289:("internal reticular apparatus"). Some doubted the discovery at first, arguing that the appearance of the structure was merely an optical illusion created by the observation technique used by Golgi. With the development of modern microscopes in the twentieth century, the discovery was confirmed. Early references to the Golgi apparatus referred to it by various names including the "Golgi–Holmgren apparatus", "Golgi–Holmgren ducts", and "Golgi–Kopsch apparatus". The term "Golgi apparatus" was used in 1910 and first appeared in the scientific literature in 1913, while "Golgi complex" was introduced in 1956.
36:
1081:
348:
605:
1947:
66:
1972:
930:. Additionally, tubular connections are not prevalent in plant cells. The roles that these connections have can be attributed to a cell-specific specialization rather than a universal trait. If the membranes are continuous, that suggests the existence of mechanisms that preserve the unique biochemical gradients observed throughout the Golgi apparatus.
1047:
474:
942:
This rapid partitioning model is the most drastic alteration of the traditional vesicular trafficking point of view. Proponents of this model hypothesize that the Golgi works as a single unit, containing domains that function separately in the processing and export of protein cargo. Cargo from the ER
744:
Vesicles contain proteins destined for extracellular release. After packaging, the vesicles bud off and are stored in the cell until a signal is given for their release. When the appropriate signal is received they move toward the membrane and fuse to release their contents. This process is known as
955:
This model cannot explain the transport kinetics of large protein cargo, such as collagen. This model falls short on explaining the observation of discrete compartments and polarized biochemistry of the Golgi cisternae. It also does not explain formation and disintegration of the Golgi network, nor
894:
The model addresses the existence of Golgi compartments, as well as differing biochemistry within the cisternae, transport of large proteins, transient formation and disintegration of the cisternae, and retrograde mobility of native Golgi proteins, and it can account for the variability seen in the
887:
of the Golgi stack, which progresses later to become mature TGN cisternae. Once matured, the TGN cisternae dissolve to become secretory vesicles. While this progression occurs, COPI vesicles continually recycle Golgi-specific proteins by delivery from older to younger cisternae. Different recycling
988:
This model does not explain morphological variations in the Golgi apparatus, nor define a role for COPI vesicles. This model does not apply well for plants, algae, and fungi in which individual Golgi stacks are observed (transfer of domains between stacks is not likely). Additionally, megavesicles
913:
This model is an extension of the cisternal progression/maturation model. It incorporates the existence of tubular connections among the cisternae that form the Golgi ribbon, in which cisternae within a stack are linked. This model posits that the tubules are important for bidirectional traffic in
456:
responsible for selectively modifying protein cargo. These modifications influence the fate of the protein. The compartmentalization of the Golgi apparatus is advantageous for separating enzymes, thereby maintaining consecutive and selective processing steps: enzymes catalyzing early modifications
981:
This model is consistent with numerous observations and encompasses some of the strengths of the cisternal progression/maturation model. Additionally, what is known of the Rab GTPase roles in mammalian endosomes can help predict putative roles within the Golgi. This model is unique in that it can
943:
move between these two domains, and randomly exit from any level of the Golgi to their final location. This model is supported by the observation that cargo exits the Golgi in a pattern best described by exponential kinetics. The existence of domains is supported by fluorescence microscopy data.
995:
Though there are multiple models that attempt to explain vesicular traffic throughout the Golgi, no individual model can independently explain all observations of the Golgi apparatus. Currently, the cisternal progression/maturation model is the most accepted among scientists, accommodating many
863:
Since the amount of COPI vesicles varies drastically among types of cells, this model cannot easily explain high trafficking activity within the Golgi for both small and large cargoes. Additionally, there is no convincing evidence that COPI vesicles move in both the anterograde and retrograde
508:
The structure and function of the Golgi apparatus are intimately linked. Individual stacks have different assortments of enzymes, allowing for progressive processing of cargo proteins as they travel from the cisternae to the trans Golgi face. Enzymatic reactions within the Golgi stacks occur
1051:
1050:
1065:
1052:
1000:. The other models are still important in framing questions and guiding future experimentation. Among the fundamental unanswered questions are the directionality of COPI vesicles and role of Rab GTPases in modulating protein cargo traffic.
1049:
1080:
683:(TGN). This area of the Golgi is the point at which proteins are sorted and shipped to their intended destinations by their placement into one of at least three different types of vesicles, depending upon the
1035:(GEFs) that mediate GTP-binding of ARFs. Treatment of cells with BFA thus disrupts the secretion pathway, promoting disassembly of the Golgi apparatus and distributing Golgi proteins to the endosomes and ER.
920:
This model encompasses the strengths of the cisternal progression/maturation model that also explains rapid trafficking of cargo, and how native Golgi proteins can recycle independently of COPI vesicles.
888:
patterns may account for the differing biochemistry throughout the Golgi stack. Thus, the compartments within the Golgi are seen as discrete kinetic stages of the maturing Golgi apparatus.
493:
or for use in the cell. In this respect, the Golgi can be thought of as similar to a post office: it packages and labels items which it then sends to different parts of the cell or to the
857:
The model explains observations of compartments, polarized distribution of enzymes, and waves of moving vesicles. It also attempts to explain how Golgi-specific enzymes are recycled.
1048:
1217:
901:
This model cannot easily explain the observation of fused Golgi networks, tubular connections among cisternae, and differing kinetics of secretory cargo exit.
399:, secretory vesicles, or the cell surface. The TGN is usually positioned adjacent to the stack, but can also be separate from it. The TGN may act as an early
313:. In experiments it is seen that as microtubules are depolymerized the Golgi apparatuses lose mutual connections and become individual stacks throughout the
509:
exclusively near its membrane surfaces, where enzymes are anchored. This feature is in contrast to the ER, which has soluble proteins and enzymes in its
355:
In most eukaryotes, the Golgi apparatus is made up of a series of compartments and is a collection of fused, flattened membrane-enclosed disks known as
367:. A mammalian cell typically contains 40 to 100 stacks of cisternae. Between four and eight cisternae are usually present in a stack; however, in some
1064:
309:. Tubular connections are responsible for linking the stacks together. Localization and tubular connections of the Golgi apparatus are dependent on
2426:
780:, or to lysosome-like storage organelles. These proteins include both digestive enzymes and membrane proteins. The vesicle first fuses with the
2004:
414:
There are structural and organizational differences in the Golgi apparatus among eukaryotes. In some yeasts, Golgi stacking is not observed.
429:
The Golgi apparatus tends to be larger and more numerous in cells that synthesize and secrete large amounts of substances; for example, the
1246:
331:, Golgi stacks are not concentrated at the centrosomal region and do not form Golgi ribbons. Organization of the plant Golgi depends on
914:
the ER-Golgi system: they allow for fast anterograde traffic of small cargo and/or the retrograde traffic of native Golgi proteins.
1071:
811:
In this model, the Golgi is viewed as a set of stable compartments that work together. Each compartment has a unique collection of
235:
inside the cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and
949:
Notably, this model explains the exponential kinetics of cargo exit of both large and small proteins, whereas other models cannot.
1158:
Fabene PF, Bentivoglio M (October 1998). "1898-1998: Camillo Golgi and "the Golgi": one hundred years of terminological clones".
1015:
used experimentally to disrupt the secretion pathway as a method of testing Golgi function. BFA blocks the activation of some
1598:
1571:
1295:
1133:
653:
560:
occurs within the TGN. Other general post-translational modifications of proteins include the addition of carbohydrates (
1032:
1997:
1976:
1202:
968:
This is the most recent model. In this model, the Golgi is seen as a collection of stable compartments defined by
2066:
840:
828:
649:
629:
514:
486:
392:
232:
89:
848:
2168:
1095:
489:, which then fuse with the Golgi apparatus. These cargo proteins are modified and destined for secretion via
1396:
Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V (June 2008).
1990:
664:. Once inside the lumen, the molecules are modified, then sorted for transport to their next destinations.
110:
94:
660:
face of the Golgi apparatus, where they fuse with the Golgi membrane and empty their contents into the
481:
The Golgi apparatus is a major collection and dispatch station of protein products received from the
2163:
422:
323:
1020:
426:
does not. In plants, the individual stacks of the Golgi apparatus seem to operate independently.
1848:
D'Souza-Schorey C, Chavrier P (May 2006). "ARF proteins: roles in membrane traffic and beyond".
371:
as many as sixty cisternae have been observed. This collection of cisternae is broken down into
2128:
2103:
1502:
907:
1238:
718:, where they fuse and release the contents into the extracellular space in a process known as
572:
that determines the final destination of the protein. For example, the Golgi apparatus adds a
2367:
2153:
2046:
1588:
668:
609:
482:
364:
336:
1956:
269:
Owing to its large size and distinctive structure, the Golgi apparatus was one of the first
2400:
593:
254:
that attach various sugar monomers to proteins as the proteins move through the apparatus.
879:
In this model, the fusion of COPII vesicles from the ER begins the formation of the first
35:
8:
2421:
2026:
1563:
1087:
573:
533:
494:
335:
cables and not microtubules. The common feature among Golgi is that they are adjacent to
217:
1952:
273:
to be discovered and observed in detail. It was discovered in 1898 by
Italian physician
1919:
1892:
1873:
1818:
1793:
1766:
1741:
1717:
1692:
1531:
1506:
1471:
1446:
1422:
1397:
1183:
805:
1982:
1171:
387:(TGN). The CGN is the first cisternal structure, and the TGN is the final, from which
2350:
2188:
2071:
1924:
1865:
1823:
1771:
1757:
1722:
1640:
1594:
1567:
1536:
1476:
1427:
1378:
1373:
1356:
1337:
1291:
1284:
1187:
1175:
1129:
844:
747:
720:
585:
225:
1877:
321:, multiple Golgi apparatuses are scattered throughout the cytoplasm (as observed in
257:
The Golgi apparatus was identified in 1898 by the
Italian biologist and pathologist
1914:
1904:
1857:
1813:
1805:
1761:
1753:
1712:
1704:
1630:
1526:
1518:
1466:
1458:
1417:
1409:
1368:
1329:
1167:
1121:
1016:
754:
613:
363:, also called "dictyosomes"), originating from vesicular clusters that bud off the
167:
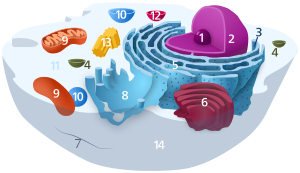
620:; 3. Rough endoplasmic reticulum (RER); 4. Smooth endoplasmic reticulum (SER); 5.
1462:
1125:
969:
926:
This model cannot explain the transport kinetics of large protein cargo, such as
839:-coated vesicles. This model proposes that COPI vesicles move in two directions:
715:
661:
565:
518:
510:
416:
2210:
1708:
1556:
784:, and the contents are then transferred to the lysosome via unknown mechanisms.
714:
release. After packaging, the vesicles bud off and immediately move towards the
2014:
1118:
The Golgi
Apparatus: State of the art 110 years after Camillo Golgi's discovery
684:
589:
569:
278:
229:
213:
1809:
1522:
1333:
962:
347:
2415:
2297:
2257:
2232:
2148:
2061:
2036:
731:
711:
679:
face, to a complex network of membranes and associated vesicles known as the
625:
581:
561:
434:
274:
258:
248:
149:
115:
28:
1413:
1398:"The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis"
908:
Model 3: Cisternal progression/maturation with heterotypic tubular transport
473:
2277:
2249:
2227:
2220:
2200:
2140:
2056:
2041:
1928:
1869:
1827:
1775:
1726:
1644:
1540:
1480:
1431:
1382:
1341:
617:
580:. Another important function of the Golgi apparatus is in the formation of
557:
449:
302:
105:
78:
46:
21:
1635:
1622:
1179:
1091:
2379:
2345:
2272:
2267:
2158:
1008:
869:
This model was widely accepted from the early 1980s until the late 1990s.
541:
310:
236:
55:
1909:
800:
461:
face cisternae, and enzymes catalyzing later modifications are found in
2374:
2314:
2307:
2292:
2287:
2282:
2237:
2183:
2173:
2123:
2118:
2108:
1012:
490:
306:
282:
210:
144:
1058:
Yeast Golgi dynamics. Green labels early Golgi, red labels late Golgi.
588:, thus creating proteoglycans. Glycosaminoglycans are long unbranched
2395:
2357:
2330:
2302:
2195:
2178:
2113:
2086:
2018:
997:
777:
773:
672:
641:
549:
537:
477:
The Golgi apparatus (salmon pink) in context of the secretory pathway
356:
314:
298:
270:
244:
221:
206:
133:
129:
73:
1861:
1027:
which regulate vesicular trafficking through the binding of COPs to
806:
Model 1: Anterograde vesicular transport between stable compartments
667:
Those proteins destined for areas of the cell other than either the
2340:
2096:
2081:
2076:
1070:
Two Golgi stacks connected as a ribbon in a mouse cell. Taken from
1028:
927:
884:
792:
788:
781:
769:
727:
621:
604:
577:
553:
525:
502:
430:
400:
396:
368:
301:. In mammals, a single Golgi apparatus is usually located near the
139:
83:
1086:
Three-dimensional projection of a mammalian Golgi stack imaged by
448:
exit face. These faces are characterized by unique morphology and
2362:
2262:
2215:
2091:
982:
explain the observation of "megavesicle" transport intermediates.
816:
529:
388:
297:
The subcellular localization of the Golgi apparatus varies among
240:
125:
120:
1792:
Marie M, Sannerud R, Avsnes Dale H, Saraste J (September 2008).
2205:
1971:
1791:
1024:
972:
936:
812:
758:
453:
251:
1320:
Nakano A, Luini A (August 2010). "Passage through the Golgi".
65:
824:
498:
408:
404:
332:
328:
318:
1445:
Day, Kasey J.; Casler, Jason C.; Glick, Benjamin S. (2018).
1116:
Pavelk M, Mironov AA (2008). "Golgi apparatus inheritance".
873:
1847:
963:
Model 5: Stable compartments as cisternal model progenitors
836:
185:
1562:(4 ed.). Menlo Park, CA: Benjamin/Cummings. pp.
182:
176:
2335:
768:
Vesicles contain proteins and ribosomes destined for the
173:
1794:"Take the 'A' train: on fast tracks to the cell surface"
261:. The organelle was later named after him in the 1910s.
2012:
1893:"COPI selectively drives maturation of the early Golgi"
1890:
1395:
485:(ER). Proteins synthesized in the ER are packaged into
239:
pathways. It is of particular importance in processing
851:
vesicles recycle Golgi-specific trafficking proteins.
532:
residues. Removal of mannose residues and addition of
1507:"A three-stage model of Golgi structure and function"
801:
Current models of vesicular transport and trafficking
437:
of the immune system have prominent Golgi complexes.
1031:
and the Golgi. BFA inhibits the function of several
179:
1210:
Bollettino della Società Medico-Chirurgica di Pavia
989:
are not established to be intra-Golgi transporters.
671:or the Golgi apparatus are moved through the Golgi
170:
1555:
1283:
1693:"Models for Golgi traffic: a critical assessment"
1586:
1500:
1447:"Budding Yeast Has a Minimal Endomembrane System"
1157:
819:cargo. Proteins are delivered from the ER to the
2413:
1891:Papanikou E, Day KJ, Austin J, Glick BS (2015).
1686:
1684:
1682:
1680:
1678:
1676:
1674:
1232:
1230:
1109:
16:Cell organelle that packages proteins for export
1843:
1841:
1839:
1837:
1787:
1785:
1672:
1670:
1668:
1666:
1664:
1662:
1660:
1658:
1656:
1654:
1496:
1494:
1492:
1490:
521:on lysosomal proteins occurs in the early CGN.
379:compartments, making up two main networks: the
1444:
1315:
1313:
1311:
1309:
1307:
1203:"Intorno alla struttura delle cellule nervose"
452:. Within individual stacks are assortments of
440:In all eukaryotes, each cisternal stack has a
1998:
1227:
1153:
1151:
1149:
1147:
1145:
1115:
517:of proteins. For example, phosphorylation of
1884:
1834:
1782:
1651:
1614:
1487:
937:Model 4: Rapid partitioning in a mixed Golgi
1438:
1389:
1348:
1319:
1304:
1236:
292:
2005:
1991:
1739:
1697:Cold Spring Harbor Perspectives in Biology
1690:
1620:
1580:
1354:
1142:
612:(orange) to Golgi apparatus (magenta). 1.
584:. Enzymes in the Golgi append proteins to
497:. The Golgi apparatus is also involved in
1918:
1908:
1817:
1765:
1716:
1634:
1530:
1470:
1421:
1372:
1277:
1275:
1273:
1271:
1269:
1267:
1265:
1263:
874:Model 2: Cisternal progression/maturation
1623:"Synthesis and sorting of proteoglycans"
1553:
1547:
603:
472:
346:
34:
1281:
710:Vesicle contains proteins destined for
536:occur in medial cisternae. Addition of
2427:Anatomy named for one who described it
2414:
1850:Nature Reviews. Molecular Cell Biology
1260:
599:
513:. Much of the enzymatic processing is
1986:
1200:
281:. After first observing it under his
70:Components of a typical animal cell:
1798:Cellular and Molecular Life Sciences
1282:Alberts, Bruce; et al. (1994).
568:). Protein modifications may form a
465:face cisternae of the Golgi stacks.
39:Diagram of a single "stack" of Golgi
1740:Wei JH, Seemann J (November 2010).
1733:
1691:Glick BS, Luini A (November 2011).
1033:guanine nucleotide exchange factors
528:are associated with the removal of
13:
1621:Prydz K, Dalen KT (January 2000).
1090:and volume surface rendered using
608:Diagram of secretory process from
14:
2438:
1938:
1593:. Nova Publishers. pp. 45–.
1120:. Berlin: Springer. p. 580.
831:. Cargo then progress toward the
1970:
1945:
1758:10.1111/j.1600-0854.2010.01114.x
1590:Biotechnology and Bioengineering
1374:10.1111/j.1600-0854.2011.01316.x
1223:from the original on 2018-04-07.
1079:
1063:
1045:
166:
64:
1511:Histochemistry and Cell Biology
1355:Suda Y, Nakano A (April 2012).
1322:Current Opinion in Cell Biology
1249:from the original on 2006-11-07
576:label to proteins destined for
515:post-translational modification
420:does have stacked Golgi, while
351:3D rendering of Golgi apparatus
277:during an investigation of the
1629:. 113. 113 Pt 2 (2): 193–205.
1194:
1003:
1:
2169:Microtubule organizing center
1742:"Unraveling the Golgi ribbon"
1505:, Glick BS (September 2013).
1402:Molecular Biology of the Cell
1286:Molecular Biology of the Cell
1172:10.1016/S0361-9230(98)00079-3
1103:
640:face of Golgi apparatus; 11.
636:face of Golgi apparatus; 10.
285:, he termed the structure as
1463:10.1016/j.devcel.2017.12.014
1245:. Florida State University.
1126:10.1007/978-3-211-76310-0_34
342:
264:
111:Smooth endoplasmic reticulum
7:
1709:10.1101/cshperspect.a005215
1357:"The yeast Golgi apparatus"
654:rough endoplasmic reticulum
468:
287:apparato reticolare interno
95:Rough endoplasmic reticulum
10:
2443:
1587:William G. Flynne (2008).
1237:Davidson MW (2004-12-13).
1038:
956:the role of COPI vesicles.
18:
2388:
2323:
2248:
2139:
2025:
1810:10.1007/s00018-008-8355-0
1554:Campbell, Neil A (1996).
1523:10.1007/s00418-013-1128-3
1334:10.1016/j.ceb.2010.05.003
632:; 8. Golgi apparatus; 9.
592:molecules present in the
63:
54:
45:
2164:Prokaryotic cytoskeleton
895:structures of the Golgi.
423:Saccharomyces cerevisiae
324:Saccharomyces cerevisiae
293:Subcellular localization
132:; with which, comprises
20:Not to be confused with
1627:Journal of Cell Science
1414:10.1091/mbc.E07-10-0998
1160:Brain Research Bulletin
656:are transported to the
644:of the Golgi apparatus.
1290:. Garland Publishing.
721:constitutive secretion
645:
478:
352:
247:, containing a set of
40:
2154:Intermediate filament
2047:Endoplasmic reticulum
1636:10.1242/jcs.113.2.193
1243:Molecular Expressions
1239:"The Golgi Apparatus"
1094:software. Taken from
776:containing many acid
730:release by activated
669:endoplasmic reticulum
610:endoplasmic reticulum
607:
483:endoplasmic reticulum
476:
365:endoplasmic reticulum
350:
337:endoplasmic reticulum
193:), also known as the
128:(fluid that contains
38:
2401:Extracellular matrix
1979:at Wikimedia Commons
996:observations across
815:that work to modify
704:Exocytotic vesicles
624:attached to RER; 6.
594:extracellular matrix
457:are gathered in the
2104:Cytoplasmic granule
1910:10.7554/eLife.13232
1088:confocal microscopy
765:Lysosomal vesicles
748:regulated secretion
738:Secretory vesicles
681:trans-Golgi network
600:Vesicular transport
574:mannose-6-phosphate
534:N-acetylglucosamine
495:extracellular space
385:trans Golgi network
218:endomembrane system
86:(dots as part of 5)
57:Animal cell diagram
2129:Weibel–Palade body
2013:Structures of the
1955:has a profile for
1451:Developmental Cell
1023:). ARFs are small
1011:(BFA) is a fungal
845:secretory proteins
646:
586:glycosaminoglycans
564:) and phosphates (
479:
391:are packaged into
353:
41:
27:For the song, see
2409:
2408:
2189:Spindle pole body
1975:Media related to
1961:
1600:978-1-60456-067-1
1573:978-0-8053-1957-6
1297:978-0-8153-1619-0
1135:978-3-211-76310-0
1053:
798:
797:
791:destined for the
444:entry face and a
381:cis Golgi network
339:(ER) exit sites.
226:packages proteins
158:
157:
2434:
2007:
2000:
1993:
1984:
1983:
1974:
1959:
1958:Golgi apparatus
1949:
1948:
1933:
1932:
1922:
1912:
1888:
1882:
1881:
1845:
1832:
1831:
1821:
1789:
1780:
1779:
1769:
1752:(11): 1391–400.
1737:
1731:
1730:
1720:
1688:
1649:
1648:
1638:
1618:
1612:
1611:
1609:
1607:
1584:
1578:
1577:
1561:
1551:
1545:
1544:
1534:
1498:
1485:
1484:
1474:
1442:
1436:
1435:
1425:
1393:
1387:
1386:
1376:
1352:
1346:
1345:
1317:
1302:
1301:
1289:
1279:
1258:
1257:
1255:
1254:
1234:
1225:
1224:
1222:
1207:
1201:Golgi C (1898).
1198:
1192:
1191:
1155:
1140:
1139:
1113:
1083:
1067:
1055:
1054:
1017:ADP-ribosylation
772:, a degradative
755:Neurotransmitter
690:
689:
614:Nuclear membrane
519:oligosaccharides
201:, or simply the
192:
191:
188:
187:
184:
181:
178:
175:
172:
102:(or, Golgi body)
68:
58:
49:
43:
42:
32:
25:
2442:
2441:
2437:
2436:
2435:
2433:
2432:
2431:
2412:
2411:
2410:
2405:
2384:
2319:
2244:
2135:
2052:Golgi apparatus
2028:
2021:
2011:
1977:Golgi apparatus
1967:
1966:
1965:
1950:
1946:
1941:
1936:
1889:
1885:
1862:10.1038/nrm1910
1846:
1835:
1804:(18): 2859–74.
1790:
1783:
1738:
1734:
1703:(11): a005215.
1689:
1652:
1619:
1615:
1605:
1603:
1601:
1585:
1581:
1574:
1552:
1548:
1499:
1488:
1457:(1): 56–72.e4.
1443:
1439:
1394:
1390:
1353:
1349:
1318:
1305:
1298:
1280:
1261:
1252:
1250:
1235:
1228:
1220:
1205:
1199:
1195:
1156:
1143:
1136:
1114:
1110:
1106:
1099:
1084:
1075:
1068:
1059:
1056:
1046:
1041:
1006:
970:Rab (G-protein)
965:
939:
910:
876:
843:vesicles carry
808:
803:
716:plasma membrane
685:signal sequence
652:that leave the
628:; 7. Transport
602:
570:signal sequence
566:phosphorylation
471:
417:Pichia pastoris
345:
305:, close to the
295:
267:
169:
165:
162:Golgi apparatus
154:
100:Golgi apparatus
56:
47:
33:
26:
19:
17:
12:
11:
5:
2440:
2430:
2429:
2424:
2407:
2406:
2404:
2403:
2398:
2392:
2390:
2386:
2385:
2383:
2382:
2377:
2372:
2371:
2370:
2365:
2355:
2354:
2353:
2348:
2343:
2333:
2327:
2325:
2324:Other internal
2321:
2320:
2318:
2317:
2312:
2311:
2310:
2305:
2300:
2295:
2290:
2285:
2280:
2275:
2270:
2260:
2254:
2252:
2246:
2245:
2243:
2242:
2241:
2240:
2235:
2225:
2224:
2223:
2218:
2213:
2208:
2198:
2193:
2192:
2191:
2186:
2181:
2176:
2166:
2161:
2156:
2151:
2145:
2143:
2137:
2136:
2134:
2133:
2132:
2131:
2126:
2121:
2116:
2111:
2101:
2100:
2099:
2094:
2089:
2084:
2079:
2074:
2064:
2059:
2054:
2049:
2044:
2039:
2033:
2031:
2023:
2022:
2010:
2009:
2002:
1995:
1987:
1981:
1980:
1951:
1944:
1943:
1942:
1940:
1939:External links
1937:
1935:
1934:
1883:
1833:
1781:
1732:
1650:
1613:
1599:
1579:
1572:
1546:
1486:
1437:
1408:(6): 2579–87.
1388:
1347:
1303:
1296:
1259:
1226:
1193:
1141:
1134:
1107:
1105:
1102:
1101:
1100:
1085:
1078:
1076:
1069:
1062:
1060:
1057:
1044:
1040:
1037:
1005:
1002:
993:
992:
991:
990:
983:
964:
961:
960:
959:
958:
957:
950:
938:
935:
934:
933:
932:
931:
921:
909:
906:
905:
904:
903:
902:
896:
875:
872:
871:
870:
867:
866:
865:
858:
807:
804:
802:
799:
796:
795:
785:
766:
762:
761:
752:
742:
735:
734:
732:plasma B cells
725:
708:
706:(constitutive)
701:
700:
697:
694:
626:Macromolecules
601:
598:
590:polysaccharide
544:occurs in the
501:transport and
470:
467:
435:plasma B cells
383:(CGN) and the
375:, medial, and
344:
341:
294:
291:
279:nervous system
266:
263:
230:membrane-bound
216:. Part of the
209:found in most
156:
155:
153:
152:
147:
142:
137:
123:
118:
113:
108:
103:
97:
92:
87:
81:
76:
69:
61:
60:
52:
51:
15:
9:
6:
4:
3:
2:
2439:
2428:
2425:
2423:
2420:
2419:
2417:
2402:
2399:
2397:
2394:
2393:
2391:
2387:
2381:
2378:
2376:
2373:
2369:
2366:
2364:
2361:
2360:
2359:
2356:
2352:
2349:
2347:
2344:
2342:
2339:
2338:
2337:
2334:
2332:
2329:
2328:
2326:
2322:
2316:
2313:
2309:
2306:
2304:
2301:
2299:
2298:Proteinoplast
2296:
2294:
2291:
2289:
2286:
2284:
2281:
2279:
2276:
2274:
2271:
2269:
2266:
2265:
2264:
2261:
2259:
2258:Mitochondrion
2256:
2255:
2253:
2251:
2250:Endosymbionts
2247:
2239:
2236:
2234:
2233:Lamellipodium
2231:
2230:
2229:
2226:
2222:
2219:
2217:
2214:
2212:
2209:
2207:
2204:
2203:
2202:
2199:
2197:
2194:
2190:
2187:
2185:
2182:
2180:
2177:
2175:
2172:
2171:
2170:
2167:
2165:
2162:
2160:
2157:
2155:
2152:
2150:
2149:Microfilament
2147:
2146:
2144:
2142:
2138:
2130:
2127:
2125:
2122:
2120:
2117:
2115:
2112:
2110:
2107:
2106:
2105:
2102:
2098:
2095:
2093:
2090:
2088:
2085:
2083:
2080:
2078:
2075:
2073:
2070:
2069:
2068:
2065:
2063:
2062:Autophagosome
2060:
2058:
2055:
2053:
2050:
2048:
2045:
2043:
2040:
2038:
2037:Cell membrane
2035:
2034:
2032:
2030:
2027:Endomembrane
2024:
2020:
2016:
2008:
2003:
2001:
1996:
1994:
1989:
1988:
1985:
1978:
1973:
1969:
1968:
1963:
1962:
1954:
1930:
1926:
1921:
1916:
1911:
1906:
1902:
1898:
1894:
1887:
1879:
1875:
1871:
1867:
1863:
1859:
1856:(5): 347–58.
1855:
1851:
1844:
1842:
1840:
1838:
1829:
1825:
1820:
1815:
1811:
1807:
1803:
1799:
1795:
1788:
1786:
1777:
1773:
1768:
1763:
1759:
1755:
1751:
1747:
1743:
1736:
1728:
1724:
1719:
1714:
1710:
1706:
1702:
1698:
1694:
1687:
1685:
1683:
1681:
1679:
1677:
1675:
1673:
1671:
1669:
1667:
1665:
1663:
1661:
1659:
1657:
1655:
1646:
1642:
1637:
1632:
1628:
1624:
1617:
1602:
1596:
1592:
1591:
1583:
1575:
1569:
1565:
1560:
1559:
1550:
1542:
1538:
1533:
1528:
1524:
1520:
1517:(3): 239–49.
1516:
1512:
1508:
1504:
1497:
1495:
1493:
1491:
1482:
1478:
1473:
1468:
1464:
1460:
1456:
1452:
1448:
1441:
1433:
1429:
1424:
1419:
1415:
1411:
1407:
1403:
1399:
1392:
1384:
1380:
1375:
1370:
1367:(4): 505–10.
1366:
1362:
1358:
1351:
1343:
1339:
1335:
1331:
1327:
1323:
1316:
1314:
1312:
1310:
1308:
1299:
1293:
1288:
1287:
1278:
1276:
1274:
1272:
1270:
1268:
1266:
1264:
1248:
1244:
1240:
1233:
1231:
1219:
1215:
1211:
1204:
1197:
1189:
1185:
1181:
1177:
1173:
1169:
1165:
1161:
1154:
1152:
1150:
1148:
1146:
1137:
1131:
1127:
1123:
1119:
1112:
1108:
1097:
1093:
1089:
1082:
1077:
1073:
1066:
1061:
1043:
1042:
1036:
1034:
1030:
1026:
1022:
1018:
1014:
1010:
1001:
999:
987:
984:
980:
977:
976:
974:
971:
967:
966:
954:
951:
948:
945:
944:
941:
940:
929:
925:
922:
919:
916:
915:
912:
911:
900:
897:
893:
890:
889:
886:
882:
878:
877:
868:
862:
859:
856:
853:
852:
850:
846:
842:
838:
834:
830:
826:
822:
818:
814:
810:
809:
794:
790:
786:
783:
782:late endosome
779:
775:
771:
767:
764:
763:
760:
757:release from
756:
753:
750:
749:
743:
741:
737:
736:
733:
729:
726:
723:
722:
717:
713:
712:extracellular
709:
707:
703:
702:
698:
695:
692:
691:
688:
686:
682:
678:
674:
670:
665:
663:
659:
655:
651:
643:
639:
635:
631:
627:
623:
619:
615:
611:
606:
597:
595:
591:
587:
583:
582:proteoglycans
579:
575:
571:
567:
563:
562:glycosylation
559:
558:carbohydrates
555:
551:
547:
543:
539:
535:
531:
527:
524:
520:
516:
512:
506:
504:
500:
496:
492:
488:
484:
475:
466:
464:
460:
455:
451:
447:
443:
438:
436:
432:
427:
425:
424:
419:
418:
412:
410:
406:
402:
398:
394:
390:
386:
382:
378:
374:
370:
366:
362:
358:
349:
340:
338:
334:
330:
326:
325:
320:
316:
312:
308:
304:
300:
290:
288:
284:
280:
276:
275:Camillo Golgi
272:
262:
260:
259:Camillo Golgi
255:
253:
250:
249:glycosylation
246:
242:
238:
234:
231:
227:
223:
219:
215:
212:
208:
204:
200:
196:
195:Golgi complex
190:
163:
151:
150:Cell membrane
148:
146:
143:
141:
138:
135:
131:
127:
124:
122:
119:
117:
116:Mitochondrion
114:
112:
109:
107:
104:
101:
98:
96:
93:
91:
88:
85:
82:
80:
77:
75:
72:
71:
67:
62:
59:
53:
50:
44:
37:
30:
29:Junta (album)
23:
2278:Gerontoplast
2228:Pseudopodium
2221:Radial spoke
2201:Undulipodium
2141:Cytoskeleton
2057:Parenthesome
2051:
1957:
1900:
1896:
1886:
1853:
1849:
1801:
1797:
1749:
1745:
1735:
1700:
1696:
1626:
1616:
1604:. Retrieved
1589:
1582:
1557:
1549:
1514:
1510:
1503:Staehelin LA
1454:
1450:
1440:
1405:
1401:
1391:
1364:
1360:
1350:
1328:(4): 471–8.
1325:
1321:
1285:
1251:. Retrieved
1242:
1213:
1209:
1196:
1166:(3): 195–8.
1163:
1159:
1117:
1111:
1007:
994:
985:
978:
952:
946:
923:
917:
898:
891:
880:
860:
854:
832:
820:
746:
739:
719:
705:
696:Description
687:they carry.
680:
676:
675:towards the
666:
657:
647:
637:
633:
618:Nuclear pore
596:of animals.
545:
522:
507:
480:
462:
458:
450:biochemistry
445:
441:
439:
428:
421:
415:
413:
395:destined to
384:
380:
376:
372:
360:
354:
322:
311:microtubules
303:cell nucleus
296:
286:
268:
256:
202:
198:
194:
161:
159:
106:Cytoskeleton
99:
48:Cell biology
22:gyrification
2380:Magnetosome
2346:Spliceosome
2273:Chromoplast
2268:Chloroplast
2159:Microtubule
1606:13 November
1009:Brefeldin A
1004:Brefeldin A
986:Weaknesses:
953:Weaknesses:
924:Weaknesses:
899:Weaknesses:
864:directions.
861:Weaknesses:
841:anterograde
823:face using
740:(regulated)
548:cisternae.
542:sialic acid
505:formation.
433:-secreting
359:(singular:
2422:Organelles
2416:Categories
2375:Proteasome
2368:Inclusions
2315:Nitroplast
2308:Apicoplast
2293:Elaioplast
2288:Amyloplast
2283:Leucoplast
2238:Filopodium
2184:Basal body
2174:Centrosome
2124:Peroxisome
2119:Glyoxysome
2109:Melanosome
2019:organelles
1253:2010-09-20
1216:(1): 316.
1104:References
1013:metabolite
998:eukaryotes
979:Strengths:
947:Strengths:
918:Strengths:
892:Strengths:
855:Strengths:
849:retrograde
787:Digestive
778:hydrolases
491:exocytosis
307:centrosome
299:eukaryotes
283:microscope
271:organelles
211:eukaryotic
199:Golgi body
145:Centrosome
130:organelles
2396:Cell wall
2358:Cytoplasm
2331:Nucleolus
2303:Tannosome
2211:Flagellum
2196:Myofibril
2179:Centriole
2114:Microbody
2087:Phagosome
1188:208785591
1096:the movie
1072:the movie
1029:endosomes
1019:factors (
789:proteases
774:organelle
673:cisternae
642:Cisternae
578:lysosomes
554:tyrosines
550:Sulfation
538:galactose
397:lysosomes
357:cisternae
343:Structure
315:cytoplasm
265:Discovery
245:secretion
237:endocytic
222:cytoplasm
207:organelle
134:cytoplasm
74:Nucleolus
2389:External
2341:Ribosome
2097:Acrosome
2082:Endosome
2077:Lysosome
1960:(Q83181)
1929:26709839
1878:19092867
1870:16633337
1828:18726174
1776:21040294
1727:21875986
1645:10633071
1541:23881164
1501:Day KJ,
1481:29316441
1432:18385516
1383:22132734
1342:20605430
1247:Archived
1218:Archived
928:collagen
885:cisterna
847:, while
835:face in
829:vesicles
827:-coated
793:lysosome
770:lysosome
728:Antibody
699:Example
650:vesicles
630:vesicles
622:Ribosome
526:cisterna
503:lysosome
487:vesicles
469:Function
431:antibody
401:endosome
393:vesicles
389:proteins
369:protists
361:cisterna
241:proteins
233:vesicles
205:, is an
140:Lysosome
84:Ribosome
2363:Cytosol
2263:Plastid
2216:Axoneme
2092:Vacuole
2072:Exosome
2067:Vesicle
2042:Nucleus
1953:Scholia
1920:4758959
1819:7079782
1767:4221251
1746:Traffic
1718:3220355
1566:, 123.
1558:Biology
1532:3779436
1472:5765772
1423:2397314
1361:Traffic
1180:9865849
1039:Gallery
1025:GTPases
973:GTPases
817:protein
813:enzymes
759:neurons
530:mannose
454:enzymes
252:enzymes
220:in the
126:Cytosol
121:Vacuole
90:Vesicle
79:Nucleus
2206:Cilium
2029:system
1927:
1917:
1876:
1868:
1826:
1816:
1774:
1764:
1725:
1715:
1643:
1597:
1570:
1539:
1529:
1479:
1469:
1430:
1420:
1381:
1340:
1294:
1186:
1178:
1132:
1092:Imaris
693:Types
409:plants
329:plants
327:). In
2351:Vault
1897:eLife
1874:S2CID
1221:(PDF)
1206:(PDF)
1184:S2CID
833:trans
825:COPII
677:trans
662:lumen
638:Trans
616:; 2.
546:trans
511:lumen
499:lipid
463:trans
446:trans
405:yeast
377:trans
333:actin
319:yeast
317:. In
228:into
224:, it
214:cells
203:Golgi
2015:cell
1925:PMID
1866:PMID
1824:PMID
1772:PMID
1723:PMID
1641:PMID
1608:2010
1595:ISBN
1568:ISBN
1537:PMID
1477:PMID
1428:PMID
1379:PMID
1338:PMID
1292:ISBN
1176:PMID
1130:ISBN
1021:ARFs
837:COPI
648:The
556:and
540:and
407:and
243:for
160:The
2336:RNA
1915:PMC
1905:doi
1858:doi
1814:PMC
1806:doi
1762:PMC
1754:doi
1713:PMC
1705:doi
1631:doi
1564:122
1527:PMC
1519:doi
1515:140
1467:PMC
1459:doi
1418:PMC
1410:doi
1369:doi
1330:doi
1168:doi
1122:doi
881:cis
821:cis
658:cis
634:Cis
552:of
523:Cis
459:cis
442:cis
403:in
373:cis
2418::
2017:/
1923:.
1913:.
1903:.
1899:.
1895:.
1872:.
1864:.
1852:.
1836:^
1822:.
1812:.
1802:65
1800:.
1796:.
1784:^
1770:.
1760:.
1750:11
1748:.
1744:.
1721:.
1711:.
1699:.
1695:.
1653:^
1639:.
1625:.
1535:.
1525:.
1513:.
1509:.
1489:^
1475:.
1465:.
1455:44
1453:.
1449:.
1426:.
1416:.
1406:19
1404:.
1400:.
1377:.
1365:13
1363:.
1359:.
1336:.
1326:22
1324:.
1306:^
1262:^
1241:.
1229:^
1214:13
1212:.
1208:.
1182:.
1174:.
1164:47
1162:.
1144:^
1128:.
1098:.
975:.
751:.
724:.
411:.
197:,
183:dʒ
2006:e
1999:t
1992:v
1964:.
1931:.
1907::
1901:4
1880:.
1860::
1854:7
1830:.
1808::
1778:.
1756::
1729:.
1707::
1701:3
1647:.
1633::
1610:.
1576:.
1543:.
1521::
1483:.
1461::
1434:.
1412::
1385:.
1371::
1344:.
1332::
1300:.
1256:.
1190:.
1170::
1138:.
1124::
1074:.
883:-
189:/
186:i
180:l
177:ɒ
174:ɡ
171:ˈ
168:/
164:(
136:)
31:.
24:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.