FCR)
1672:(FDR)
80:Tukey
5070:Test
4270:Sign
4122:Wald
3195:Mode
3133:Mean
2768:and
2701:and
2635:and
2562:xkcd
2520:PMID
2473:PMID
2369:link
2351:PMID
2306:link
2252:PMID
2234:ISSN
2150:PMID
2106:PNAS
2054:PMID
2036:ISSN
1975:2020
1946:PMID
1895:ISBN
1834:PMID
1797:ISBN
1545:and
1512:and
1358:>
670:0.05
137:drug
94:and
82:and
4250:BIC
4245:AIC
2559:An
2512:doi
2465:hdl
2457:doi
2410:doi
2341:PMC
2333:doi
2329:325
2325:BMJ
2286:doi
2242:PMC
2226:doi
2179:doi
2140:PMC
2122:doi
2110:100
2044:PMC
1997:doi
1936:PMC
1928:doi
1891:744
1826:doi
1576:".
1014:0.3
1002:.05
986:.05
761:If
538:In
157:or
47:or
35:In
5374::
2661::
2526:.
2518:.
2510:.
2496:.
2479:.
2471:.
2463:.
2453:17
2451:.
2408:.
2396:59
2394:.
2365:}}
2361:{{
2349:.
2339:.
2327:.
2323:.
2302:}}
2298:{{
2280:.
2276:.
2250:.
2240:.
2232:.
2222:27
2220:.
2216:.
2193:.
2185:.
2175:96
2173:.
2148:.
2138:.
2130:.
2120:.
2108:.
2104:.
2079:57
2077:.
2052:.
2042:.
2032:33
2030:.
2026:.
2003:.
1995:.
1987:.
1973:.
1967:.
1944:.
1934:.
1924:86
1922:.
1918:.
1862:.
1840:.
1832:.
1822:52
1820:.
1588:A
1481:.
758:.
593:.
581:,
577:,
244:)
237:)
98:.
43:,
4195:G
4169:F
4161:t
4149:Z
3868:V
3863:U
3065:e
3058:t
3051:v
2848:)
2844:(
2836:)
2832:(
2592:e
2585:t
2578:v
2534:.
2514::
2504::
2498:9
2487:.
2467::
2459::
2416:.
2412::
2402::
2371:)
2357:.
2335::
2308:)
2294:.
2288::
2282:8
2258:.
2228::
2201:.
2181::
2156:.
2124::
2116::
2089:.
2060:.
2011:.
1999::
1991::
1981::
1952:.
1930::
1903:.
1866:.
1848:.
1828::
1805:.
1646:q
1496:m
1469:)
1466:1
1463:+
1460:i
1454:m
1451:(
1447:/
1438:=
1432:}
1429:n
1426:o
1423:s
1420:i
1417:r
1414:a
1411:p
1408:m
1405:o
1402:c
1396:r
1393:e
1390:p
1387:{
1361:1
1355:i
1335:1
1332:=
1329:i
1299:m
1295:/
1291:1
1286:)
1275:1
1272:(
1264:1
1261:=
1256:}
1248:{
1220:}
1217:n
1214:o
1211:s
1208:i
1205:r
1202:a
1199:p
1196:m
1193:o
1190:c
1184:r
1181:e
1178:p
1175:{
1149:m
1129:m
1125:/
1116:=
1110:}
1107:n
1104:o
1101:s
1098:i
1095:r
1092:a
1089:p
1086:m
1083:o
1080:c
1074:r
1071:e
1068:p
1065:{
1011:=
1008:6
994:6
990:)
980:1
977:(
971:1
968:=
938:,
933:}
925:{
914:m
844:.
839:m
834:)
828:}
820:{
809:1
805:(
797:1
794:=
763:m
736:.
702:.
690:m
667:=
641:.
587:V
583:U
579:T
575:S
571:R
555:0
551:m
540:m
522:S
519:+
516:V
513:=
510:R
495:U
486:T
476:S
466:V
445:0
441:m
434:m
406:0
402:m
389:m
380:m
363:0
359:m
352:m
329:0
325:m
298:R
292:m
281:T
276:U
266:R
261:S
256:V
242:A
235:0
222:i
220:H
212:.
209:m
205:H
201:2
198:H
194:1
191:H
186:m
31:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.